Abstract
Background: Animal studies suggest that early-life lead exposure influences gene expression and production of proteins associated with Alzheimer’s disease (AD).
Objectives: We attempted to assess the relationship between early-life lead exposure and potential biomarkers for AD among young men and women. We also attempted to assess whether early-life lead exposure was associated with changes in expression of AD-related genes.
Methods: We used sandwich enzyme-linked immunosorbent assays (ELISA) to measure plasma concentrations of amyloid β proteins Aβ40 and Aβ42 among 55 adults who had participated as newborns and young children in a prospective cohort study of the effects of lead exposure on development. We used RNA microarray techniques to analyze gene expression.
Results: Mean plasma Aβ42 concentrations were lower among 13 participants with high umbilical cord blood lead concentrations (≥ 10 μg/dL) than in 42 participants with lower cord blood lead concentrations (p = 0.08). Among 10 participants with high prenatal lead exposure, we found evidence of an inverse relationship between umbilical cord lead concentration and expression of ADAM metallopeptidase domain 9 (ADAM9), reticulon 4 (RTN4), and low-density lipoprotein receptor-related protein associated protein 1 (LRPAP1) genes, whose products are believed to affect Aβ production and deposition. Gene network analysis suggested enrichment in gene sets involved in nerve growth and general cell development.
Conclusions: Data from our exploratory study suggest that prenatal lead exposure may influence Aβ-related biological pathways that have been implicated in AD onset. Gene network analysis identified further candidates to study the mechanisms of developmental lead neurotoxicity.
Keywords: Alzheimer’s disease, children, fetal basis of adult disease, human, lead
The weight of evidence supports an association between early-life exposure to lead and impaired cognitive function in children (Bellinger 2004; Lanphear et al. 2005; Needleman 2004). Even at low levels, childhood lead exposure results in cognitive dysfunction that persists into adulthood (Mazumdar et al. 2011). In older adults, chronic environmental exposure to lead is associated with accelerated cognitive decline (van Wijngaarden et al. 2009; Weisskopf et al. 2004, 2007; Weuve et al. 2009; Wright et al. 2003). Whether early exposure to lead has latent effects that contribute to neurodegenerative disease in old age is unknown.
Barker and Osmond (1986) demonstrated an inverse relationship between birth weight and the incidence of cardiovascular disease. The Barker hypothesis, also known as the fetal basis of adult disease (FeBAD) hypothesis, states that many adult diseases have a fetal origin (Barker 1995). According to FeBAD, injury occurring at a critical period of development could result in changes in gene expression or gene imprinting, leading to deficits that become apparent later in life.
Alzheimer’s disease (AD) is a progressive, neurodegenerative disorder that results in dementia and death. The two classical lesions of AD are a) neuritic plaques containing extracellular deposits of the amyloid β (Aβ) proteins and b) neurofibrillary tangles, which are bundles of paired, helically wound filaments inside neurons (Selkoe 2008). The leading hypothesis for AD pathogenesis suggests that accumulation of Aβ in the brain is the primary influence driving its development (Hardy and Selkoe 2002). Animal and cell culture studies have shown that lead exposure affects Aβ production (Gu et al. 2011; Huang et al. 2011).
Recent animal studies suggest the expression of AD-related genes is altered in rodents and primates exposed to lead as infants. Basha et al. (2005) exposed newborn rats to lead; at 20 months of age, the gene encoding β-amyloid precursor protein (APP) exhibited a delayed overexpression (Basha et al. 2005). The increase in APP gene expression in old age was accompanied by an elevation in Aβ in brain tissue. These changes were not seen in rats exposed to lead as adults, suggesting that early timing of lead exposure is an important determinant of gene expression and protein production. The same group also found that cynomolgus monkeys exposed to lead as infants had more neuritic plaques in their brains and exhibited higher levels of APP and Aβ than did monkeys not exposed to lead (Wu et al. 2008). In humans, case studies have reported that children who died of acute lead poisoning had neurofibrillary tangles in their brains at autopsy (Hess and Straub 1974; Niklowitz 1975; Wisniewski et al. 1979).
