Abstract
Clinical isolates of Klebsiella pneumoniae producing NDM-1 carbapenemase from India (n = 22), the United Kingdom (n = 13), and Sweden (n = 4) were subjected to multilocus sequence typing (MLST), automated repetitive sequence-based PCR (rep-PCR), serotyping, virulence gene screening, and plasmid replicon typing. The most frequently detected MLST sequence types (STs) were ST14 (n = 13; all serotype K2), ST11, ST149, ST231, and ST147. The correlation between MLST and automated rep-PCR was excellent. IncA/C was the most frequently detected plasmid replicon type (n = 14). ST14, ST11, and other successful clones may be important for the dissemination of blaNDM-1.
TEXT
Since its first description in late 2009, New Delhi metallo-β-lactamase 1 (NDM-1) has become established as a major public health threat, epicentral to the Indian subcontinent but with considerable international dissemination (9).
The genetic context of blaNDM-1 varies, and the gene is associated with a variety of plasmid types, although commonly with broad-range IncA/C elements (4). NDM-1 has been acquired by Escherichia coli sequence type 131 (ST131), a clone notorious for its role in the dissemination of blaCTX-M genes, particularly blaCTX-M-15, in the community, though this is not a frequent host of the carbapenemase gene as yet (5). There is also some evidence of an association between NDM enzymes and E. coli ST101, but overall, the phylogeny of NDM-positive E. coli strains is diverse (5).
Klebsiella pneumoniae strains carry blaNDM more frequently than does E. coli. Little has been done systematically to type carriers belonging to K. pneumoniae by multilocus sequence typing (MLST); however, several case reports, including the first from 2009, note host strains belonging to ST14 (6). Likewise, there have been no previous attempts to analyze the serotypes and virulence genes of K. pneumoniae isolates with the NDM-1 enzyme, and potential connections between sequence types and plasmid replicon types remain to be explored. Such linkages are potentially important; it is well known that one lineage of K. pneumoniae, ST258, has played a major role in the global spread of KPC carbapenemases (10).
The aim of this study therefore was to determine the clonal and serotype diversity of a collection of K. pneumoniae isolates with the NDM-1 enzyme from India, the United Kingdom, and Sweden and to profile the virulence genes of these isolates. Further, we explored the potential for using automated repetitive sequence-based PCR (rep-PCR) as a proxy for MLST to identify rapidly successful clones.
A total of 39 K. pneumoniae isolates from Chennai, India (n = 16), Haryana, India (n = 6), the United Kingdom (n = 13), and Sweden (n = 4) were included. The isolates from Chennai and Haryana were derived from single hospitals in each city, sampled over periods of around 9 months. Clinical information was limited, although all of these isolates were from human sources, mainly from urinary and lower respiratory tract samples. Previous pulsed-field gel electrophoresis (PFGE) typing had suggested that the Chennai isolates were diverse, whereas several earlier isolates from Haryana appeared to be clonally related (4). The 17 isolates from the United Kingdom and Sweden were all from different hospitals and cities. A PCR method (8) was used to predict the expression of capsular serotypes K1, K2, K5, K20, K54, and K57, all of which are strongly associated with invasive disease. The virulence genes allS (which promotes growth in iron-deficient media), rmpA (which regulates extracapsular polysaccharide synthesis), and wcaG (which increases the capacity to escape phagocytosis by synthesizing fucose) were sought according to Brisse et al. (1), while PCR-based replicon typing of plasmids was performed according to Carattoli et al. (3). Sequence typing was performed according to the K. pneumoniae MLST website (http://www.pasteur.fr/recherche/genopole/PF8/mlst/Kpneumoniae.html). Based on the allelic profiles, the evolutionary relationship among isolates was assessed by the minimal spanning tree (MST) algorithm in Bionumerics (Applied Maths, NV St-Martens-Latem, Belgium). A stringent definition of 6/7 shared alleles was used to define clonal complexes (i.e., single locus variants [SLV] only). Automated rep-PCR was conducted with the DiversiLab (DL) system (bioMérieux, Marcy l'Etoile, France), as described previously (2). Isolates were assigned to the same DL type if they had >93% similarity and ≤2 peak differences in the electropherogram overlay analysis.
