Abstract
The presence of SXT/R391-related integrating conjugative elements (ICEs) in bacterial strains isolated from fish obtained from marine aquaculture environments in 2001 to 2010 in the northwestern Iberian Peninsula was studied. ICEs were detected in 12 strains taxonomically related to Vibrio scophthalmi (3 strains), Vibrio splendidus (5 strains), Vibrio alginolyticus (1 strain), Shewanella haliotis (1 strain), and Enterovibrio nigricans (2 strains), broadening the known host range able to harbor SXT/R391-like ICEs. Variable DNA regions, which confer element-specific properties to ICEs of this family, were characterized. One of the ICEs encoded antibiotic resistance functions in variable region III, consisting of a tetracycline resistance locus. Interestingly, hot spot 4 included genes providing resistance to rifampin (ICEVspPor2 and ICEValPor1) and quaternary ammonium compounds (QACs) (ICEEniSpa1), and variable region IV included a mercury resistance operon (ICEVspSpa1 and ICEEniSpa1). The S exclusion group was more represented than the R exclusion group, accounting for two-thirds of the total ICEs. Mating experiments allowed ICE mobilization to Escherichia coli strains, showing the corresponding transconjugants' rifampin, mercury, and QAC resistance. These results show the first evidence of ICEs providing rifampin and QAC resistances, suggesting that these mobile genetic elements contribute to the dissemination of antimicrobial, heavy metal, and QAC resistance determinants in aquaculture environments.
INTRODUCTION
Unlike eukaryotes, which evolve mainly by modifying existing genetic information, bacteria acquire a significant proportion of their genetic diversity from unrelated organisms by means of horizontal transfer (25, 43). As a result, considerable proportions of most known bacterial genomes consist of horizontally acquired genes (24). Horizontal gene transfer is important in microbial ecology, especially regarding the relationships between microbes and other organisms, where these genes play a major role in health and disease (44).
The most important contributors to horizontal gene transfer in bacteria are mobile genetic elements such as plasmids, transposons, integrons, phages, genomic islands, and integrating conjugative elements (4, 9, 13, 18). These elements may contain genes encoding proteins conferring an advantageous phenotype under particular environmental conditions (7, 14, 17). Horizontal gene acquisition is likely to cause neutral or deleterious effects on the chromosome of recipient bacterial cells (35), limiting gene transfer to the population. However, mobile genetic elements that encode their own transfer and maintenance functions or those conferring an advantageous phenotype to the recipient strain have the potential to spread rapidly among the population (35).
Integrating conjugative elements (ICEs) are mobile elements that can be excised from their host's chromosome and self-transferred by conjugation to a new host to be reintegrated into its chromosome (8, 43). The SXT/R391 family of ICEs includes more than 40 elements detected in different clinical and environmental isolates, and all of these elements share a conserved integrase that mediates site-specific integration into the prfC gene of the host chromosome (42). This family of ICEs has an important role in the dissemination of antibiotic resistance genes in pathogens such as Vibrio cholerae, which causes cholera in humans. The SXTMO10 ICE was described in Vibrio cholerae O139, and it carries the genes encoding resistance to four antibiotics: sulfamethoxazole, trimethoprim, chloramphenicol, and streptomycin (39). The R391 ICE from Providencia rettgeri mediates resistance to kanamycin and mercury (11).
SXT/R391 ICEs share a structure containing a set of 52 conserved genes involved in integration/excision, conjugative transfer, and regulation (42). Additionally, all the known elements contain variable DNA inserted into intergenic regions that confers element-specific properties (8). The size of the variable DNA ranges from approximately 30 to 60 kb per ICE and is found mostly in 5 hotspots (HSs), dubbed HS1 to HS5. In addition, some of these ICEs also contain variable DNA inserted outside the 5 hotspots in up to four variable regions (VRs), named VRI to VRIV (42). Among the functions encoded within the variable DNA of these mobile elements are resistance to antibiotics and heavy metals, the regulation of biofilm formation and motility (5), and toxin-antitoxin systems preventing the loss of ICEs (41). Since the number of known ICEs of this family is limited, and only 13 of them have been fully sequenced (42), additional yet-unknown functions are likely to be encoded by these elements. For instance, it was suggested previously that several genes encoding putative restriction-modification systems, helicases and endonucleases, might act as barriers against invasion by foreign DNA or promote ICE integrity during transfer (42).
Our knowledge of the distribution and history of SXT/R391 ICEs is also limited and biased (8). Most of the screenings searching for the presence of SXT/R391 were performed on antibiotic-resistant clinical isolates, mainly of the species V. cholerae (8), and thus, several ecosystems are almost unexplored. One of these unexplored ecosystems is the marine aquaculture environment, where only Photobacterium damselae subsp. piscicida was found to harbor an ICE of this family to date (26). Beyond the ecological roles that bacteria play in marine ecosystems, they also fuel food webs in marine aquaculture systems and influence the health of cultured marine organisms (19, 36), thus reinforcing the interest in studying the prevalence of these elements in the above-mentioned ecosystems. Furthermore, in these antibiotic-impacted environments, bacterial communities may serve as reservoirs of antibiotic resistance that could potentially come into contact with human populations (31).
In the present study, we analyzed a relatively large set of novel SXT/R391-like ICEs detected in bacterial strains isolated from fish collected from different marine aquaculture environments in the northwestern Iberian Peninsula (Spain and Portugal) in the 2001-2010 period. Twelve novel ICEs were genetically and functionally analyzed, with special attention being paid to variable DNA regions that contain element-specific genes. The comparative analysis of these elements included the study of the host range and the exclusion group within this family of ICEs.
