Abstract
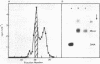
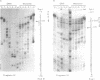
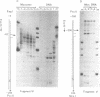
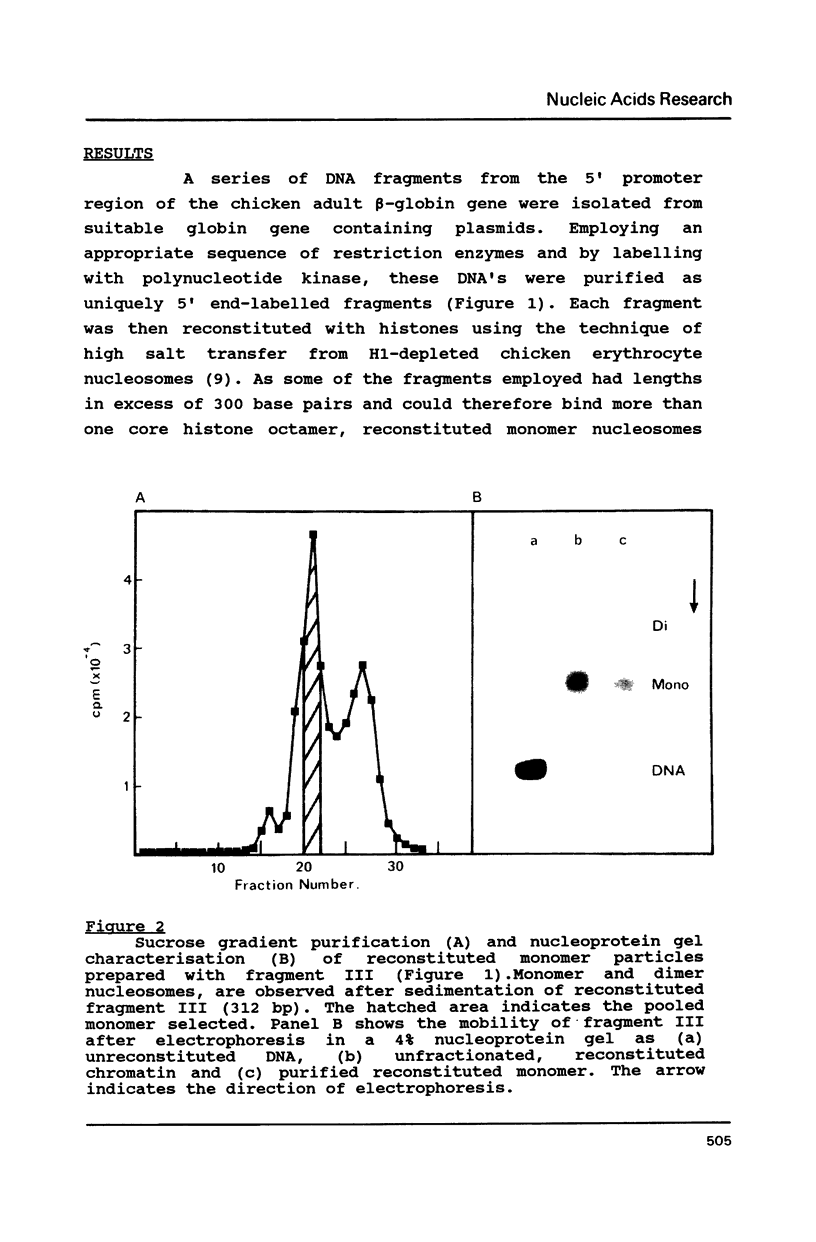
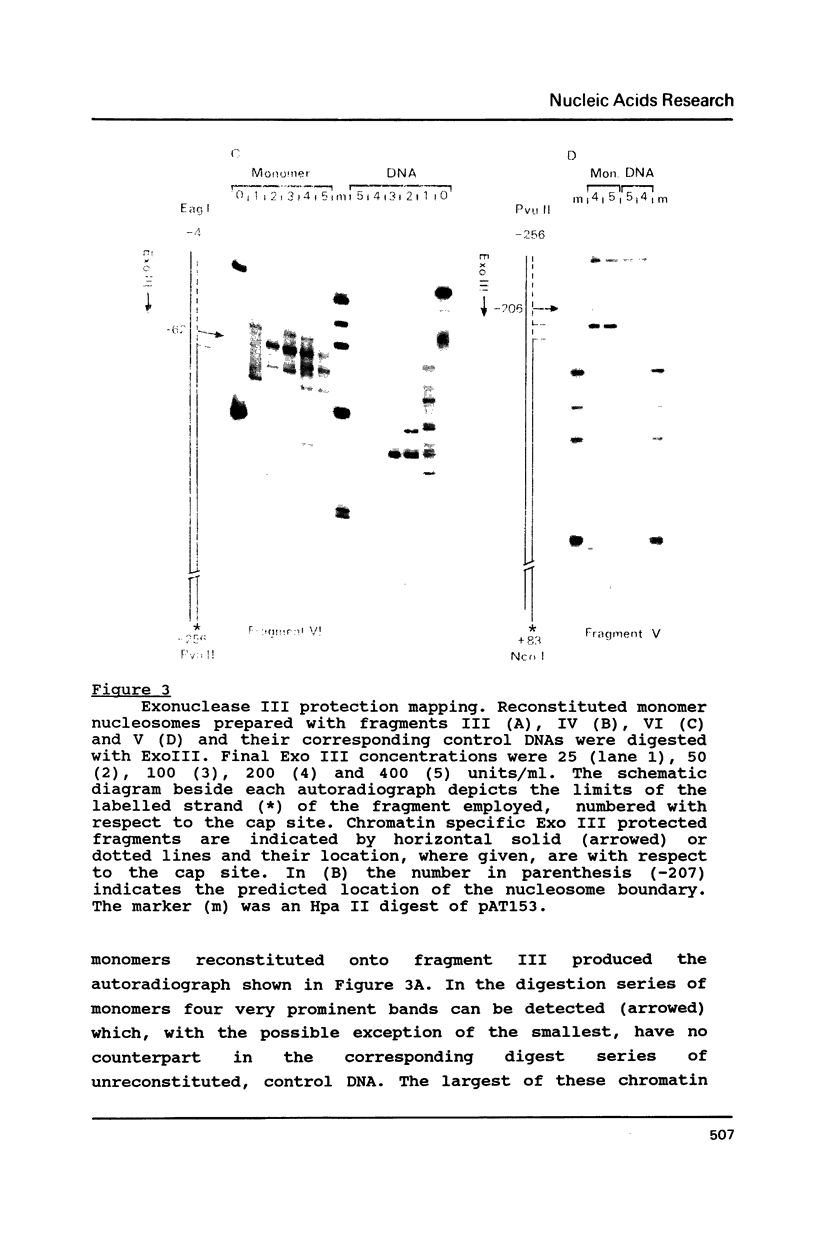
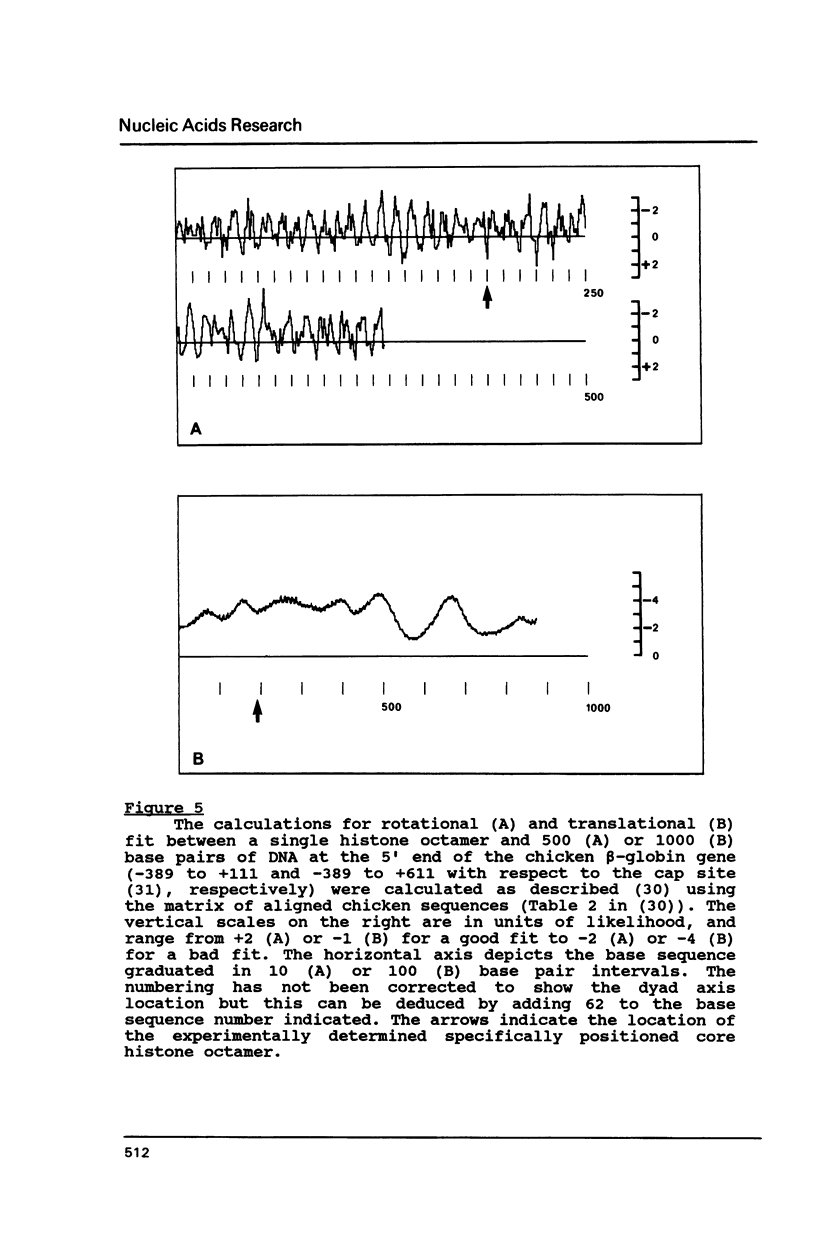
Histone octamers were reconstituted onto 5' end-labelled DNA fragments derived from the promoter region of the chicken beta A globin gene. The location of the reconstituted histone octamer with respect to the DNA sequence of each fragment was assessed by Exonuclease III digestion of purified nucleosome monomers. By this approach we have found a strong preference for histone octamers to be positioned over nucleotides -206 to -62 relative to the gene cap site. This stretch of DNA contains all those 5' beta globin sequences which, by DNase footprinting, bind specific protein factors and incorporates three promoter consensus sequence motifs. The upstream terminal 32 base pairs of this DNA segment contains the binding sites for the erythrocyte specific G-string binding protein and transcription factor Spl and appears to be relatively weakly bound to the histone octamer.
Full text
PDF













Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allan J., Staynov D. Z., Gould H. Reversible dissociation of linker histone from chromatin with preservation of internucleosomal repeat. Proc Natl Acad Sci U S A. 1980 Feb;77(2):885–889. doi: 10.1073/pnas.77.2.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almer A., Rudolph H., Hinnen A., Hörz W. Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction releases additional upstream activating DNA elements. EMBO J. 1986 Oct;5(10):2689–2696. doi: 10.1002/j.1460-2075.1986.tb04552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calladine C. R., Drew H. R. Principles of sequence-dependent flexure of DNA. J Mol Biol. 1986 Dec 20;192(4):907–918. doi: 10.1016/0022-2836(86)90036-7. [DOI] [PubMed] [Google Scholar]
- Cordingley M. G., Riegel A. T., Hager G. L. Steroid-dependent interaction of transcription factors with the inducible promoter of mouse mammary tumor virus in vivo. Cell. 1987 Jan 30;48(2):261–270. doi: 10.1016/0092-8674(87)90429-6. [DOI] [PubMed] [Google Scholar]
- Dierks P., van Ooyen A., Cochran M. D., Dobkin C., Reiser J., Weissmann C. Three regions upstream from the cap site are required for efficient and accurate transcription of the rabbit beta-globin gene in mouse 3T6 cells. Cell. 1983 Mar;32(3):695–706. doi: 10.1016/0092-8674(83)90055-7. [DOI] [PubMed] [Google Scholar]
- Dolan M., Dodgson J. B., Engel J. D. Analysis of the adult chicken beta-globin gene. Nucleotide sequence of the locus, microheterogeneity at the 5'-end of beta-globin mRNA, and aberrant nuclear RNA species. J Biol Chem. 1983 Mar 25;258(6):3983–3990. [PubMed] [Google Scholar]
- Drew H. R., Calladine C. R. Sequence-specific positioning of core histones on an 860 base-pair DNA. Experiment and theory. J Mol Biol. 1987 May 5;195(1):143–173. doi: 10.1016/0022-2836(87)90333-0. [DOI] [PubMed] [Google Scholar]