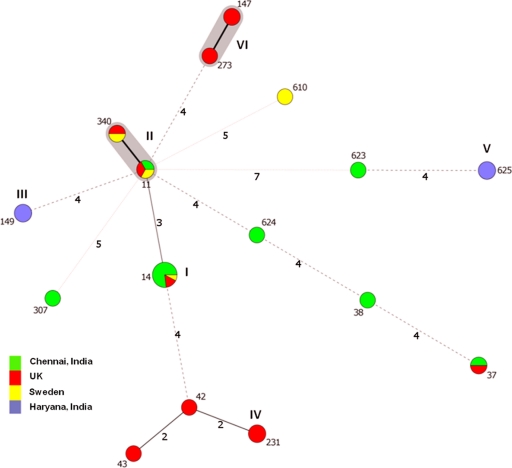
The population structure of the 39 K. pneumoniae isolates is presented in Fig. 1, with six clusters identifiable. ST14 was the most common sequence type, with 13 representatives, all of them also belonging to serotype K2; types ST11 (the SLV ST340 comprised two isolates; hence, clonal complex 11 [CC11] comprised five isolates), ST149, ST231, and ST625 had three representatives each, while a further three isolates belonged to ST147 (n = 2) and ST273 (n = 1), which fall into the same clonal complex, thus forming the sixth cluster. The rest of the isolates either were singletons or belonged to pairs of the same sequence type. ST14 was represented by 10 of the 16 isolates from Chennai (Table 1), two isolates from the United Kingdom, and one from Sweden. The six other Chennai isolates represented six other STs, clearly demonstrating polyclonality within a single hospital. ST11 was the only other type represented by isolates from all three countries, although ST37 was found in India (Chennai) and the United Kingdom, and ST340 was found in Sweden and the United Kingdom. MLST showed that the Haryana isolates belonged to two clones, ST149 (n = 3) and ST625 (n = 3), neither of which was seen among isolates from the United Kingdom or Sweden. Previous PFGE data on other NDM-positive K. pneumoniae isolates from Haryana had indicated considerable clonality (4), but we included only two representatives of this PFGE pattern (both ST625); the rest of the isolates from Haryana had not been typed previously. Among the 13 United Kingdom isolates, ST231 was the single most common type (n = 3), but the clonal complex formed by ST147 and ST273 comprised an equal number of isolates; the seven other United Kingdom isolates belonged to six STs (Table 1). The isolates from Sweden belonged to four different sequence types, reflecting the fact that they were imported from various geographic areas (India, Iraq, and the Balkan region).
Fig 1.
Minimal spanning tree (MST) of K. pneumoniae isolates producing NDM-1 carbapenemase, showing STs versus country of isolation. Each circle corresponds to an ST. The area of each circle is proportional to the number of isolates. The six clusters have been marked with Roman numerals (I to VI). Gray zones surround STs that belong to the same clonal complex (six common alleles). The numbers on the connecting lines between STs correspond to the numbers of allelic differences.
Table 1.