MATERIALS AND METHODS
Bacterial strain isolation and screening for the presence of SXT/R391-like ICEs.
A collection of bacterial strains was obtained from liver, kidney, spleen, and gut of fish collected from different marine aquaculture environments in Galicia (northwest Spain) and Portugal during the years 2001 to 2010. Pure cultures were obtained on Trypticase soy agar (TSA) (Panreac, Spain) plates, and taxonomic classification was carried out by conventional biochemical tests. A total of 498 strains assigned to the genera Vibrio and Shewanella were screened for the presence of SXT/R391-like ICEs by targeting the int gene, encoding a conserved integrase within this family of ICEs. PCR using int-specific primers (Table 1) was performed directly on lysed colonies. Freshly grown colonies were resuspended in 25 μl of sterile water and heated at 94°C for 5 min, immediately prior to PCR. PCR amplification of 25-μl reaction mixtures containing 0.4 μM each primer, 0.8 mM deoxynucleoside triphosphates (dNTPs) (GenScript, NJ), 1 U of KAPATaq DNA polymerase (KAPA Biosystems, MA), and KAPA reaction buffer was performed on My Cycler thermal cyclers (Bio-Rad, CA). PCR steps were as follows: an initial denaturation step at 94°C for 3 min, followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 1 min, with a final 10-min elongation step at 72°C. The size (expected, 1,034 bp in length) of the PCR products was determined by agarose gel electrophoresis using a 1-kb O'gene ruler DNA size standard (Fermentas, Lithuania).
Table 1.
Primers used in this study
| Primer | Sequence (5′–3′) | Targeted gene and/or region |
|---|---|---|
| PA forward | AGAGTTTGATCCTGGCTCAG | 16S rRNA gene |
| PH reverse | AAGGAGGTGATCCAGCCGCA | |
| Anti-KK forward | CGTGCCAGCAGCCGCGGTAAT | |
| attP-B forward | GCCAATTACGATTAACACGACGG | attP site |
| attP-F reverse | TGCTGTCATCTGCATTCTCCT | |
| sxt-286 forward | CTGTGGCCAATCATCAACTC | int |
| sxt-287 reverse | CGACCGAGATGGGCTAAGTG | |
| HI5-1 forward | AGCGTGTTGTATAAGCGGGA | rumBA (variable region III) |
| rumA-3-1 reverse | CCTTCACCAGCGCAGGATTA | |
| S043-5-1 forward | ACCTGGAAGATTAAGAAGAC | s043-traB (hot spot 1) |
| TraB-5-2 reverse | TCGCAGCCAAACTATCGACC | |
| HS2-1F forward | GGTGATGCTTTCACCGATGT | traA-s054 (hot spot 2) |
| s054-3-1 reverse | GGACCAGGCATCATAGAGCG | |
| HS4-1F forward | TGTGAAGGTTTGACCGCGAG | traN-s063 (hot spot 4) |
| HS4-1R reverse | TGTTGATGGCAACTACGCAT | |
| traGs forward | GATGGGTTAAAAGCCGCCGTTGATGC | traG-eex (variable region IV and eexa) |
| eex-seq-1 reverse | GCAATCGCATATTGGCAAGC |
The eex gene sequence defines R/S exclusion groups.
Taxonomic characterization of ICE-harboring strains.
Bacterial strains presenting int gene amplification underwent taxonomic assignment to the species level on the basis of the 16S rRNA gene sequence using primers PA forward (corresponding to Escherichia coli 16S rRNA gene positions 8 to 27) (6), PH reverse (E. coli 16S rRNA gene positions 1522 to 1541), and Anti-KK forward (E. coli 16S rRNA gene positions 514 to 534) (Table 1). PCR conditions were as described above, with 1.5 min of elongation time. For strains related to Vibrio scophthalmi, Vibrio splendidus, and Shewanella haliotis, 16S rRNA gene-based identification was completed with additional tests. Growth in 8% NaCl and in galactose as the sole carbon source allowed the identification of Vibrio scophthalmi and Vibrio splendidus, and growth in 6% NaCl together with hemolysis on blood agar allowed the identification of Shewanella haliotis.
Antibiotic, mercury, and QAC resistance.
Antibiotic resistance profiles of ICE-harboring strains were obtained by culturing the strains on TSA plates supplemented with tetracycline (4 and 12 μg ml−1), streptomycin (200 μg ml−1), trimethoprim (32 μg ml−1), kanamycin (50 μg ml−1), chloramphenicol (20 μg ml−1), nalidixic acid (40 μg ml−1), and spectinomycin (50 μg ml−1).
Resistance to mercury, quaternary ammonium compounds (QACs), and rifampin, suggested by the presence of genetic determinants in variable regions, was tested on ICE-harboring strains and E. coli transconjugants (see “Mating assays” below). Strains were transferred onto TSA plates supplemented with 300 μM HgCl2 (Panreac, Spain), 60 μg ml−1 of benzalkonium chloride (Fluka, MO), or rifampin (50 μg ml−1).
DNA sequencing and analysis.
Variable DNA insertions were amplified by using PCR primers targeting conserved core genes (Table 1). The S/R exclusion group, determined by the sequences of the TraG and Eex proteins (21), was defined by a partial sequence analysis of the eex gene (Table 1).
Genomic DNA was extracted by employing the Easy-DNA kit (Invitrogen, CA). Variable DNA insertions were amplified by using the Long PCR enzyme mix kit (Fermentas, Lithuania). PCR conditions were set according to the manufacturer's instructions, using 55°C as the annealing temperature and 17 min as the extension time. PCR products were purified (GeneJET PCR purification kit; Fermentas, Lithuania) and quantified (NanoDrop ND-1000 spectrophotometer; Thermo Fisher Scientific, MA).
