Abstract
The energetically demanding process of translation is linked to multiple signalling events through mTOR mediated regulation of eukaryotic initiation factor (eIF) 4F complex assembly. Disrupting mTOR constraints on eIF4F activity can be oncogenic and alter chemotherapy response, making eIF4F an attractive anti-neoplastic target. Here we combine a newly developed inducible RNAi platform and pharmacological targeting of eIF4F activity to define a critical role for endogenous eIF4F in Myc-dependent tumor initiation. We find elevated Myc levels are associated with deregulated eIF4F activity in the prelymphomatous stage of the Eμ-Myc lymphoma model. Inhibition of eIF4F is synthetic lethal with elevated Myc in pre-malignant preB/B cells resulting in reduced numbers of cycling preB/B cells and delayed tumor onset. At the organismal level, eIF4F suppression affected a subset of normal regenerating cells but this was well tolerated and rapidly and completely reversible. Therefore, eIF4F is a key Myc client that represents a tumor-specific vulnerability.
INTRODUCTION
The heterotrimeric eukaryotic initiation factor (eIF)4F complex catalyzes the rate-limiting step of translation initiation by stimulating ribosome recruitment to mRNA templates. This is achieved through the coordinated action of the eIF4E subunit (required for binding to mRNA 5' cap [5'm7GpppN3'] structures), ATP hydrolysis (mediated by two eIF4A isoforms [eIF4AI and eIF4AII]), and interaction between the eIF4G subunit and the 43S pre-initiation complex. Different mRNAs show varying dependencies on eIF4F for ribosome recruitment – a feature attributed to accessibility of the mRNA 5' cap (Dever, 2002). Altered signalling flux through the PI3K/Akt/mTOR pathway in human cancers is associated with changes in eIF4F levels and modification of the cancer cell proteome due to selective translational effects (Rajasekhar et al., 2003). When elevated, eIF4E, the rate limiting subunit of eIF4F (Duncan et al., 1987), antagonizes Myc induced apoptosis and cooperates with Myc in tumorigenesis (Li et al., 2003; Ruggero et al., 2004; Wendel et al., 2004). As well, Myc exerts profound effects on protein synthesis through regulation of ribosome biogenesis (van Riggelen et al., 2010) and transcriptional control of the three eIF4F subunits (Jones et al., 1996; Lin et al., 2008). Increases in eIF4F activity have been shown to selectively stimulate the expression of malignancy-related mRNAs by augmenting nucleo/cytoplasmic transport of Cyclin D1 (Rousseau et al., 1996) and translation of Mcl-1 (Wendel et al., 2007) and Myc (Lin et al., 2008). The recent description of a Myc/eIF4F transcription/translation-coupled mitogenic loop (Lin et al., 2008) prompted us to develop a trackable mouse model to assess the in vivo contribution of eIF4F to Myc-dependent tumor initiation.
RESULTS
Transient Suppression of eIF4E Delays Myc-dependent Tumor Initiation
Mouse models provide valuable platforms for identifying and characterizing lesions that promote tumorigenesis and for testing the significance of effector pathways downstream of known oncogenes and/or tumor suppressors for their contribution to cancer development and/or maintenance (Schmitt and Lowe, 2002). In the Eμ-Myc lymphoma model, Myc expression is driven by the lymphoid-specific IgH enhancer (Eμ) and becomes elevated in the pre-B/B cell compartment (Adams et al., 1985). Significant insight into Myc biology has been obtained using this model - from the finding that tumor-derived Myc mutants uncouple proliferation from apoptosis (Hemann et al., 2005) to the identification/characterization of ornithine decarboxylase, the rate limiting enzyme for polyamine biosynthesis, as a Myc effector (Nilsson et al., 2005).
Sensitive transplantation assays of ostensibly healthy Eμ-Myc mice have shown that the majority (~90%) of Eμ-Myc donors, between 4–6 weeks of age do not harbor malignant lymphoma cells (Langdon et al., 1986) (Figure 1A). In this prelymphomatous stage, there is a polyclonal expansion of morphologically distinct pre B cells (Figure 1B) that is offset by an increased apoptotic index (Jacobsen et al., 1994; Langdon et al., 1986). Subsequent acquisition of genetic lesions that block the cell death program trigger tumorigenesis (Strasser et al., 1990). Indeed, we have previously shown that enforced expression of eIF4E can drive aggressive cancers by attenuating apoptosis in this setting (Wendel et al., 2004). The Eμ-Myc mouse is thus a powerful model to study the contribution of Myc network components to tumor initiation, leading us to ask if perturbed eIF4F activity is a pre-malignant feature of Eμ-Myc pre B/B cells.
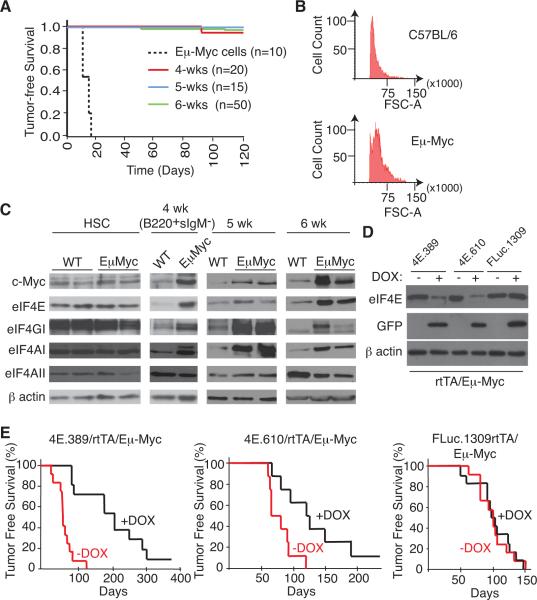
Figure 1. Transient Suppression of eIF4E Delays Myc-dependent Tumor Initiation.
A. Kaplan-Meier plot illustrating tumor-free survival of recipient C57BL/6 mice transplanted with 20 Eμ-Myc lymphoma cells or 2 × 106 bone marrow cells from 4, 5, and 6 wk old Eμ-Myc mice. B. Forward light scatter from a flow cytometer of B220+ splenocytes from 6 wk old C57BL/6 and Eμ-Myc mice. This assay was used as an independent validation to confirm the prelymphomatous nature of Eμ-Myc cells used in the transplantation experiments in (A). C. Western blot analysis of c-Myc transcriptional targets (eIF4E, eIF4AI, eIF4GI) in C57BL/6 (WT) and Eμ-Myc HSCs, B220+sIgM− pre-B cells, and splenocytes from mice of the indicated ages. D. Suppression of eIF4E expression in B220+ cells isolated from 6 wk old mice of the indicated genotype pre-treated with vehicle or DOX for 2 weeks. E. Kaplan-Meier plot showing lymphoma-free survival of 4E.389/rtTA/Eμ-Myc (n=12), 4E.610/rtTA/Eμ-Myc (n=10) and FLuc.1309/rtTA/Eμ-Myc (n=14) mice that had been treated with vehicle (red) or with DOX (black) for 21 days starting at 4 wk of age.