Sequence types, replicon types, DiversiLab types, and places of isolation for the clinical isolates
| Isolate | MLST allelic profile |
ST | DiversiLab type | Replicon type (Inc) | K2 serotype | Virulence genesa | Place of isolation | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| gapA | infB | mdh | pgi | phoE | rpoB | tonB | |||||||
| IR11K | 1 | 6 | 1 | 1 | 1 | 1 | 1 | ST14 | K | FrepB+/AC+ | + | − | Chennai, India |
| IR18K | 1 | 6 | 1 | 1 | 1 | 1 | 1 | ST14 | K | FrepB+/AC+ | + | − | Chennai, India |
| IR19E | 1 | 6 | 1 | 1 | 1 | 1 | 1 | ST14 | K | FrepB+/AC+/HI1+ | + | − | Chennai, India |
| IR19K | 1 | 6 | 1 | 1 | 1 | 1 | 1 | ST14 | K | FrepB+/AC+ | + | − | Chennai, India |
| IR20E | 1 | 6 | 1 | 1 | 1 | 1 | 1 | ST14 | K | FrepB+ | + | − | Chennai, India |
| IR20K | 1 | 6 | 1 | 1 | 1 | 1 | 1 | ST14 | K | FrepB+ | + | − | Chennai, India |
| IR21 | 1 | 6 | 1 | 1 | 1 | 1 | 1 | ST14 | K | FrepB+/AC+ | + | − | Chennai, India |
| IR35 | 1 | 6 | 1 | 1 | 1 | 1 | 1 | ST14 | K | N+ | + | − | Chennai, India |
| IR37 | 1 | 6 | 1 | 1 | 1 | 1 | 1 | ST14 | K | L/M+ | + | − | Chennai, India |
| IR62E | 1 | 6 | 1 | 1 | 1 | 1 | 1 | ST14 | K | FrepB+/AC+ | + | − | Chennai, India |
| N6 | 1 | 6 | 1 | 1 | 1 | 1 | 1 | ST14 | K | FrepB+/AC+ | + | − | UK |
| N7 | 1 | 6 | 1 | 1 | 1 | 1 | 1 | ST14 | K | FrepB+/AC+ | + | − | UK |
| OS506 | 1 | 6 | 1 | 1 | 1 | 1 | 1 | ST14 | K | FrepB+/AC+ | + | − | Sweden |
| IR8 | 3 | 3 | 1 | 1 | 1 | 1 | 4 | ST11 | I | HI1+ | − | − | Chennai, India |
| N17 | 3 | 3 | 1 | 1 | 1 | 1 | 4 | ST11 | I | FII+ | − | − | UK |
| ED502873 | 3 | 3 | 1 | 1 | 1 | 1 | 4 | ST11 | I | N+ | − | − | Sweden |
| K1 | 4 | 34 | 1 | 1 | 21 | 1 | 35 | ST149 | E | NDb | + | − | Haryana, India |
| K6 | 4 | 34 | 1 | 1 | 21 | 1 | 35 | ST149 | E | ND | − | − | Haryana, India |
| K9 | 4 | 34 | 1 | 1 | 21 | 1 | 35 | ST149 | E | ND | − | − | Haryana, India |
| N12 | 2 | 6 | 1 | 3 | 26 | 1 | 77 | ST231 | A | HI1+ | − | − | UK |
| N13 | 2 | 6 | 1 | 3 | 26 | 1 | 77 | ST231 | A | HI1+ | − | − | UK |
| N14 | 2 | 6 | 1 | 3 | 26 | 1 | 77 | ST231 | A | HI1+ | − | − | UK |
| HR10 | 50 | 19 | 82 | 20 | 113 | 18 | 148 | ST625 | N | ND | − | − | Haryana, India |
| HR4 | 50 | 19 | 82 | 20 | 113 | 18 | 148 | ST625 | N | ND | − | − | Haryana, India |
| K16 | 50 | 19 | 82 | 20 | 113 | 18 | 148 | ST625 | N | ND | − | − | Haryana, India |
| IR25 | 2 | 9 | 2 | 1 | 13 | 1 | 16 | ST37 | M | ND | − | − | Chennai, India |
| N21 | 2 | 9 | 2 | 1 | 13 | 1 | 16 | ST37 | L | AC+ | − | − | UK |
| N11 | 3 | 4 | 6 | 1 | 7 | 4 | 38 | ST147 | D | AC+ | − | − | UK |
| N26 | 3 | 4 | 6 | 1 | 7 | 4 | 38 | ST147 | D | AC+ | − | − | UK |
| B383 | 3 | 3 | 1 | 1 | 1 | 1 | 18 | ST340 | I | N+ | − | − | UK |
| ED501570 | 3 | 3 | 1 | 1 | 1 | 1 | 18 | ST340 | I | ND | − | − | Sweden |
| IR15 | 2 | 1 | 2 | 1 | 2 | 2 | 2 | ST38 | F | ND | − | − | Chennai, India |
| B335 | 2 | 6 | 1 | 3 | 8 | 1 | 15 | ST42 | C | AC+ | − | − | UK |
| B357 | 2 | 6 | 1 | 5 | 11 | 1 | 15 | ST43 | G | ND | − | − | UK |
| N27 | 3 | 4 | 6 | 1 | 7 | 4 | 4 | ST273 | D | AC+ | − | − | UK |
| IR54 | 4 | 1 | 2 | 52 | 1 | 1 | 7 | ST307 | O | FrepB+ | − | − | Chennai, India |
| ED501927 | 4 | 5 | 1 | 29 | 1 | 4 | 23 | ST610 | H | L/M+ | − | − | Sweden |
| IR27 | 17 | 19 | 39 | 20 | 117 | 18 | 156 | ST623 | B | ND | − | − | Chennai, India |
| IR34 | 2 | 1 | 1 | 1 | 9 | 1 | 157 | ST624 | J | L/M+ | − | − | Chennai, India |
The following genes were sought: allS, rmpA, and wcaG.