The nucleotide sequence was obtained by using the GenomeLab Dye Terminator cycle sequencing kit (Beckman Coulter, CA), and samples were loaded into an automated CEQ8000 genetic analysis system (Beckman Coulter, CA). Sequences were analyzed against other SXT/R391-like ICEs in GenBank by employing BLASTN and BLASTP tools. Putative transposase genes were analyzed by using ISfinder (http://www-is.biotoul.fr/is.html).
The phylogenetic tree for int sequences was obtained by employing CLUSTALX for sequence alignment (37) and PHYLO WIN to build trees according to the maximum parsimony method (15). Bootstrap analysis was performed based on 500 replications with the Kimura two-parameter correction for the distances.
Mating assays.
ICE mobility was tested in conjugation experiments using kanamycin-resistant E. coli CAG18420 and nalidixic acid-resistant E. coli BI533 as recipients, as previously described (26). Exponentially growing cells of donor and recipient strains were mixed in a 1:1 ratio, concentrated 10 times by centrifugation, spotted onto LB agar plates (Sigma-Aldrich, MO), and incubated at 25°C for 5 to 15 h. After incubation, mating spots were collected and resuspended to make serial dilutions. Transconjugants were selected at 37°C on LB agar plates containing tetracycline, trimethoprim, or streptomycin together with kanamycin or nalidixic acid, depending on the recipient strain used. The ICE transfer frequency was expressed as the number of transconjugants per recipient cell.
The excision ability of the ICEs was verified by PCR targeting of the circular intermediate in ICE-harboring strains (i.e., the reconstituted attP site), using primers attP-B forward and attP-F reverse (Table 1).
Nucleotide sequence accession numbers.
The GenBank/EMBL/DDBJ accession numbers for the ICE sequences are HE577619 to HE577630.
RESULTS
Strains diversity and characteristics.
SXT/R391-like ICEs were detected in 12 of the 498 strains studied (i.e., 2.4%). These strains belonged to 5 different bacterial species: V. splendidus, V. scophthalmi, V. alginolyticus, Shewanella haliotis, and Enterovibrio nigricans (Table 2). Neither these species nor the genus Enterovibrio had been previously related with the SXT/R391 family of ICEs.
Table 2.
SXT/R391 ICEs detected in this study and characteristics of ICE-harboring strainsb
| ICEa | Strain | Phylogenetically closest species | Fish species | Location, yr of isolation | Strain resistance(s) | Resistance(s) mobilized to E. coli transconjugantsc | ICE exclusion group |
|---|---|---|---|---|---|---|---|
| ICEVscSpa1 | ACC7 | Vibrio scophthalmi | Scophthalmus maximus | Lugo, Spain, 2010 | SPT | — | S |
| ICEVscSpa2 | NC1 | Vibrio scophthalmi | Solea solea | A Coruña, Spain, 2006 | Not found | — | R |
| ICEVscSpa3 | YF7 | Vibrio scophthalmi | Solea solea | Lugo, Spain, 2009 | TET, STR | — | S |
| ICEVspPor1 | ZD4 | Vibrio splendidus | Dicentrarchus labrax | Porto, Portugal, 2009 | TET, STR,TMP, NAL, SPT | TET, STR,TMP | S |
| ICEVspSpa1 | ZF2 | Vibrio splendidus | Scophthalmus maximus | Lugo, Spain, 2009 | TET, Hg | TET, Hg | R |
| ICEVspPor2 | ZD5 | Vibrio splendidus | Dicentrarchus labrax | Porto, Portugal, 2009 | TMP, NAL, SPT, RIF | TMP, RIF | S |
| ICEVspSpa2 | EB5 | Vibrio splendidus | Scophthalmus maximus | A Coruña, Spain, 2004 | STR | STR | S |
| ICEVspSpa3 | 14-3 | Vibrio splendidus | Scophthalmus maximus | Lugo, Spain, 2010 | TET, STR, TMP | — | S |
| ICEEniSpa1 | VA8 | Enterovibrio nigricans | Scophthalmus maximus | Pontevedra, Spain, 2008 | TET, STR, Hg | TET, STR, Hg, BC | R |
| ICEEniSpa2 | SH5 | Enterovibrio nigricans | Scophthalmus maximus | A Coruña, Spain, 2007 | Not found | — | S |
| ICEValPor1 | HI5 | Vibrio alginolyticus | Solea solea | Porto, Portugal, 2005 | TET, STR, TMP, CHL, SPT, RIF | TET, TMP, CHL, RIF | S |
| ICEShaPor1 | AC6 | Shewanella haliotis | Scophthalmus maximus | Aveiro, Portugal, 2002 | TET, TMP | — | R |
ICEs are named after the initials of the host bacterial species followed by the location from where the isolate was obtained.
TET, tetracycline; STR, streptomycin; TMP, trimethoprim; CHL, chloramphenicol; NAL, nalidixic acid; SPT, spectinomycin; RIF, rifampin; Hg, HgCl2; BC, benzalkonium chloride.
—, ICEs for which mobilization to E. coli could not be detected (E. coli recipients were unable to grow in antibiotic-supplemented medium).
Most ICE-harboring strains were isolated from turbot (Scophthalmus maximus) and additionally from common sole (Solea solea) and European sea bass (Dicentrarchus labrax). ICEs were detected in strains isolated almost every year from 2002 to 2010 but especially in the recent years 2009 and 2010. One-half of the total number of ICE-harboring strains were isolated these 2 years. Interestingly, strains isolated in the years 2009 to 2010 represented only 16.6% of the 498 strains examined. These results suggest the spread of SXT/R391-like ICEs in Vibrio, Enterovibrio, and Shewanella sp. strains inhabiting marine aquaculture environments of the northwestern Iberian Peninsula.