To this end, we analyzed extracts from wild-type (WT) and Eμ-Myc hematopoietic stem cells (HSCs), 4 wk B220+sIgM−, and 5–6 wk splenic cells (Figure 1C). Myc expression is not elevated in Eμ-Myc HSCs but is appreciably increased in Eμ-Myc B220+sIgM− B cells and splenocytes isolated from 4–6 wk old mice compared to WT controls (Figure 1C). The expression of all three eIF4F subunits is also increased in Eμ-Myc B220+sIgM− B cells and splenocytes from 4–6 week old mice relative to WT controls (Figure 1C). These results are consistent with previous studies indicating that eIF4E, eIF4AI, and eIF4GI, but not eIF4AII, are transcriptional targets of Myc (Jones et al., 1996; Lin et al., 2008) and demonstrate that eIF4F upregulation is a signature of pre-malignant Eμ-Myc pre-B/B lymphocytes.
To address whether elevated levels of eIF4F in Eμ-Myc pre B/B cells represents an epigenetic change essential for tumor initiation, we took advantage of a powerful platform that combines optimized GFP-coupled shRNA technology with a Flp/FRT recombinase-mediated cassette exchange (RMCE) strategy to generate mice that conditionally express potent shRNAs targeting eIF4E (McJunkin et al., 2011; Premsrirut et al., 2011). Two independent miR-based shRNAs (shRNAmir) that target the eIF4E coding region, 4E.389 and 4E.610, and one that expresses a neutral control shRNAmir targeting Firefly Luciferase, FLuc.1309 (Premsrirut et al., 2011), were introduced into the FRT-hygro-pA “homing cassette” at the ColA1 locus of KH2 ES cells (Figure S1A). These cells also harbor a reverse tet-transactivator (rtTA2) targeted to the Rosa26 locus (referred to herein as rtTA) enabling potent, doxycycline (DOX)-inducible suppression of eIF4E (Figure S1B). Mice generated from these ES cells show inducible suppression of eIF4E in a wide spectrum of cells and tissues, including, liver, spleen, skin, intestine, components of the hematopoietic system and embryo-derived fibroblasts (Premsrirut et al., 2011) (Figures S1C–D and below).
We crossed shRNAmir/rtTA and Eμ-Myc mice to generate triple transgenic progeny in which eIF4E expression was suppressed during the prelymphomatous stage between 4–7 wks of age. Robust induction of GFP expression and potent suppression of eIF4E was apparent in pre-B/B B220+ cells from 4E.389/rtTA/Eμ-Myc and 4E.610/rtTA/Eμ-Myc mice on DOX (Figure 1D). DOX-treated 4E.389/rtTA/Eμ-Myc and 4E.610/rtTA/Eμ-Myc mice showed a significant delay in lymphoma onset compared to either untreated controls or to DOX-treated FLuc.1309/rtTA/Eμ-Myc mice (Figure 1E; p<0.001 for 4E.389/rtTA/Eμ-Myc mice and p<0.01 for 4E.610/rtTA/Eμ-Myc). We attempted to assess the consequences of eIF4E suppression on tumor cell maintenance by administering DOX to 4 month old lymphoma-bearing 4E.389/rtTA/Eμ-Myc mice, but no stable response to disease was noted and GFP induction was detected in only a minority of tumor cells (~8–20%) (data not shown). This either reflects a collapse of the rtTA-inducible system as noted in other settings (Podsypanina et al., 2008), alterations in the DOX- or sheIF4E-responsiveness of the target cell population, and/or weak or mosaic sheIF4E expression – the latter having been documented in this shRNAmir/rtTA transgenic system (McJunkin et al., 2011). None-the-less, our results indicate that endogenous eIF4E is required for conversion of Eμ-Myc preB/B cells to malignant lymphomas.
Suppression of eIF4E Reduces the B220+ Eμ-Myc pre-B/B Cell Compartment by Impairing Cell Division and Apoptosis
To elucidate the mechanism(s) responsible for the delayed tumor onset, we analyzed the consequences of eIF4E suppression on the B cell population in the different transgenic settings (Figure 2). We observed a reduction in splenic (Figure 2A) and bone marrow derived (Figure S2A) B220+ cells (1.8 – 2.5 fold) in DOX-treated 4E.389/rtTA/Eμ-Myc and 4E.610/rtTA/Eμ-Myc mice relative to vehicle-treated controls. As well, no significant change in the pre B/B cell population was observed in DOX-treated FLuc.1309/rtTA/Eμ-Myc mice and a modest effect (<1.4 fold) was observed in DOX-treated 4E.389/rtTA mice (Figure 2A and S2A). A significant reduction in spleen mass (>2 fold, *, p<0.001; **, p<0.01) in DOX-treated 4E.389/rtTA/Eμ-Myc and 4E.610/rtTA/Eμ-Myc mice was noted compared to vehicle-treated controls, DOX-treated FLuc.1309/rtTA/Eμ-Myc mice, or 4E.389/rtTA mice (Figure 2B). The CD4+ (T cells), CD11b+ (monocyte/macrophages, granulocytes), and Gr-1+ (granulocytes) cell populations were not depleted upon eIF4E suppression and even increased in some cases (Figure 2C). These results indicate that Eμ-Myc pre B/B cells are exquisitely sensitive to eIF4E attenuation.
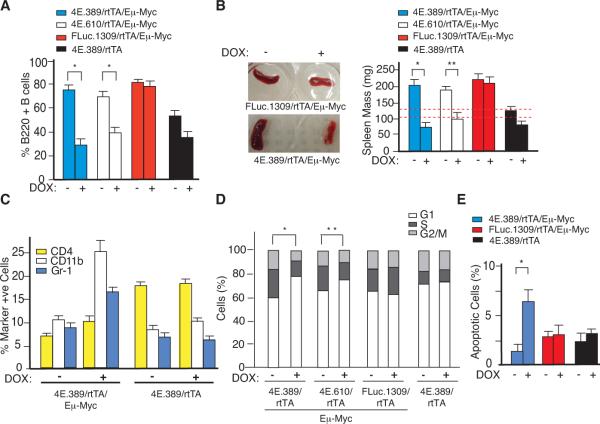
Figure 2. Inducible Suppression of eIF4E Reduces the B220+ B Cell Compartment, Impairs Cell Division, and Augments Apoptosis in Pre-malignant B Cells.