The long latent period between exposure and outcome poses unique challenges to the study of the FeBAD hypothesis. Most previous research on early-life environmental influences on neurodegenerative diseases has been cross-sectional or retrospective in design, thus limited in its ability to collect data on early exposures. Use of an existing cohort with banked data, ideally with biological samples, is a promising method for studying whether early-life environmental exposures result in neurodegeneration.
The Boston prospective study was one of several cross-sectional and cohort studies initiated in the late 1970s and early 1980s to study the relationship between lead and early development. Follow-up of the Boston cohort showed that the association noted between blood lead concentration and IQ at 2 years of age continued at 10 years of age (Bellinger et al. 1991). A recent report suggests that the association between early-life lead exposure and cognitive function persists into adulthood (Mazumdar et al. 2011).
The objective of this exploratory study was to assess the relationship between early-life environmental lead exposure and potential biomarkers for AD among a group of young adults who were enrolled in the Boston prospective cohort study as newborns. We also attempted to assess whether developmental exposure to lead was associated with changes in expression of genes believed to be involved in AD pathogenesis.
Materials and Methods
Study population. Between August 1979 and April 1981, a cohort of 249 infants was established among babies born at the Brigham and Women’s Hospital in Boston, Massachusetts. Umbilical cord blood lead concentrations were measured, and postnatal blood lead concentrations and development were assessed at 6, 12, 18, 24, and 57 months, and again at 10 years. Follow-up of this cohort at 10 years of age included 148 children (87.6% of those considered eligible; 59.4% of the original cohort) (Bellinger et al. 1992).
In January 2009, members of the original cohort were mailed an introductory letter explaining a new study regarding early-life lead exposure and health outcomes in adulthood. Names and last known addresses were available for only the 148 participants who took part in the 10-year follow-up study. The Committee for Clinical Investigation at Children’s Hospital Boston approved the study, and each participant provided written informed consent.
Measures of exposure, birth to 10 years. Blood samples were obtained from umbilical cords at birth, from participants using capillary tubes at ages 6, 12, 18, and 24 months, and via venipuncture at 57 months and 10 years. Blood lead concentrations were measured in duplicate using graphic furnace atomic absorption spectrometry (Bellinger et al. 1987, 1992).
Measurement of covariates. At the time of enrollment in the present study, participants completed a questionnaire that gathered information on demographics, medical history, family medical history, concurrent medications, and alcohol and tobacco use. Information about other potentially important variables, including maternal medication and alcohol and tobacco use, was available in records from earlier assessments.
Plasma Aβ subspecies isolation. Blood used for plasma Aβ analysis was collected from the present cohort via venipuncture in EDTA tubes, processed immediately (centrifuged and aliquotted as plasma, buffy coat, and red blood cells), and then stored at –80°C. Plasma samples were sent to and analyzed in the Selkoe Laboratory at Brigham and Women’s Hospital.
Aβ40 and Aβ42 were assayed by sandwich enzyme-linked immunosorbent assays. Plates were coated with capture antibodies (2G3 for Aβ40 and 21F12 for Aβ42) in phosphate-buffered saline (PBS), incubated for 4 hr at room temperature, and then blocked with 4% Block Ace (BA; AbD Serotec, Raleigh, NC) at 4°C. Plates were washed three times with PBS containing Tween 20, and samples were freshly diluted in 0.4% BA, loaded into the wells, and incubated with antibodies for 2 hr at room temperature. Samples were then reincubated in solution containing detector biotinylated antibody (266B) for 2 hr at room temperature. Finally, samples were incubated with streptavidin alkaline phosphatase (Promega, Madison, WI) in PBS for 1 hr at room temperature and washed three times with Tris-buffered saline. After adding AttoPhos (Promega), the signal was amplified and measured with a Victor2 microtiter plate reader (PerkinElmer, Boston, MA).