Resistance to the tested antibiotics was widely distributed among ICE-harboring isolates (Table 2). Most resistant strains (70%) could grow in the presence of tetracycline. Resistance to streptomycin and trimethoprim was found in 60 and 50% of resistant strains, respectively, and less abundant were spectinomycin (40%), nalidixic acid (20%), and chloramphenicol (10%) resistances. All strains were sensitive to kanamycin. Different antibiotic resistance patterns were observed for strains belonging to the same bacterial species (Table 2), suggesting that some of these antibiotic resistances could be encoded in the ICEs or in other mobile genetic elements.
Comparative analysis of ICE variable regions.
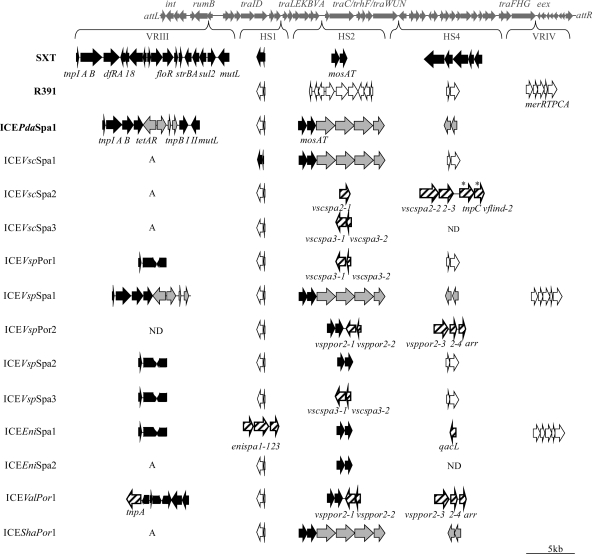
Different regions potentially containing variable DNA were analyzed in the 12 detected ICEs, including variable regions III and IV and hotspots 1, 2, and 4. These regions were selected on the basis of previous reports showing that functionally important traits, such as antibiotic resistance or toxin-antitoxin systems, were often encoded in these regions. Three previously sequenced elements, SXT, R391, and ICEPdaSpa1, were used for comparative purposes (Fig. 1).
Fig 1.
Comparative analysis of analyzed variable regions (VRs) and hotspots (HSs) between the studied ICEs and others from the SXT/R391 family. Genes originally described for SXT (GenBank accession number AY055428), R391 (accession number AY090559), and ICEPdaSpa1 (accession number AJ870986) are shown in black, white, and gray, respectively. Genes unrelated to previous ICEs are shown with hatched arrows. A, absence; ND, not determined. The asterisks indicate that proteins encoded by these two genes shared 85 and 83% similarities to those encoded by tnpC and vflind-2 of ICEVflInd1 (accession number GQ463144), respectively. Specific genes (and the corresponding predicted proteins) were as follows: arr (rifampin ADP-ribosyl transferase), enispa1-1 (putative iron-dependent peroxidase), enispa1-2 (putative Zn peptidase), enispa1-3 (ISSod5 transposase), tnpC (putative transposase, IS21 family), qacL (quaternary ammonium compound resistance protein), tnpA (putative transposase, IS4 family), vscspa2-1 (putative Prop osmoprotectant transporter), vscspa2-2, (putative ATP-dependent endonuclease), vscspa2-3 (putative UvrD/REP helicase), vscspa3-1 (conserved hypothetical protein), vscspa3-2 (conserved hypothetical protein), vsppor2-1 (putative antitoxin component), vsppor2-2 (hypothetical protein), vsppor2-3 (putative ISSod25 integrase), vsppor2-4 (putative transposase), and vflind-2 (putative transposition helper protein).
VRIII, inserted into the rumB gene, was present in at least 6 of the ICEs. The remaining ICEs showed the integrity of the rumB gene, and for ICEVspPor2, the region could not be amplified, suggesting either the presence of a large insertion unable to be amplified under the PCR conditions used or variations in the rumB sequence preventing primer annealing. Although antibiotic resistance determinants were often present within this region in ICEs characterized in previous studies, our results showed tetracycline resistance genes only in ICEVspSpa1 (Fig. 1). The ICEVspSpa1 tetA and tetR genes encoded proteins showing 100% identity to the tetracycline resistance protein TetA and the tetracycline repressor TetR, respectively, of ICEPdaSpa1, previously described for Photobacterium damselae subsp. piscicida (26). The VRIII sequences of these two ICEs (ICEVspSpa1 and ICEPdaSpa1) shared 99% nucleotide identity. Four ICEs presented a truncated version of VRIII previously described for ICEs such as SXT or ICEPdaSpa1 (Fig. 1). Finally, ICEValPor1 presented a different truncated form of V. cholerae SXT VRIII (Fig. 1), including a putative transposase encoded by tnpA, which showed 99% nucleotide identity to ISEc29 of the IS4 family.
HS1 was amplified in all studied ICEs, including in 10 of these elements contents identical to that of HS1 of the previously described R391 and ICEPdaSpa1 (Fig. 1), consisting of genes encoding two conserved hypothetical proteins. ICEVscSpa1 showed a content similar to that of SXT, consisting of the s044 and s045 genes, encoding a putative toxin-antitoxin-stabilizing system (42). ICEEniSpa1 contained three genes in HS1 encoding proteins showing 53% similarity to a putative iron-dependent peroxidase of Edwardsiella tarda, 75% similarity to a putative Zn peptidase of Aeromonas veronii, and 83% similarity to the ISSod5 transposase of Shewanella oneidensis, respectively, by a BLASTP search (Fig. 1). These three genes have not been described for SXT/R391 ICEs to date.