- Drew H. R., Travers A. A. DNA bending and its relation to nucleosome positioning. J Mol Biol. 1985 Dec 20;186(4):773–790. doi: 10.1016/0022-2836(85)90396-1. [DOI] [PubMed] [Google Scholar]
- Dynan W. S., Tjian R. The promoter-specific transcription factor Sp1 binds to upstream sequences in the SV40 early promoter. Cell. 1983 Nov;35(1):79–87. doi: 10.1016/0092-8674(83)90210-6. [DOI] [PubMed] [Google Scholar]
- Edwards C. A., Firtel R. A. Site-specific phasing in the chromatin of the rDNA in Dictyostelium discoideum. J Mol Biol. 1984 Nov 25;180(1):73–90. doi: 10.1016/0022-2836(84)90431-5. [DOI] [PubMed] [Google Scholar]
- Emerson B. M., Felsenfeld G. Specific factor conferring nuclease hypersensitivity at the 5' end of the chicken adult beta-globin gene. Proc Natl Acad Sci U S A. 1984 Jan;81(1):95–99. doi: 10.1073/pnas.81.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson B. M., Lewis C. D., Felsenfeld G. Interaction of specific nuclear factors with the nuclease-hypersensitive region of the chicken adult beta-globin gene: nature of the binding domain. Cell. 1985 May;41(1):21–30. doi: 10.1016/0092-8674(85)90057-1. [DOI] [PubMed] [Google Scholar]
- Linxweller W., Hörz W. Reconstitution experiments show that sequence-specific histone-DNA interactions are the basis for nucleosome phasing on mouse satellite DNA. Cell. 1985 Aug;42(1):281–290. doi: 10.1016/s0092-8674(85)80123-9. [DOI] [PubMed] [Google Scholar]
- Palen T. E., Cech T. R. Chromatin structure at the replication origins and transcription-initiation regions of the ribosomal RNA genes of Tetrahymena. Cell. 1984 Apr;36(4):933–942. doi: 10.1016/0092-8674(84)90043-6. [DOI] [PubMed] [Google Scholar]