ND, not detected.
As with NDM-positive E. coli (5), this collection of NDM-1-positive K. pneumoniae isolates showed relatively high clonal diversity. This is in contrast to the situation for K. pneumoniae producing KPC enzymes, where one clone (ST258) is predominant globally (10). ST14 was the most frequent type observed and was represented by isolates from all three countries sampled. Nevertheless, 10 of its 13 representatives were from one hospital in Chennai, and we cannot exclude the possibility that our results are biased by local prevalence; this possibility can be confirmed or refuted only once India-wide surveys of prevalence are undertaken. K. pneumoniae ST14 has previously been described as a host lineage for the NDM-1 enzyme and is also a frequent host of CTX-M enzymes (10). Further, ST14 is a single locus variant (SLV) of ST15, which frequently carries CTX-M extended-spectrum β-lactamases (ESBLs) (10). Serotype K2 has been described in one previous publication as frequently linked to ST14 (1).
Production of the NDM-1 enzyme by isolates belonging to ST11 and to the ST147-ST273 complex is notable in the same way as the finding of the NDM-1 enzyme in E. coli ST131, mentioned earlier. ST11 is a frequent host of CTX-M and KPC (mainly in China) and is also an SLV of the notorious ST258, pivotal for its international dissemination of KPC enzymes (10), while ST147 was recently described by Samuelsen et al. as hosting VIM enzymes among K. pneumoniae isolates imported to Scandinavia, mainly from Greece (7). These findings show that blaNDM-1 has transferred into successful epidemic clones of K. pneumoniae, as well as those of E. coli, though it is unclear to what extent this underpins its rapid dissemination on the Indian subcontinent and to what extent these associations will facilitate its future spread.
The association between blaNDM and particular plasmid types was not sought. Nevertheless, the most commonly detected plasmid replicon type in the isolates was IncA/C (n = 14). In most cases (9/14), these IncA/C plasmids were hosted by strains that also carried IncFrepB plasmids, which comprised the second most common replicon type (n = 12). Five isolates hosted IncHI1 plasmids, in one case together with both IncA/C and IncFrepB. None of the known replicon types were detected in 11 isolates. ST14 was in many, but not all, cases associated with IncA/C (Fig. 1). All of the ST231 isolates contained IncHI1 plasmids, and the ST147-ST273 cluster was always associated with IncA/C elements. No clear relationship between replicon type and sequence type was observed among the other STs.
DiversiLab assigned all 13 ST14 isolates from three countries to the same type, with >93% (95.4%) similarity (Table 1). DiversiLab also found >93% similarity within all five other lineages identified by MLST, whereas none of the STs represented by singletons or pairs clustered with any of the frequently occurring STs; moreover, all of the larger ST clusters were clearly distinct by DiversiLab. Similar observations have been made previously for ESBL-producing K. pneumoniae and E. coli, and automated rep-PCR may represent a valuable tool for rapid identification of strains with high epidemic potential but may not offer the discrimination within an ST that can be achieved by PFGE and that may be needed for local outbreak investigations (2).