HS2 was also amplified in all the elements studied, and for the ICEVscSpa1, ICEVspSpa1, and ICEShaPor1 sequences, it showed 100% identity to HS2 of the previously described ICEPdaSpa1 (Fig. 1), ICEVchBan9, and ICEVchMoz10 (42). In HS2, eight ICEs presented the mosAT genes, encoding a toxin-antitoxin-stabilizing system (41). In addition to mosAT, ICEVspPor2 and ICEValPor1 contained a gene encoding a protein showing 63% similarity to an antitoxin component of Neisseria polysaccharea (Fig. 1). This antitoxin component shows similarity to the helix-turn-helix Xre family of transcription factors, which also includes MosA (22), and may represent an additional antitoxin component protecting against the loss of ICEs. Also in this hot spot, a gene encoding a protein showing 49% similarity to the Francisella tularensis subsp. holarctica ProP osmoprotectant transporter (ICEVscSpa2) as well as others encoding conserved hypothetical proteins (ICEVscSpa3, ICEVspPor1, and ICEVspSpa3) were identified (Fig. 1).
HS4 was detected in all ICEs except ICEVscSpa3 and ICEEniSpa2, where it could not be amplified by PCR. Half of the elements showed gene contents identical to those previously described for either R391 or ICEPdaSpa1 in this hot spot (Fig. 1). Genes encoding putative resistances to antibiotics and biocides were found in HS4 in some elements. One of these genes, encoding a protein showing 88% similarity to the Spirosoma linguale Arr rifampin ADP-ribosyl transferase, was found in both ICEVspPor2 and ICEValPor1 and conferred resistance to rifampin to its Vibrio hosts (Table 2). Another gene, encoding a protein showing 88% similarity to the QacE quaternary ammonium compound (QAC) resistance protein of Enterobacter aerogenes plasmid R751, was found in ICEEniSpa1 (Fig. 1). We verified that this gene conferred resistance to the QAC benzalkonium chloride (Table 2). On the basis of this phenotype and the low level of similarity to other previously described qac genes, this gene was named qacL. These findings constitute the first report of rifampin and QAC resistance determinants carried by ICEs of the SXT/R391 family.
Moreover, two novel genes were found in ICEVscSpa2 HS4, whose predicted proteins showed similarities of 74% to a Pseudomonas syringae putative ATP-dependent endonuclease and 63% to an Escherichia coli helicase. This element also contains two additional genes encoding proteins showing 85 and 83% similarity to the TnpC putative transposase and vflind-2 transposition helper protein of ICEVflInd1, respectively, in HS4 (Fig. 1). The tnpC gene of ICEVscSpa2 shared 70% nucleotide identity with IS1421 of the IS21 family.
VRIV was present in two ICEs (Fig. 1). ICEVspSpa1 and ICEEniSpa1 presented gene contents in this region identical to that of R391, consisting of a narrow-spectrum mercury resistance operon (Fig. 1). The presence of a mer operon was previously described for R391 and related ICEs; however, the simultaneous presence of this operon and a tetA resistance locus, as in ICEVspSpa1, is a novelty.
ICE exclusion groups.
S and R exclusion groups were represented among the ICEs studied. Eight ICEs belonged to the S exclusion group, and 4 ICEs belonged to the R exclusion group (Table 2). The 3′ end of the Eex amino acid sequence (i.e., amino acid positions 102 to 143) was highly conserved with respect to previously described ICEs of the S exclusion group (21). However, variations in the Eex sequence were observed for two ICEs belonging to the R exclusion group. ICEVscSpa2 presented an arginine instead of a histidine at position 103 of Eex, and ICEShaPor1 presented a phenylalanine instead of a leucine at position 137.
int gene-based phylogeny.
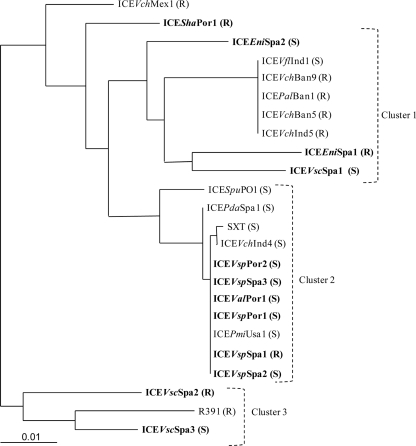
Relationships between the studied ICEs and others from the SXT/R391 family were investigated by a phylogenetic analysis of the int gene, encoding the integrase. Three different clusters could be observed in the phylogenetic tree (Fig. 2), each of them including both previously described ICEs and ICEs detected in this study. Cluster 1 involved a high number of ICEs belonging to the R exclusion group (i.e., 62%) (Fig. 2). Also, in this cluster, all ICEs described in previous studies had been detected in bacterial strains isolated in Southeast Asia (i.e., India and Bangladesh) (Fig. 2). Cluster 2 included mostly ICEs belonging to the S exclusion group (i.e., 90%). In this cluster, ICEs described in previous studies had diverse origins (Fig. 2). Finally, cluster 3 involved ICEs of the S and R types and R391 detected in a South African bacterial isolate (Fig. 2).
Fig 2.
Maximum likelihood phylogenetic tree, based on int gene sequences, showing the relationships between the ICEs described in the present study (shown in boldface type) and others from the SXT/R391 family. Bar, 0.01 substitutions per nucleotide position. S/R indicates the ICE exclusion group.
ICE excision, mobility, and phenotypes transferred.