A. Flow cytometry analysis of B220+ B cells isolated from 6 wk old transgenic mice that had been treated with vehicle or DOX for 2 weeks. Error bars denote standard error of the mean (SEM); n=3, *; p<0.002, as determined by the Student's t-test. B. Representative photographs of spleens and average spleen mass of indicated 6 wk old triple transgenic mice that had been treated with vehicle or DOX for 2 weeks. Red dotted lines represent the range of spleen masses from four 6 wk old control C57BL/6 mice. Error bars denote SEM; n=4, *; p<0.001, **; p<0.01. C. Flow cytometry analysis of spleen cells isolated from 4E.389/rtTA/Eμ-Myc mice and stained for the indicated cells surface markers. Each marker positive population was also > 90% GFP+. Error bars are SEM; n=3. D. Cell cycle distribution of B220+ splenic B cells of the indicated genotype and DOX treatment cohorts. Results are expressed as the average of three independent experiments (n=3). *, p < 0.01; **, p<0.05 for % G1 and S phase cells. E. In situ TUNEL analysis on freshly isolated splenic cells from 5 wk transgenic mice that had been treated with vehicle or DOX for 6 days. Error bars denote SEM; n=3; *, p<0.05.
The differential effects of eIF4E inhibition on WT versus Myc over-expressing B cells appeared to be a consequence of impaired cell cycle progression and increased apoptosis. The cell cycle distribution of B220+ cells was similar among all untreated triple transgenic mouse strains but altered in B220+ cells derived from DOX-treated 4E.389/rtTA/Eμ-Myc and 4E.610/rtTA/Eμ-Myc mice, both of which showed an accumulation in G1 phase and reductions in S/G2 phase populations (Figure 2D). Moreover, the percentage of apoptotic B220+ cells in 4E.389/rtTA/Eμ-Myc mice was increased upon DOX-treatment compared to cells from vehicle-treated controls, DOX-treated FLuc.1309/rtTA/Eμ-Myc, or 4E.389/rtTA mice (Figure 2E). Similar results were obtained in other settings - notably in murine 3T3 fibroblasts and human hTert-BJ cells, where co-expression of Myc and sheIF4E was associated with a proliferative disadvantage (Figures S2B – C) and increased apoptosis (Fig. S2D). Taken together, these results indicate that eIF4E suppression and Myc over-expression share a synthetic lethal relationship that affects cell cycle progression and survival.
Suppression of eIF4E Impairs eIF4F Complex Formation and Activity
Alterations in eIF4E levels leads to selective effects on translational output mediated through the eIF4F complex (Dever, 2002). To determine if eIF4F levels are perturbed in DOX-treated 4E.389/rtTA/Eμ-Myc mice, B220+ cells were isolated and the eIF4F complex purified by m7GDP affinity chromatography (Figure 3A). Reductions in all three eIF4F subunits were noted in DOX-treated 4E.389/rtTA/Eμ-Myc B220+ cells compared to cells from untreated 4E.389/rtTA/Eμ-Myc or DOX-treated control FLuc.1309/rtTA/Eμ-Myc mice (Figure 3A). Metabolic labelling revealed only a modest reduction of ~20% in global protein synthesis in B220+ cells from DOX-treated 4E.389/rtTA/Eμ-Myc and 4E.610/rtTA/Eμ-Myc mice (Figure 3B), with no alteration in the global profile of newly synthesized proteins (Figure S3A). Such results are consistent with what has been previously documented upon anti-sense suppression of eIF4E expression (Graff et al., 2007) and imply that the consequences on translation of altering eIF4E levels are due to its impact on specific mRNAs.
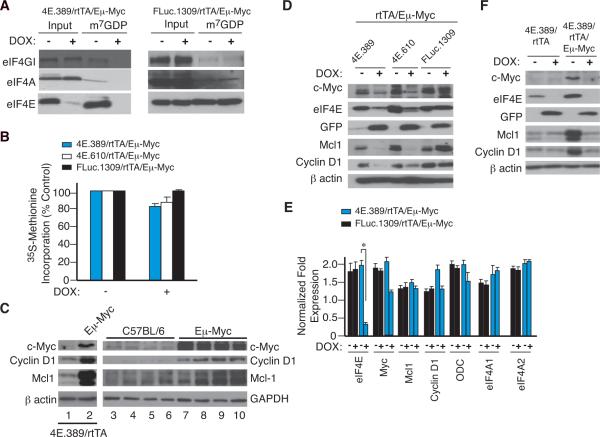
Figure 3. Inducible Suppression of eIF4E Impairs eIF4F Levels, Selectively Blocks Protein Synthesis, and Affects Production of Cyclin D1, Mcl-1, and Myc.
A. Relative abundance of the eIF4F complex in B220+ cells from mice of the indicated genotype. B. Protein synthesis rates in B220+ cells assessed by [35S]methionine incorporation and TCA precipitation. Error bars denote SEM; n=3. C. Western blot analysis of eIF4E responsive targets in B220+ cells isolated from 6 wk old mice. Cells were isolated from 4E.389/rtTA (lane 1), 4E.389/rtTA/Eμ-Myc (lane 2), and 4 independent C57BL/6 (lanes 3–6) or Eμ-Myc (lanes 7–10) mice. D. Western blot analysis showing reductions in Mcl-1, Cyclin D1, and Myc levels upon suppression of eIF4E in B220+ cells in vivo. E. qRT-PCR quantitation of the indicated mRNA levels from B220+ splenocytes. N=6; *, p = 0.003. F. Western blot analysis indicating levels of eIF4E targets upon eIF4E suppression in B220+ cells.
The effects of eIF4E suppression on preB/B cell cycle progression and apoptosis (Fig. 2) are consistent with previous observations that Cyclin D1, Myc, and Mcl-1 are particularly sensitive to eIF4E levels (Lin et al., 2008; Rosenwald et al., 1993; Wendel et al., 2007). Indeed, premalignant Eμ-Myc preB/B cells showed elevated levels of all three proteins (Figure 3C). Suppressing eIF4E expression led to reductions in the levels of all three eIF4E-responsive targets in splenic (Figure 3D) or bone marrow (Figure S3B) derived B220+ cells isolated from DOX-treated 4E.389/rtTA/Eμ-Myc and 4E.610/rtTA/Eμ-Myc, but not from vehicle- or DOX-treated FLuc.1309/rtTA/Eμ-Myc mice. Reductions in Myc, Mcl-1, and Cyclin D1 protein levels observed upon eIF4E suppression are not a consequence of decreased transcript levels (Figure 3E) and were fully reversible upon removal of DOX (Figure S3C). In addition, eIF4E suppression had no consequences on expression from the eIF4E-insensitive β-actin, IRES-driven GFP (Balvay et al., 2007), and p27Kip1 mRNAs (Miskimins et al., 2001) (Figures 3D and S3D). Reductions in Mcl-1, Cyclin D1, and Myc upon eIF4E suppression were Myc-context dependent in vivo since they were not observed in DOX-treated 4E.389/rtTA cells (Figure 3F). These findings define eIF4E as a Myc client responsible for augmenting pro-survival and proliferative capacity through effectors, such as Mcl-1 and Cyclin D1.