- Plumb M. A., Nicolas R. H., Wright C. A., Goodwin G. H. Multiple sequence-specific DNA binding activities are eluted from chicken nuclei at low ionic strengths. Nucleic Acids Res. 1985 Jun 11;13(11):4047–4065. doi: 10.1093/nar/13.11.4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay N., Felsenfeld G., Rushton B. M., McGhee J. D. A 145-base pair DNA sequence that positions itself precisely and asymmetrically on the nucleosome core. EMBO J. 1984 Nov;3(11):2605–2611. doi: 10.1002/j.1460-2075.1984.tb02181.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley D., Weintraub H. Nucleosomal DNA is digested to repeats of 10 bases by exonuclease III. Cell. 1978 Feb;13(2):281–293. doi: 10.1016/0092-8674(78)90197-6. [DOI] [PubMed] [Google Scholar]
- Satchwell S. C., Drew H. R., Travers A. A. Sequence periodicities in chicken nucleosome core DNA. J Mol Biol. 1986 Oct 20;191(4):659–675. doi: 10.1016/0022-2836(86)90452-3. [DOI] [PubMed] [Google Scholar]
- Simpson R. T., Künzler P. Cromatin and core particles formed from the inner histones and synthetic polydeoxyribonucleotides of defined sequence. Nucleic Acids Res. 1979 Apr;6(4):1387–1415. doi: 10.1093/nar/6.4.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma F., Simpson R. T. Local protein-DNA interactions may determine nucleosome positions on yeast plasmids. Nature. 1985 May 16;315(6016):250–252. doi: 10.1038/315250a0. [DOI] [PubMed] [Google Scholar]