Mobility tests were attempted for 10 of the studied ICEs that presented resistance to at least one of the tested antibiotics in order to be used as selectable markers. Prior to conjugative transfer experiments, the excision ability of the ICEs was confirmed by positive PCR amplification of the reconstituted attP site in all ICEs except ICEVscSpa3. The ability to be transferred to an E. coli strain was demonstrated for 6 ICEs by the mobilization of antibiotic resistances from the donor to the recipient strain (Table 2) and by the PCR detection of the ICE integrase gene in the recipients (data not shown). The transfer rates ranged from 1.2 × 10−5 to 2.3 × 10−8 transconjugants per recipient cell.
In order to corroborate the functionality of the two novel determinants for rifampin and QAC resistances as well as of the mercury resistance operon, we also examined the resistance phenotypes conferred to the E. coli transconjugants. Recipient E. coli strains of ICEVspPor2 and ICEValPor1, containing the arr rifampin ADP-ribosyl transferase gene in HS4, were resistant to this antibiotic (Table 2). Similarly, transconjugants of ICEVspSpa1 and ICEEniSpa1, containing a narrow-spectrum mercury resistance operon in VRIV, were resistant to mercury II chloride (Table 2). Finally, transconjugants harboring ICEEniSpa1 that contained the qacL quaternary ammonium compound resistance gene in HS2 were resistant to benzalkonium chloride (Table 2).
DISCUSSION
The present study revealed an increased ICE prevalence in Vibrio, Enterovibrio, and Shewanella sp. strains inhabiting different marine aquaculture environments. Few previous surveys analyzed the prevalence of this family of ICEs, and they were limited to clinical isolates related to V. cholerae. Those studies reported the spread of ICE-harboring V. cholerae strains in Eastern Africa during the late 1990s (29) and also in India from 1994 to 2005 (10). Therefore, our results and previous data exhibited similar trends, indicating a gradual emergence of SXT/R391-like ICEs in bacteria of different environments and locations.
ICEs of this family were previously detected in only 8 species of gammaproteobacteria, which included V. cholerae (39), Vibrio fluvialis (1), Vibrio parahaemolyticus (33), Providencia alcalifaciens (20), P. rettgeri (11), Proteus mirabilis (27), Shewanella putrefaciens (28), and Photobacterium damselae subsp. piscicida (26). This study allowed the identification of 5 new species harboring ICEs in aquaculture environments (V. splendidus, V. scophthalmi, V. alginolyticus, Shewanella haliotis, and Enterovibrio nigricans), contributing notably to the broadening of the known host range for these elements. These results show that SXT/R391 ICEs have the potential to be transferred and, hence, to shape the genomes of an important number of bacterial species. The presence of ICEs in two species of Enterovibrio shows that the horizontal transfer of these elements occurs beyond the genera Vibrio, Providencia, Proteus, Shewanella, and Photobacterium within the gammaproteobacteria. The detection of the excision ability of the ICEs in nearly all the elements and the demonstration of conjugative transfer to E. coli for an important fraction of them suggest that they remain functional. Since ICEs can also mobilize genomic islands (12), the increased host range also suggests that the variety of candidate genomic islands that could be mobilized by these elements might be greater than expected.
The spread of SXT/R391-like ICEs could be related to the potential ecological advantages that they may provide. Element-specific properties, such as antibiotic and heavy metal resistance or the regulation of motility, are encoded in variable DNA insertions (5, 8). Antibiotic resistance determinants are often encoded in the resistance cluster, also referred to as VRIII (2, 34, 41). Our study showed that VRIII included antibiotic resistance determinants in only one element (ICEVspSpa1). Conversely, the sequence analysis of further variable regions in the present study, such as HS4, revealed rifampin resistance genes in two ICEs (ICEVspPor2 and ICEValPor1).
Wozniak et al. (42) had shown previously that antibiotic resistance determinants were not limited to the resistance cluster in SXT/R391-like ICEs. Thus, we cannot exclude the presence of other antibiotic resistance determinants, such as those observed for E. coli transconjugants, in unexplored variable regions of these ICEs. Interestingly, this was the first evidence of rifampin-resistant determinants in this family of mobile elements. The acquisition of this gene (arr) by these ICEs appeared to be mediated by the integrase, as suggested by the nearby presence of the ISSod25 integrase gene. Furthermore, rifampin resistance by means of resistance genes is frequent in nonclinical bacterial isolates (40), with resistance due to rpoB mutations being more common in clinical isolates (38). This suggests a nonclinical origin for SXT/R391 ICE-encoded rifampin resistance.
Other resistance determinants protecting against QACs and heavy metals were found in this study. While a similar narrow-spectrum mercury resistance operon was found previously in R391 and related ICEs (3), there is no previous evidence of qacL-related genes in SXT/R391 ICEs. QACs are used in hospitals and by food industries, including fish farms, to control unwanted microorganisms (23, 30). Despite this, previous studies failed to find QAC-resistant bacteria in fish farms (32). The resistance determinants encoded in the ICEs reported in the present study, including QACs, rifampin, and mercury II chloride, were shown to be functional. The transfer of the corresponding ICEs to E. coli provided resistance to these chemicals. Thus, SXT/R391-like ICEs contribute to the spread of QAC, antibiotic, and mercury resistances in aquaculture environments.
Whether the ecological success of SXT/R391 ICEs is related exclusively to the presence of resistance determinants is still unclear. Bani et al. (2) suggested previously that the disappearance of ICEVchVie0 from V. cholerae isolates in Vietnam may be explained by the lack of resistance determinants. However, fully sequenced ICEs without any antibiotic resistances have been described (42), and other biological functions were proven to confer selective advantages to the ICE-harboring cell (5). Determinants for antibiotic resistance were not found in most ICEs in this study. Other genes found in variable regions may confer further advantages to the recipient strain. Genes encoding putative helicases and endonucleases (HS4 of ICEVscSpa2) might play a role against invasion by foreign DNA or promote the integrity of the ICE genome during transfer between hosts (42).