Pharmacological Suppression of eIF4F Activity Blocks Eμ-Myc-driven Tumor Initiation
To confirm and extend these genetic observations using a chemical biology approach, we took advantage of silvestrol, a small molecular inhibitor of the helicase activity of eIF4A, another essential component of the 4F complex (Bordeleau et al., 2008). Treatment of prelymphomatous Eμ-Myc mice for 3 weeks with silvestrol significantly delayed tumor onset (Figure 4A; p<0.01). As noted for shRNA mediated inhibition of eIF4E, silvestrol inhibited proliferation of Eμ-Myc preB/B cells, but not Eμ-Myc CD4+ or CD11b+ cells (Figures 4B, C). This was associated with a prolongation of G1 and shortening of S/G2 cell cycle phases (Figure 4D), increased apoptosis (Figure 4E), and reductions in Mcl-1 and Cyclin D1 protein levels (Figure 4F). Whether the growth inhibitory effects of silvestrol were Myc context-dependent was investigated utilizing NIH/3T3 cells ectopically expressing Myc/ER, a chimeric protein in which a mutant estrogen receptor (ER) ligand binding doman is fused to the carboxy terminal domain of c-Myc. In this system, Myc/ER is constitutively expressed but only becomes active when 4-hydroxytamoxifen (4-OHT) is supplied (Littlewood et al., 1995). Indeed, NIH/3T3 cells were found to be more sensitive to silvestrol upon induction of Myc/ER by 4-OHT (Figure 4G). The convergence of genetic and pharmacological phenotypes confirms that Myc-expressing cells are sensitive to eIF4F inhibition.
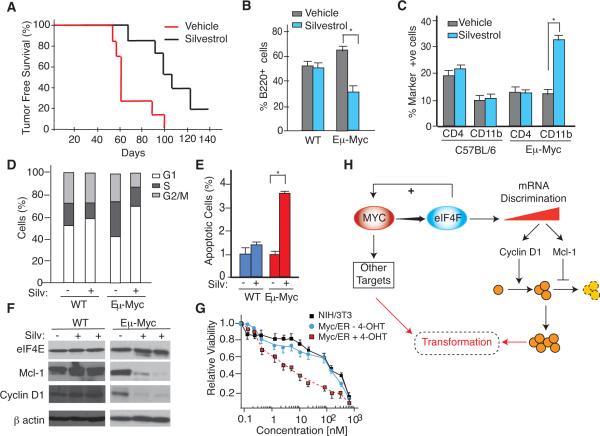
Figure 4. Blocking Eμ-Myc-driven Tumor Initiation by Pharmacological Suppression of eIF4F Activity.
A. Kaplan-Meier plot showing lymphoma-free survival of 4 wk old Eμ-Myc mice treated with or without 0.2 mg/kg silvestrol (n=7; p< 0.01) for 23 days. B. Percent B220+ cells isolated from 6 wk old C57BL/6 or Eμ-Myc mice that had been treated with vehicle or silvestrol for 2 wks. Error bar represent SEM, n=3. *, p<0.01. C. Percent CD4+ and CD11b+ cells isolated from 6 wk old C57BL/6 or Eμ-Myc mice that had been treated with vehicle or silvestrol for 2 weeks. Error bar represent SEM, n=3. D. Cell cycle distribution of B220+ splenic B cells of the indicated genotype and drug treatments. Results are expressed as the average of three independent experiments. E. In situ TUNEL analysis on freshly isolated splenic cells from 5 wk old mice that had been treated with vehicle or silvestrol for 6 days. n= 3 mice; *, p<0.001 F. Western blot analysis of eIF4F targets in splenic B220+ cells isolated from untreated or silvestrol-treated Eμ-Myc or C57BL/6 mice. G. Sensitivity of NIH/3T3 cells to silvestrol. NIH/3T3 or Myc/ER NIH 3T3 cells were exposed to vehicle or 250 nM 4-OHT for 18 hrs and then to silvestrol for an additional 24 hrs at the indicated concentrations. Cell viability was determined using the Sulforhodamine B (SRB) colorimetric assay and is set relative to vehicle-treated cells. Values represent the average of 3 biological replicates and error bars denote SEM. H. Relationship between Myc, eIF4F, and eIF4E effectors leading to increased cell cycle progression and survival advantage during the pre-malignant phase of lymphomagenesis. The + sign indicates that increased eIF4F levels also stimulate c-Myc mRNA translation (Lin et al., 2008).
Although the R26-rtTA allele is not ubiquitously expressed in the adult, it is well expressed in the gut and hematopoietic compartments, allowing us to assess toxicity in settings often most prone to drug induced toxicities (Beard et al., 2006; Premsrirut et al., 2011). We noticed that when 4 wk old 4E.389/rtTA/Eμ-Myc mice were treated with DOX for 2 weeks, a reduction in body weight was apparent within ~10 days (Figure S4A). Analysis of tissues with low proliferative indices that express GFP and sh4E.389 (i.e. liver [Figure S1C]) showed no discernable histological changes (data not shown). In contrast, a significant increase in apoptotic bodies in crypt epithelium, loss of goblet cells, and the presence of immature/undifferentiated crypts was apparent in intestines of DOX-treated 4E.389/rtTA/Eμ-Myc mice (Figures S4B, C) – a tissue that displays strong GFP induction and efficient shRNA-mediated attenuation of expression by R26-rtTA (Figures S1C and S4B) (McJunkin et al., 2011; Premsrirut et al., 2011). These phenotypic changes were completely reversed upon DOX withdrawal (Figure S4B). No differences in Ki-67 staining in intestines from DOX or vehicle treated 4E.389/rtTA/Eμ-Myc mice nor discernible histological changes in intestines from DOX or vehicle treated FLuc.1309/rtTA/Eμ-Myc mice were noticed (data not shown). Thus, although suppression of eIF4E has profound effects on some proliferating somatic tissues such as the intestine, these are well tolerated on the short term and are completely reversible without any discernible long-term negative impact on the animal's well being.
DISCUSSION
Myc amplifications are frequent somatic copy-number alterations in human cancers (Beroukhim et al., 2010). The effectiveness of suppressing Myc as an anticancer therapeutic approach at the organismal level has been shown using dominant negative forms of Myc (Soucek et al., 2008). Here, using both genetic and pharmacologic approaches, we demonstrate that the eIF4F complex functions as an essential Myc client during the initial phases of tumorigenesis. This relationship is likely in place for supporting normal B-cell development where Mcl-1 has been shown essential for development of pro-B cells and later on, for maintenance of mature B lymphocytes (Opferman et al., 2003), in contrast to Bcl2, which appears largely dispensable for early pro-B, pre-B and immature cell development (Kelly et al., 2007). The pre-B cell origin of sporadic Eμ-Myc lymphomas (Adams et al., 1985) is consistent with eIF4F being a Myc client - leading to up regulation of Mcl-1 expression.
