Abstract
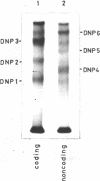
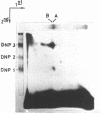
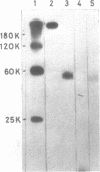
A method of affinity labelling proteins specifically associated with DNA target sequences is proposed. The method utilizes covalent UV-crosslinking of proteins to highly labelled DNA (e.g. in crude cell or nuclear extracts) followed by degradation of the DNA to short oligonucleotides. Proteins selectively labelled by attached residual oligonucleotides are readily amenable to molecular mass determination. Using this approach, we have characterized a HeLa polypeptide specifically bound to a short segment of the adenovirus-2 major late promoter (Ad2 MLP). A molecular mass value (approximately 51 kD) and precise location of the crosslinking site(s) of the protein within the MLP (-55 with respect to the cap site) were determined.
Full text
PDF















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baker C. C., Ziff E. B. Biogenesis, structures, and sites of encoding of the 5' termini of adenovirus-2 mRNAs. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 1):415–428. doi: 10.1101/sqb.1980.044.01.045. [DOI] [PubMed] [Google Scholar]
- Carthew R. W., Chodosh L. A., Sharp P. A. An RNA polymerase II transcription factor binds to an upstream element in the adenovirus major late promoter. Cell. 1985 Dec;43(2 Pt 1):439–448. doi: 10.1016/0092-8674(85)90174-6. [DOI] [PubMed] [Google Scholar]
- Chatterjee P. K., Vayda M. E., Flint S. J. Identification of proteins and protein domains that contact DNA within adenovirus nucleoprotein cores by ultraviolet light crosslinking of oligonucleotides 32P-labelled in vivo. J Mol Biol. 1986 Mar 5;188(1):23–37. doi: 10.1016/0022-2836(86)90477-8. [DOI] [PubMed] [Google Scholar]
- Chodosh L. A., Carthew R. W., Sharp P. A. A single polypeptide possesses the binding and transcription activities of the adenovirus major late transcription factor. Mol Cell Biol. 1986 Dec;6(12):4723–4733. doi: 10.1128/mcb.6.12.4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dretzen G., Bellard M., Sassone-Corsi P., Chambon P. A reliable method for the recovery of DNA fragments from agarose and acrylamide gels. Anal Biochem. 1981 Apr;112(2):295–298. doi: 10.1016/0003-2697(81)90296-7. [DOI] [PubMed] [Google Scholar]
- Dynan W. S., Tjian R. Control of eukaryotic messenger RNA synthesis by sequence-specific DNA-binding proteins. 1985 Aug 29-Sep 4Nature. 316(6031):774–778. doi: 10.1038/316774a0. [DOI] [PubMed] [Google Scholar]
- Dynan W. S., Tjian R. Isolation of transcription factors that discriminate between different promoters recognized by RNA polymerase II. Cell. 1983 Mar;32(3):669–680. doi: 10.1016/0092-8674(83)90053-3. [DOI] [PubMed] [Google Scholar]
- Egly J. M., Miyamoto N. G., Moncollin V., Chambon P. Is actin a transcription initiation factor for RNA polymerase B? EMBO J. 1984 Oct;3(10):2363–2371. doi: 10.1002/j.1460-2075.1984.tb02141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried M., Crothers D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Dec 11;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton M. D., Whiteley H. R. UV cross-linking of the Bacillus subtilis RNA polymerase to DNA in promoter and non-promoter complexes. J Biol Chem. 1985 Jul 5;260(13):8121–8127. [PubMed] [Google Scholar]
- Hu S. L., Manley J. L. DNA sequence required for initiation of transcription in vitro from the major late promoter of adenovirus 2. Proc Natl Acad Sci U S A. 1981 Feb;78(2):820–824. doi: 10.1073/pnas.78.2.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jove R., Manley J. L. In vitro transcription from the adenovirus 2 major late promoter utilizing templates truncated at promoter-proximal sites. J Biol Chem. 1984 Jul 10;259(13):8513–8521. [PubMed] [Google Scholar]
- Karch F., Török I., Tissières A. Extensive regions of homology in front of the two hsp70 heat shock variant genes in Drosophila melanogaster. J Mol Biol. 1981 May 25;148(3):219–230. doi: 10.1016/0022-2836(81)90536-2. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K., Johnson R. A. Maturation of the head of bacteriophage T4. II. Head-related, aberrant tau-particles. J Mol Biol. 1973 Nov 15;80(4):601–611. doi: 10.1016/0022-2836(73)90199-x. [DOI] [PubMed] [Google Scholar]
- Manley J. L., Fire A., Cano A., Sharp P. A., Gefter M. L. DNA-dependent transcription of adenovirus genes in a soluble whole-cell extract. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3855–3859. doi: 10.1073/pnas.77.7.3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Moncollin V., Miyamoto N. G., Zheng X. M., Egly J. M. Purification of a factor specific for the upstream element of the adenovirus-2 major late promoter. EMBO J. 1986 Oct;5(10):2577–2584. doi: 10.1002/j.1460-2075.1986.tb04537.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M. Gene regulation by proteins acting nearby and at a distance. Nature. 1986 Aug 21;322(6081):697–701. doi: 10.1038/322697a0. [DOI] [PubMed] [Google Scholar]
- Sawadogo M., Roeder R. G. Interaction of a gene-specific transcription factor with the adenovirus major late promoter upstream of the TATA box region. Cell. 1985 Nov;43(1):165–175. doi: 10.1016/0092-8674(85)90021-2. [DOI] [PubMed] [Google Scholar]
- Shenk T. Transcriptional control regions: nucleotide sequence requirements for initiation by RNA polymerase II and III. Curr Top Microbiol Immunol. 1981;93:25–46. doi: 10.1007/978-3-642-68123-3_3. [DOI] [PubMed] [Google Scholar]
- Wiederrecht G., Shuey D. J., Kibbe W. A., Parker C. S. The Saccharomyces and Drosophila heat shock transcription factors are identical in size and DNA binding properties. Cell. 1987 Feb 13;48(3):507–515. doi: 10.1016/0092-8674(87)90201-7. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]