The presence of both exclusion groups suggested that these elements have different origins. This was also supported by the phylogeny of the int gene, which differentiated Southeast Asian ICEs, S-group ICEs with multiple origins, and R391-related ICEs. Even though most previous surveys were performed with clinical isolates, ICEs of the SXT/R391 family have been detected in bacteria isolated worldwide (8). Five of the twelve ICEs analyzed in this study presented int gene similarity and belonged to the same exclusion group as that of ICEPdaSpa1, which was detected previously in the aquaculture environment of the Iberian Peninsula (26). Therefore, ICEPdaSpa1 appears to be related to at least some of these ICEs. The analysis of variable regions showed novel gene combinations for all ICEs in this study (Fig. 1). Extensive recombination between tandem ICEs that coexist transiently in the same cell are supposed to shape the ICE genomes and to contribute to their mosaicism (9, 16, 42). The presence of ICEs of both the R and S exclusion groups in aquaculture environments is supposed to allow the transient formation of tandem ICEs and the subsequent genetic reorganization of their variable DNAs.
In conclusion, the present study revealed an increasing prevalence of SXT/R391 ICEs in marine aquaculture environments, where they may be important reservoirs of resistance to antibiotics, QACs, and heavy metals, along with plasmids, transposons, and integrons. The results also showed the SXT/R391 ICE host range to be broader than expected, suggesting a more important role of these elements as contributors to horizontal gene transfer.
ACKNOWLEDGMENTS
This work was partially supported by grant INCITE08PXIB235028PR from Xunta de Galicia and by grants AGL2009-12266-C02-01 and CSD2007-00002 (Consolider Aquagenomics) (both cofunded by the FEDER Programme of the European Union) from the Ministry of Science and Innovation (MICINN) of Spain.
Footnotes
Published ahead of print 6 February 2012
REFERENCES
- 1. Ahmed AM, Shinoda S, Shimamoto T. 2005. A variant type of Vibrio cholerae SXT element in a multidrug-resistant strain of Vibrio fluvialis. FEMS Microbiol. Lett. 242:241–247 [DOI] [PubMed] [Google Scholar]
- 2. Bani S, et al. 2007. Molecular characterization of ICEVchVie0 and its disappearance in Vibrio cholerae O1 strains isolated in 2003 in Vietnam. FEMS Microbiol. Lett. 266:42–48 [DOI] [PubMed] [Google Scholar]
- 3. Böltner D, Osborn AM. 2004. Structural comparison of the integrative and conjugative elements R391, pMERPH, R997, and SXT. Plasmid 51:12–23 [DOI] [PubMed] [Google Scholar]
- 4. Böltner D, MacMahon C, Pembroke JT, Strike P, Osborn AM. 2002. R391: a conjugative integrating mosaic comprised of phage, plasmid, and transposon elements. J. Bacteriol. 184:5158–5169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bordeleau E, Brouillette E, Robichaud N, Burrus V. 2010. Beyond antibiotic resistance: integrating conjugative elements of the SXT/R391 family that encode novel diguanylate cyclases participate to c-di-GMP signalling in Vibrio cholerae. Environ. Microbiol. 12:510–523 [DOI] [PubMed] [Google Scholar]
- 6. Brosius J, Dull TJ, Sleeter DD, Noller HF. 1981. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J. Mol. Biol. 148:107–127 [DOI] [PubMed] [Google Scholar]
- 7. Burrus V, Bontemps C, Decaris B, Guédon G. 2001. Characterization of a novel type II restriction-modification system, Sth368I, encoded by the integrative element ICESt1 of Streptococcus thermophilus CNRZ368. Appl. Environ. Microbiol. 67:1522–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Burrus V, Marrero J, Waldor MK. 2006. The current ICE age: biology and evolution of SXT-related integrating conjugative elements. Plasmid 55:173–183 [DOI] [PubMed] [Google Scholar]
- 9. Burrus V, Waldor MK. 2004. Shaping bacterial genomes with integrative and conjugative elements. Res. Microbiol. 155:376–386 [DOI] [PubMed] [Google Scholar]
- 10. Ceccarelli D, et al. 2011. ICEVchInd5 is prevalent in epidemic Vibrio cholerae O1 El Tor strains isolated in India. Int. J. Med. Microbiol. 301:318–324 [DOI] [PubMed] [Google Scholar]
- 11. Coetzee JN, Datta N, Hedges RW, Appelbaum PC. 1972. Transduction of R factors in Proteus mirabilis and P. rettgeri. J. Gen. Microbiol. 76:355–368 [DOI] [PubMed] [Google Scholar]
- 12. Daccord A, Ceccarelli D, Burrus V. 2010. Integrating conjugative elements of the SXT/R391 family trigger the excision and drive the mobilization of a new class of Vibrio genomic islands. Mol. Microbiol. 78:576–588 [DOI] [PubMed] [Google Scholar]
- 13. De la Cruz F, Davies J. 2000. Horizontal gene transfer and the origin of species: lessons from bacteria. Trends Microbiol. 8:128–133 [DOI] [PubMed] [Google Scholar]
- 14. Frost LS, Koraimann G. 2010. Regulation of bacterial conjugation: balancing opportunity with adversity. Future Microbiol. 5:1057–1071 [DOI] [PubMed] [Google Scholar]
- 15. Galtier N, Gouy M, Gautier C. 1996. SEAVIEW and PHYLO_WIN, two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biosci. 12:543–548 [DOI] [PubMed] [Google Scholar]