We find that inhibition of eIF4F significantly delayed Myc-induced lymphomagenesis without overt toxicity to normal hematopoietic cells and other tissues. The transgenic model that we established did not allow us to probe the Myc-eIF4F relationship in tumor maintenance due to apparent collapse of the rtTA-inducible system (see above), however we expect such a relationship to also be relevant in established tumors since curtailing eIF4F helicase activity with small molecule inhibitors (silvestrol or hippuristanol) (Cencic et al., 2009; Lucas et al., 2009; Tsumuraya et al., 2011) or suppression of eIF4E by systemic delivery of eIF4E antisense oligonucleotides (Graff et al., 2007) in xenograft models all show significant anti-cancer activity whilst not displaying overt cytotoxicity to non-transformed cells, As well, Myc lymphoma cells expressing shRNAs to eIF4E have a selective growth disadvantage (Mills et al., 2008), over-expression of eIF4E antagonizes Myc-dependent apoptosis (Li et al., 2003; Ruggero et al., 2004), and gene amplification along the Myc-eIF4F axis can evade mTOR- and PI3K-targeted therapy (Ilic et al., 2011; Wendel et al., 2004),
We have not observed a loss in body weight with the use of silvestrol even when administered daily for 4 weeks (data not shown) consistent with previous studies documenting that long-term administration of this compound is well tolerated - affecting neither spleen or liver weights, nor altering liver aminotransferase activity, and showing little effect on cells of the hematopoietic lineage (Cencic et al., 2009). This may pertain to differences in the degree/extent of translation inhibition achieved in vivo with single daily doses of silvestrol versus chronic suppression of eIF4E by DOX. As silvestrol has a short serum half-life (~ 6 hrs) (Saradhi et al., 2011), treatment with this compound would result in cycles consisting of a period of translation inhibition followed by recovery. This is in contrast to the chronic suppression of eIF4E expected from continuous administration of DOX to sheIF4E-expressing mice. Alternatively, differences in the downstream consequences on translation of inhibiting eIF4A versus eIF4E in vivo may also be contributing to the differences in toxicity. Hence, a key advantage of the shRNA system used herein is to predict on-target toxicities. From the work presented here, we anticipate that eIF4F targeting drugs with increased potency may have increased intestinal toxicity, but such toxicities would be manageable.
While previous studies have indicated silvestrol can have anticancer effects (Bordeleau et al., 2008; Cencic et al., 2009; Lucas et al., 2009), the remarkable similarity between results produced through RNAi-mediated or chemical inhibition strongly suggest that silvestrol acts to limit cancer progression through targeting eIF4F. Of note, other recent efforts informed by RNAi or chemical biology based screens have identified chromatin- and SUMOylation-modifiers required for Myc-driven tumorigenesis (Kessler et al., 2012; Mertz et al., 2011; Zuber et al., 2011). Consequently, while Myc has long been considered “undruggable”, there are now several therapeutically viable options for selectively targeting Myc oncogenic functions in different contexts. Indeed combination strategies using these approaches might achieve greater potency and potentially bypass resistance.
Regardless of these therapeutic possibilities, our studies demonstrate that Myc can exert a role in tumor maintenance via selective effects on the translation of specific mRNAs. The eIF4F complex preferentially stimulates the translation of a subset of mRNAs that harbor highly structured 5' untranslated regions and that encode regulators of cell growth, proliferation, and apoptosis (Koromilas et al., 1992; Larsson et al., 2007; Mamane et al., 2007). Large scale changes to the cellular transcriptome, as executed upon Myc elevation, is expected to disproportionately alter the competitive ability of individual cellular mRNAs for limiting eIF4F levels, leading to distinctive translational changes (Dever, 2002). Our data indicates that Myc exploits eIF4F's mRNA discriminatory ability to selectively upregulate such effectors (Fig. 4H). In this manner Myc coordinates changes in protein synthesis with its well established nuclear activities, a feature that may be important for normal Myc function but that also creates an unnatural dependency under conditions of aberrant and/or sustained Myc levels.
EXPERIMENTAL PROCEDURES
Reagents, Cell Culture and Mice
Short hairpin (sh)RNAs targeting eIF4E were designed and tested as previously described (Paddison et al., 2004). Two of these, sh4E.389 (5'TTAAATTACTAGACAACTGGA3') and sh4E.610 (5'TTTAGCTCTAACATTAACAAC3'), as well as shFLuc.1309 (a neutral shRNA targeting Firefly luciferase (Premsrirut et al., 2011)) were targeted to the murine Col1A1 locus using a Flp/FRT recombinase-mediated cassette exchange (RMCE) strategy in pre-engineered ES cells (Hochedlinger et al., 2005). All ES cells were selected and maintained on irradiated (40 Gy) MEFs derived from the DR4 mouse strain. ES cells were cultured in knockout Dulbecco's modified Eagle's medium (Cellgro Mediatech) supplemented with 10% fetal bovine serum (FBS), L-glutamine, penicillin-streptomycin, non-essential amino acid, LIF (leukemia inhibitory factor), and 55μM β-mercaptoethanol. Electroporations with KH2 ES cells were performed with 50 μg pColTGM (aka CTGM) and 25 μg pCAGGS-FLPe at 400V and 125 μF as previously described (Premsrirut et al., 2011). ColAI-targeted clones were selected in hygromycin, tested for GFP inducibility, and transgenic mice derived using tetraploid embryo complementation.
Eμ-Myc mice were crossed to 4E.389/rtTA, 4E.610/rtTA, or FLuc.1309/rtTA mice to generate triple transgenic mice. Genotypes were obtained at the expected Mendelian inheritance ratios. The Eμ-Myc transgene was detected by genomic PCR amplification of a 600-bp product using the primers 5'-GGACAGTGCTTAGATCCAAGGTGA-3' and 5'-CCTCTGTCTCTCGCTGGAATTACT-3'. Genotyping for R26-rtTA was performed using the primers 5'-AAAGTCGCTCTGAGTTGTTAT-3', 5'-GCGAAGAGTTTGTCCTCAACC-3', and 5'-GGAGCGGGAGAAATGGATATG-3'. Genotyping for R26-rtTA yields two PCR products of ~500 bp (wild-type ROSA26 allele) and ~300 bp (R26-rtTA allele). Genotyping for 4E.389 by PCR used the primers 5'-AATTACTAGACAACTGGATTGCCT-3' and 5'-GAAGAACAATCAAGGGTCC-3' (~200 bp product), whereas genotyping for 4E.610 by PCR used the primers 5'-GCCACAGATGTATTTAGCTCTAAC-3' and 5'-GAAGAACAATCAAGGGTCC-3' (~200 bp product). Genotyping for FLuc.1309 used the primers 5'-CACCCTGAAAACTTTGCCCC-3' and 5'-AAGCCACAGATGTATTAATCAGAGA-3' (~300 bp product).