In conclusion, this study demonstrates clearly that the dissemination of NDM-1 is associated with diverse STs, which included ST14 and ST11, in all three countries studied. Worryingly, all of the ST14 isolates carried genes determining serotype K2, which is one of the K. pneumoniae serotypes associated with invasive disease. Further, we identified blaNDM in other K. pneumoniae types with high epidemic potential, namely, ST11 and ST147-ST273. Lastly, this study supports the view that DiversiLab could be used as a proxy for MLST for identifying clones of K. pneumoniae.
ACKNOWLEDGMENTS
We thank Makaoui Matallah for assistance with the minimal spanning tree analysis.
Mark Toleman is funded by EU FP7 grant PAR.
C.G.G. has received conference support from Calixa, Inc., and AB Biodisk, speaker's honoraria from Wyeth and Meda, and chairperson honorarium from AstraZeneca. D.M.L. has shareholdings in AstraZeneca, Dechra, Eco Animal Health GlaxoSmithKline, and Merck; has accepted grants, speaking invitations, and conference invitations from GSK, Merck, Pfizer, Novartis, and AstraZeneca; has recent or ongoing consultancies with Achaogen, Astellas, AstraZeneca, Basilea, Bayer, Cubist, Kalidex, and Tetraphase; and has ongoing contract work for Achaogen, Basilea, Cubist, Meiji, and Merck. N.W. has received research grants or conference support from most major pharmaceutical companies.
Footnotes
Published ahead of print 21 February 2012
REFERENCES
- 1. Brisse S, et al. 2009. Virulent clones of Klebsiella pneumoniae: identification and evolutionary scenario based on genomic and phenotypic characterization. PLoS One 4:e4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brolund A, et al. 2010. The DiversiLab system versus pulsed-field gel electrophoresis: characterisation of extended spectrum β-lactamase producing Escherichia coli and Klebsiella pneumoniae. J. Microbiol. Methods 83:224–230 [DOI] [PubMed] [Google Scholar]
- 3. Carattoli A, et al. 2005. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63:219–228 [DOI] [PubMed] [Google Scholar]
- 4. Kumarasamy KK, et al. 2010. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect. Dis. 10:597–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mushtaq S, et al. 2011. Phylogenetic diversity of Escherichia coli strains producing NDM-type carbapenemases. J. Antimicrob. Chemother. 66:2002–2005 [DOI] [PubMed] [Google Scholar]
- 6. Poirel L, Al Maskari Z, Al Rashdi F, Bernabeu S, Nordmann P. 2011. NDM-1-producing Klebsiella pneumoniae isolated in the Sultanate of Oman. J. Antimicrob. Chemother. 66:304–306 [DOI] [PubMed] [Google Scholar]
- 7. Samuelsen O, et al. 2011. Molecular characterization of VIM-producing Klebsiella pneumoniae from Scandinavia reveals genetic relatedness with international clonal complexes encoding transferable multidrug resistance. Clin. Microbiol. Infect. 17:1811–1816 [DOI] [PubMed] [Google Scholar]
- 8. Turton JF, Baklan H, Siu LK, Kaufmann ME, Pitt TL. 2008. Evaluation of a multiplex PCR for detection of serotypes K1, K2 and K5 in Klebsiella sp. and comparison of isolates within these serotypes. FEMS Microbiol. Lett. 284:247–252 [DOI] [PubMed] [Google Scholar]
- 9. Walsh TR. 2010. Emerging carbapenemases: a global perspective. Int. J. Antimicrob. Agents 36(Suppl. 3):S8–S14 [DOI] [PubMed] [Google Scholar]
- 10. Woodford N, Turton JF, Livermore DM. 2011. Multiresistant Gram-negative bacteria: the role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol. Rev. 35:736–755 [DOI] [PubMed] [Google Scholar]