- 16. Garriss G, Waldor MK, Burrus V. 2009. Mobile antibiotic resistance encoding elements promote their own diversity. PLoS Genet. 5(12):e1000775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hacker J, Carniel E. 2001. Ecological fitness, genomic islands and bacterial pathogenicity—a Darwinian view of the evolution of microbes. EMBO Rep. 2:376–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hall RM, Stokes HW. 1993. Integrons: novel DNA elements which capture genes by site-specific recombination. Genetica 90:115–132 [DOI] [PubMed] [Google Scholar]
- 19. Hansen GH, Olafsen JA. 1999. Bacterial interactions in early life stages of marine cold water fish. Microb. Ecol. 38:1–26 [DOI] [PubMed] [Google Scholar]
- 20. Hochhut B, et al. 2001. Molecular analysis of antibiotic resistance gene clusters in Vibrio cholerae O139 and O1 SXT constins. Antimicrob. Agents Chemother. 45:2991–3000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Marrero J, Waldor MK. 2007. The SXT/R391 family of integrative conjugative elements is composed of two exclusion groups. J. Bacteriol. 189:3302–3305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McDonnell GE, McConnell DJ. 1994. Overproduction, isolation, and DNA-binding characteristics of Xre, the repressor protein from the Bacillus subtilis defective prophage PBSX. J. Bacteriol. 176:5831–5834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Naik BM, Venugopal MN, Karunasagar I, Karunasagar K. 2005. Effect of sanitizer treatment on bacteriology of microcosm simulating shrimp pond ecosystem. Asian Fish Sci. 18:127–137 [Google Scholar]
- 24. Nakamura Y, Itoh T, Matsuda H, Gojobori T. 2004. Biased function of horizontally transferred genes in prokaryotic genomes. Nat. Genet. 36:760–766 [DOI] [PubMed] [Google Scholar]
- 25. Ochman H, Lawrence JG, Groisman EA. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299–304 [DOI] [PubMed] [Google Scholar]
- 26. Osorio CR, et al. 2008. Genomic and functional analysis of ICEPdaSpaI, a fish pathogen derived SXT-related integrating conjugative element that can mobilize a virulence plasmid. J. Bacteriol. 190:3353–3361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pearson MM, et al. 2008. Complete genome sequence of uropathogenic Proteus mirabilis, a master of both adherence and motility. J. Bacteriol. 190:4027–4037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pembroke JT, Piterina AV. 2006. A novel ICE in the genome of Shewanella putrefaciens W3-18-1: comparison with the SXT/R391 ICE-like elements. FEMS Microbiol. Lett. 264:80–88 [DOI] [PubMed] [Google Scholar]
- 29. Pugliese N, Maimone F, Scrascia M, Materu SF, Pazzani C. 2009. SXT-related integrating conjugative element and IncC plasmids in Vibrio cholerae O1 strains in Eastern Africa. J. Antimicrob. Chemother. 63:438–442 [DOI] [PubMed] [Google Scholar]
- 30. Russell AD. 2002. Introduction of biocides into clinical practice and the impact on antibiotic-resistant bacteria. Symp. Ser. Soc. Appl. Microbiol. 31:121S–135S [PubMed] [Google Scholar]
- 31. Seyfried EE, Newton RJ, Rubert KF, IV, Pedersen JA, McMahon KD. 2010. Occurrence of tetracycline resistance genes in aquaculture facilities with varying use of oxytetracycline. Microb. Ecol. 59:799–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sidhu MS, Sørum H, Holck A. 2002. Resistance to quaternary ammonium compounds in food-related bacteria. Microb. Drug Resist. 8:393–399 [DOI] [PubMed] [Google Scholar]
- 33. Taviani E, et al. 2008. Environmental Vibrio spp., isolated in Mozambique, contain a polymorphic group of integrative conjugative elements and class 1 integrons. FEMS Microbiol. Ecol. 64:45–54 [DOI] [PubMed] [Google Scholar]
- 34. Taviani E, Grim CJ, Chun J, Huq A, Colwell RR. 2009. Genomic analysis of a novel integrative conjugative element in Vibrio cholerae. FEBS Lett. 583:3630–3636 [DOI] [PubMed] [Google Scholar]
- 35. Thomas CM, Nielsen KM. 2005. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 3:711–721 [DOI] [PubMed] [Google Scholar]
- 36. Thompson FL, Abreu PC, Wasielesky W. 2002. Importance of biofilm for water quality and nourishment in intensive shrimp culture. Aquaculture 203:263–278 [Google Scholar]
- 37. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tribuddharat C, Fennewald M. 1999. Integron-mediated rifampin resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 43:960–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Waldor MK, Tschäpe H, Mekalanos JJ. 1996. A new type of conjugative transposon encodes resistance to sulfamethoxazole, trimethoprim, and streptomycin in Vibrio cholerae O139. J. Bacteriol. 178:4157–4165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Williams DL, et al. 1994. Characterization of rifampin-resistance in pathogenic mycobacteria. Antimicrob. Agents Chemother. 38:2380–2386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wozniak RA, Waldor MK. 2009. A toxin-antitoxin system promotes the maintenance of an integrative conjugative element. PLoS Genet. 5:e1000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wozniak RA, et al. 2009. Comparative ICE genomics: insights into the evolution of the SXT/R391 family of ICEs. PLoS Genet. 5:e1000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wozniak RA, Waldor MK. 2010. Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat. Rev. Microbiol. 8:552–563 [DOI] [PubMed] [Google Scholar]
- 44. Zoetendal EG, Vaughan EE, de Vos WM. 2006. A microbial world within us. Mol. Microbiol. 59:1639–1650 [DOI] [PubMed] [Google Scholar]