All mice strains were maintained on a C57BL/6 background. Activation of shRNAmir production in mice was performed in 4 wk old transgenic mice by supplying DOX (1mg/ml) in the drinking water (+5% sucrose) for the indicated periods of time. DOX-supplemented water was changed every 4 days. To assess the impact of silvestrol on lymphoma onset, 4 wk old Eμ-Myc mice were treated with vehicle (5.2% PEG 400/5.2% Tween-80) or 0.2 mg/kg silvestrol (daily intraperitoneal injections) for 23 consecutive days. All mice were monitored twice a week for signs of morbidity and lymphoma development, the latter scored by peripheral lymph node palpation. Tumor-free survival is defined as the time from birth to the time of appearance of a palpable lymphoma. Data were analysed in the Kaplan-Meier format using the log-rank (Mantel-Cox) test for statistical significance. All animal studies were approved by the McGill University Faculty of Medicine Animal Care Committee.
Flow cytometry
Fresh cell suspensions were isolated in PBS+ 2% FBS. Erythrocytes were removed by lysis in ACK buffer (150 mM NH4Cl, 10 mM KHCO3 and 0.1 mM EDTA). Remaining cells were collected by centrifugation and resuspended in 1 ml PBS + 2% FBS. Blocking was performed by incubating samples with purified anti-CD16/CD32 antibody (clone: 2.4G2; BD Biosciences) for 5 min on ice before labelling cells with fluorochrome conjugated substrate specific antibodies (see Supplemental Experimental Procedures). The forward and side light-scatter gate excluded small apoptotic cells and granular cells, whereas large cells were included.
Antibodies used to identify monocytes and granulocytes were: Ly-6G (Gr-1) PECy7 (clone 1A8; BD Biosciences) and CD11b PE (clone M1/70; BD Biosciences). Antibodies used to identify T and B lymphocytes were: CD4 PE (clone RM4–5; BD Biosciences) and CD45R/B220 PE (clone RA3–6B2; BD Biosciences). Incubations were performed in the dark on ice for 20 min before data acquisition and analysis were conducted on a FACSAria II (BD Biosciences). Erythrocytes, dead cells and debris were excluded with gating based on forward/side scatter characteristics. The percent B and T lymphocytes, monocytes and granulocytes for each sample was expressed as a percentage of total gated cells analyzed.
To measure apoptosis in vivo, TUNEL assays were performed on freshly isolated splenic cells from indicated transgenic mice treated with vehicle or DOX for 6 days following the manufacturer's instructions (In Situ Cell Death Detection Kit, TMR red, Roche).
For cell cycle analysis, freshly isolated splenic B220+ cells from vehicle or DOX-treated transgenic mice were incubated with 1 ml DNA staining buffer (0.3% Triton-X 100, 50 μg/mL propidium iodine, 20 μg/ml RNAase A and 4mM sodium citrate). Cell cycle distribution was analyzed by flow cytometry using a Guava EasyCyte (Millipore).
For silvestrol-treated C57BL/6 and Eμ-Myc mice, freshly isolated splenic B220+ cells (106 cells/ml) were washed, fixed in 75% ethanol solution for 1 hour at 4°C and stained with propidium iodide (Sigma) (50 μg/ml propidium iodide, 3.8 mM sodium citrate, and 500 μg/ml RNase A) for 3 hours at 4°C. Cells were then analyzed for DNA content using a FACScan (BD Biosciences).
Expression analysis
For Western blotting, cells were lysed in RIPA lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM DTT, 0.1% SDS, 1% NP-40, 0.5% sodium deoxycholate, 0.1 mM phenylmethylsulfonyl fluoride (PMSF), 1 μg/ml each of leupeptin, pepstatin, and aprotinin). Protein concentrations were determined using the Bio-Rad Protein assay. Total protein lysates (30 μg) were resolved by SDS-PAGE, transferred to PVDF membranes (Millipore), probed with the indicated antibodies, and visualized using enhanced chemiluminescence (ECL) detection (Amersham). Primary antibodies were: anti-cyclin D1 was from Cell Signaling Technology (#2926, Beverly, MA), and anti-Mcl-1 was purchased from AbD Serotec (#AHP1249, Oxford, UK). Anti-GFP (#sc-9996), anti-eIF4E(#sc-9976), anti-p27 (#sc-528) and anti-c-Myc (# sc-764) antibodies were obtained from Santa Cruz Biotechnology. Anti-β-actin (#A5316) and anti-tubulin (#T5268) antibodies were purchased from Sigma.
For metabolic studies, 2 × 105 B220+ cells were isolated from vehicle or DOX-treated triple transgenic mice and seeded in 24 well plates. Cells from DOX-treated mice were maintained in 1μg/ml DOX. Cells were cultured for 45 min in methionine-free medium, followed by 60 min in [35S]methionine-containing medium (150–220 μCi/ml) supplemented with 10% dialyzed FCS, washed and lysed in RIPA buffer. Proteins were TCA precipitated onto 3MM Whatman paper and the amount of radioactivity quantitated by scintillation counting. Values were normalized to total protein levels as determined by the Bradford assay.
For m7GTP Sepharose pull-down assays, freshly isolated cells were harvested in 300 μl of Lysis Buffer (20 mM Hepes7.5, 100 mM KCl, 1.0 mM EDTA, 1 mM DTT, 1 mM PMSF and 0.2% Tween 20, 10 mM NaF and 20 mM β-glycerophosphate), and then subjected to 3 cycles of freeze-thaw. The lysate was then incubated with 50 μl of 50% slurry of m7GTP-Sepharose 4B (GE Healthcare, United Kingdom) for 2 hrs at 4°C. The resin was washed three times with 1 ml of Lysis Buffer and one time with buffer A containing 200 μM GDP. Finally, proteins bound to the resin were eluted with 80 μl of m7GDP (1 mM) for 10 min on ice. Aliquots of the eluted fractions (25 μl) were resolved by SDS-PAGE (10% polyacrylamide) and analyzed by Western blotting.
Immunohistochemistry and TUNEL staining
Tissues were harvested, fixed in 10% formalin, and embedded in paraffin. Sections (5 μm) were then dewaxed and rehydrated through a graded series of alcohol washes to water. They were placed in 10 mM citric acid buffer (pH 6.0) and subjected to antigen retrieval by boiling for 15 minutes. Immunohistochemistry was performed using HRP/DAB Detection Kit (ab64261, Abcam) according to the manufacturer's instruction. Briefly, after incubation with blocking buffer for 1 hour and 3% hydrogen peroxide for 10 minutes, rabbit anti-eIF4E (Cat# 9742, Cell Signaling) or rabbit anti-GFP (Cat# 2555, Cell Signaling) was applied overnight at 4°C. Sections were washed with TBS-T (1 M Tris-HCl, pH 7.5, 1.5 M NaCl, and 1% Tween-20) and incubated with biotinylated goat anti-rabbit IgG for 30 minutes at room temperature. After washing with TBS-T, streptavidin peroxidase was added for 30 minutes at room temperature. The signals were developed using DAB chromogen as substrate at room temperature for 5 minutes. Sections were counterstained with hematoxylin, dehydrated and mounted with permount. Tissue sections were analyzed using an Aperio Scanscope XT (Aperio Technologies, Inc, Vista, CA, USA).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Patrick Sénéchal and Marilyn Carrier for excellent technical assistance. Chen-Ju Lin was supported by a McGill Faculty of Medicine Internal Studentship. This work is supported by the Canadian Institutes of Health Research to JP (MOP-106530), the NIH (GM-073855 to J.A.P., Jr.) and a NCI-program project grant (CA087497-11 to S.W.L.) and the Mouse Models of Human Cancer Consortium (S.W.L).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, Cory S, Palmiter RD, Brinster RL. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318:533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- Balvay L, Lopez Lastra M, Sargueil B, Darlix JL, Ohlmann T. Translational control of retroviruses. Nat Rev Microbiol. 2007;5:128–140. doi: 10.1038/nrmicro1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beard C, Hochedlinger K, Plath K, Wutz A, Jaenisch R. Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis. 2006;44:23–28. doi: 10.1002/gene.20180. [DOI] [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordeleau ME, Robert F, Gerard B, Lindqvist L, Chen SM, Wendel HG, Brem B, Greger H, Lowe SW, Porco JA, Jr., et al. Therapeutic suppression of translation initiation modulates chemosensitivity in a mouse lymphoma model. J Clin Invest. 2008;118:2651–2660. doi: 10.1172/JCI34753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cencic R, Carrier M, Galicia-Vazquez G, Bordeleau ME, Sukarieh R, Bourdeau A, Brem B, Teodoro JG, Greger H, Tremblay ML, et al. Antitumor activity and mechanism of action of the cyclopenta[b]benzofuran, silvestrol. PLoS ONE. 2009;4:e5223. doi: 10.1371/journal.pone.0005223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever TE. Gene-specific regulation by general translation factors. Cell. 2002;108:545–556. doi: 10.1016/s0092-8674(02)00642-6. [DOI] [PubMed] [Google Scholar]
- Duncan R, Milburn SC, Hershey JW. Regulated phosphorylation and low abundance of HeLa cell initiation factor eIF-4F suggest a role in translational control. Heat shock effects on eIF-4F. J Biol Chem. 1987;262:380–388. [PubMed] [Google Scholar]
- Graff JR, Konicek BW, Vincent TM, Lynch RL, Monteith D, Weir SN, Schwier P, Capen A, Goode RL, Dowless MS, et al. Therapeutic suppression of translation initiation factor eIF4E expression reduces tumor growth without toxicity. J Clin Invest. 2007;117:2638–2648. doi: 10.1172/JCI32044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemann MT, Bric A, Teruya-Feldstein J, Herbst A, Nilsson JA, Cordon-Cardo C, Cleveland JL, Tansey WP, Lowe SW. Evasion of the p53 tumour surveillance network by tumour-derived MYC mutants. Nature. 2005;436:807–811. doi: 10.1038/nature03845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochedlinger K, Yamada Y, Beard C, Jaenisch R. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell. 2005;121:465–477. doi: 10.1016/j.cell.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Ilic N, Utermark T, Widlund HR, Roberts TM. PI3K-targeted therapy can be evaded by gene amplification along the MYC-eukaryotic translation initiation factor 4E (eIF4E) axis. Proc Natl Acad Sci U S A. 2011;108:E699–708. doi: 10.1073/pnas.1108237108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen KA, Prasad VS, Sidman CL, Osmond DG. Apoptosis and macrophage-mediated deletion of precursor B cells in the bone marrow of E mu-myc transgenic mice. Blood. 1994;84:2784–2794. [PubMed] [Google Scholar]
- Jones RM, Branda J, Johnston KA, Polymenis M, Gadd M, Rustgi A, Callanan L, Schmidt EV. An essential E box in the promoter of the gene encoding the mRNA cap-binding protein (eukaryotic initiation factor 4E) is a target for activation by c-myc. Mol Cell Biol. 1996;16:4754–4764. doi: 10.1128/mcb.16.9.4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly PN, Puthalakath H, Adams JM, Strasser A. Endogenous bcl-2 is not required for the development of Emu-myc-induced B-cell lymphoma. Blood. 2007;109:4907–4913. doi: 10.1182/blood-2006-10-051847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler JD, Kahle KT, Sun T, Meerbrey KL, Schlabach MR, Schmitt EM, Skinner SO, Xu Q, Li MZ, Hartman ZC, et al. A SUMOylation-dependent transcriptional subprogram is required for Myc-driven tumorigenesis. Science. 2012;335:348–353. doi: 10.1126/science.1212728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koromilas AE, Lazaris-Karatzas A, Sonenberg N. mRNAs containing extensive secondary structure in their 5' non-coding region translate efficiently in cells overexpressing initiation factor eIF-4E. EMBO J. 1992;11:4153–4158. doi: 10.1002/j.1460-2075.1992.tb05508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdon WY, Harris AW, Cory S, Adams JM. The c-myc oncogene perturbs B lymphocyte development in E-mu-myc transgenic mice. Cell. 1986;47:11–18. doi: 10.1016/0092-8674(86)90361-2. [DOI] [PubMed] [Google Scholar]
- Larsson O, Li S, Issaenko OA, Avdulov S, Peterson M, Smith K, Bitterman PB, Polunovsky VA. Eukaryotic translation initiation factor 4E induced progression of primary human mammary epithelial cells along the cancer pathway is associated with targeted translational deregulation of oncogenic drivers and inhibitors. Cancer Res. 2007;67:6814–6824. doi: 10.1158/0008-5472.CAN-07-0752. [DOI] [PubMed] [Google Scholar]
- Li S, Takasu T, Perlman DM, Peterson MS, Burrichter D, Avdulov S, Bitterman PB, Polunovsky VA. Translation factor eIF4E rescues cells from Myc-dependent apoptosis by inhibiting cytochrome c release. J Biol Chem. 2003;278:3015–3022. doi: 10.1074/jbc.M208821200. [DOI] [PubMed] [Google Scholar]
- Lin CJ, Cencic R, Mills JR, Robert F, Pelletier J. c-Myc and eIF4F are components of a feedforward loop that links transcription and translation. Cancer Res. 2008;68:5326–5334. doi: 10.1158/0008-5472.CAN-07-5876. [DOI] [PubMed] [Google Scholar]
- Littlewood TD, Hancock DC, Danielian PS, Parker MG, Evan GI. A modified oestrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins. Nucleic Acids Res. 1995;23:1686–1690. doi: 10.1093/nar/23.10.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas DM, Edwards RB, Lozanski G, West DA, Shin JD, Vargo MA, Davis ME, Rozewski DM, Johnson AJ, Su BN, et al. The novel plant-derived agent silvestrol has B-cell selective activity in chronic lymphocytic leukemia and acute lymphoblastic leukemia in vitro and in vivo. Blood. 2009;113:4656–4666. doi: 10.1182/blood-2008-09-175430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamane Y, Petroulakis E, Martineau Y, Sato TA, Larsson O, Rajasekhar VK, Sonenberg N. Epigenetic activation of a subset of mRNAs by eIF4E explains its effects on cell proliferation. PLoS ONE. 2007;2:e242. doi: 10.1371/journal.pone.0000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McJunkin K, Mazurek A, Premsrirut PK, Zuber J, Dow LE, Simon J, Stillman B, Lowe SW. Reversible suppression of an essential gene in adult mice using transgenic RNA interference. Proc Natl Acad Sci U S A. 2011;108:7113–7118. doi: 10.1073/pnas.1104097108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertz JA, Conery AR, Bryant BM, Sandy P, Balasubramanian S, Mele DA, Bergeron L, Sims RJ., 3rd Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci U S A. 2011;108:16669–16674. doi: 10.1073/pnas.1108190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills JR, Hippo Y, Robert F, Chen SM, Malina A, Lin CJ, Trojahn U, Wendel HG, Charest A, Bronson RT, et al. mTORC1 promotes survival through translational control of Mcl-1. Proc Natl Acad Sci U S A. 2008;105:10853–10858. doi: 10.1073/pnas.0804821105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miskimins WK, Wang G, Hawkinson M, Miskimins R. Control of cyclin-dependent kinase inhibitor p27 expression by cap-independent translation. Mol Cell Biol. 2001;21:4960–4967. doi: 10.1128/MCB.21.15.4960-4967.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson JA, Keller UB, Baudino TA, Yang C, Norton S, Old JA, Nilsson LM, Neale G, Kramer DL, Porter CW, et al. Targeting ornithine decarboxylase in Myc-induced lymphomagenesis prevents tumor formation. Cancer Cell. 2005;7:433–444. doi: 10.1016/j.ccr.2005.03.036. [DOI] [PubMed] [Google Scholar]
- Opferman JT, Letai A, Beard C, Sorcinelli MD, Ong CC, Korsmeyer SJ. Development and maintenance of B and T lymphocytes requires antiapoptotic MCL-1. Nature. 2003;426:671–676. doi: 10.1038/nature02067. [DOI] [PubMed] [Google Scholar]
- Paddison PJ, Caudy AA, Sachidanandam R, Hannon GJ. Short hairpin activated gene silencing in mammalian cells. Methods Mol Biol. 2004;265:85–100. doi: 10.1385/1-59259-775-0:085. [DOI] [PubMed] [Google Scholar]
- Podsypanina K, Politi K, Beverly LJ, Varmus HE. Oncogene cooperation in tumor maintenance and tumor recurrence in mouse mammary tumors induced by Myc and mutant Kras. Proc Natl Acad Sci U S A. 2008;105:5242–5247. doi: 10.1073/pnas.0801197105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premsrirut PK, Dow LE, Kim SY, Camiolo M, Malone CD, Miething C, Scuoppo C, Zuber J, Dickins RA, Kogan SC, et al. A rapid and scalable system for studying gene function in mice using conditional RNA interference. Cell. 2011;145:145–158. doi: 10.1016/j.cell.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasekhar VK, Viale A, Socci ND, Wiedmann M, Hu X, Holland EC. Oncogenic Ras and Akt signaling contribute to glioblastoma formation by differential recruitment of existing mRNAs to polysomes. Mol Cell. 2003;12:889–901. doi: 10.1016/s1097-2765(03)00395-2. [DOI] [PubMed] [Google Scholar]
- Rosenwald IB, Lazaris-Karatzas A, Sonenberg N, Schmidt EV. Elevated levels of cyclin D1 protein in response to increased expression of eukaryotic initiation factor 4E. Mol Cell Biol. 1993;13:7358–7363. doi: 10.1128/mcb.13.12.7358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau D, Kaspar R, Rosenwald I, Gehrke L, Sonenberg N. Translation initiation of ornithine decarboxylase and nucleocytoplasmic transport of cyclin D1 mRNA are increased in cells overexpressing eukaryotic initiation factor 4E. Proc Natl Acad Sci U S A. 1996;93:1065–1070. doi: 10.1073/pnas.93.3.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggero D, Montanaro L, Ma L, Xu W, Londei P, Cordon-Cardo C, Pandolfi PP. The translation factor eIF-4E promotes tumor formation and cooperates with c-Myc in lymphomagenesis. Nat Med. 2004;10:484–486. doi: 10.1038/nm1042. [DOI] [PubMed] [Google Scholar]
- Saradhi UV, Gupta SV, Chiu M, Wang J, Ling Y, Liu Z, Newman DJ, Covey JM, Kinghorn AD, Marcucci G, et al. Characterization of silvestrol pharmacokinetics in mice using liquid chromatography-tandem mass spectrometry. AAPS J. 2011;13:347–356. doi: 10.1208/s12248-011-9273-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt CA, Lowe SW. Apoptosis and chemoresistance in transgenic cancer models. J Mol Med (Berl) 2002;80:137–146. doi: 10.1007/s00109-001-0293-3. [DOI] [PubMed] [Google Scholar]
- Soucek L, Whitfield J, Martins CP, Finch AJ, Murphy DJ, Sodir NM, Karnezis AN, Swigart LB, Nasi S, Evan GI. Modelling Myc inhibition as a cancer therapy. Nature. 2008;455:679–683. doi: 10.1038/nature07260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser A, Harris AW, Bath ML, Cory S. Novel primitive lymphoid tumours induced in transgenic mice by cooperation between myc and bcl-2. Nature. 1990;348:331–333. doi: 10.1038/348331a0. [DOI] [PubMed] [Google Scholar]
- Tsumuraya T, Ishikawa C, Machijima Y, Nakachi S, Senba M, Tanaka J, Mori N. Effects of hippuristanol, an inhibitor of eIF4A, on adult T-cell leukemia. Biochem Pharmacol. 2011;81:713–722. doi: 10.1016/j.bcp.2010.12.025. [DOI] [PubMed] [Google Scholar]
- van Riggelen J, Yetil A, Felsher DW. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer. 2010;10:301–309. doi: 10.1038/nrc2819. [DOI] [PubMed] [Google Scholar]
- Wendel HG, De Stanchina E, Fridman JS, Malina A, Ray S, Kogan S, Cordon-Cardo C, Pelletier J, Lowe SW. Survival signalling by Akt and eIF4E in oncogenesis and cancer therapy. Nature. 2004;428:332–337. doi: 10.1038/nature02369. [DOI] [PubMed] [Google Scholar]
- Wendel HG, Silva RL, Malina A, Mills JR, Zhu H, Ueda T, Watanabe-Fukunaga R, Fukunaga R, Teruya-Feldstein J, Pelletier J, et al. Dissecting eIF4E action in tumorigenesis. Genes Dev. 2007;21:3232–3237. doi: 10.1101/gad.1604407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.