RNA preparation. Blood used for RNA microarray analysis was collected from the present cohort via venipuncture into PAXgene tubes (QIAGEN Inc., Valencia, CA). Blood tubes were stored at –80°C until RNA extraction. Total RNA was isolated according to manufacturer protocols and purified using the PAXgene Blood RNA kit (QIAGEN Inc.). RNA extraction was performed at the General Clinical Research Center (GCRC) laboratories at Children’s Hospital Boston for the initial 12 samples. The GCRC laboratory closed during the study, and RNA extraction was performed on the remaining 37 samples at the Molecular Genetics Core Facility of the Children’s Hospital Boston Intellectual and Developmental Disabilities Research Center (IDDRC).
Microarray hybridization. Gene transcript expression was analyzed using Affymetrix Human Genome 1.0 ST array (Affymetrix, Santa Clara, CA). All RNA samples were analyzed at the Microarray Core Facility of the Dana-Farber Cancer Institute. Array data were retrieved and processed using the BioConductor framework, version 2.9 (Gentleman et al. 2004; http://www.bioconductor.org/), and tested for quality using the arrayQualityMetrics package (Kauffmann et al. 2009).
Statistical analysis. Statistical methods focused on estimating the association between early-life blood lead concentrations and plasma Aβ subspecies concentrations. We calculated correlation coefficients and fitted linear regression models with individual Aβ subspecies as the response and lead concentrations as the predictors. We fitted separate models for each measurement of lead concentration, and the average lead concentrations over the first 10 years of life were modeled as simple continuous variables and as log-transformed variables to account for heteroskedasticity and a possible nonlinear relationship between lead exposure and plasma Aβ subspecies. Outcomes were plasma Aβ42 concentration, Aβ40 concentration, and the ratio of Aβ42 and Aβ40 concentrations, which also were modeled before and after log transformation. We also stratified analyses by high and low prenatal lead levels, where high lead level was defined as > 10 μg/dL. We chose 10 μg/dL as our cutoff because this definition for high lead level was used in the original cohort study and was part of the initial sampling and recruitment strategy (Bellinger et al. 1987).
Microarray data analysis. RNA expression data from 49 samples were evaluated using standard quality control criteria to assess the variability across samples, for example, batch effects that may have resulted from experiments being performed on different days or by different individuals, as well as the spatial distribution of probe intensities across chips. Arrays were then background corrected and normalized with robust multichip analysis. IsoGene (Lin et al. 2007), an R statistical software package (http://www.r-project.org/), was used to identify genes with significant changes of gene expression levels in response to an increase in cord blood lead concentration at a 5% false discovery rate (FDR; determined using the Benjamini–Hochberg correction for multiple testing). A total of 245 probes mapping to 196 unique genes were considered significant using this criteria. To identify the subset of genes exhibiting monotonous changes in expression levels with increasing prenatal lead levels (at |r| ≥ 0.9), the 10 samples with blood prenatal blood levels > 10 μg/dL were binned into groups so that each group had approximately the same number of samples (lead exposures levels 10–12, 12–15, 15–18, 18–20 μg/dL) and analyzed in GATE (Grid Analysis of Time Series Expression) software (MacArthur et al. 2010; http://amp.pharm.mssm.edu/maayan-lab/gate.htm). Genes identified in the IsoGene analysis that passed the |r| ≥ 0.9 correlation filter were subsequently tested for Gene Ontology enrichment using the GeneMANIA Cytoscape plugin (Warde-Farley et al. 2010; http://www.genemania.org/plugin/). GeneMANIA extends a provided gene list with functionally similar and interacting genes to generate a functional association network. Association data include protein and genetic interactions, pathways, coexpression, colocalization, and protein domain similarity. In addition, we reviewed the literature to determine if any of the identified genes were reported to be implicated in AD biology.
Results
We located 89 (60%) of the 148 cohort members who participated in the 10-year follow-up study. Of these, 58 enrolled in the present study. Fifty-five blood samples were available for plasma studies. The study population generally consisted of white, college-educated children with college-educated mothers. Participants were similar to members of the original cohort in terms of demographic factors, measures of socioeconomic status, blood lead history, and IQ scores in early childhood (Table 1).
Table 1.
Characteristics of the participants at 28–30 years of age and comparison with nonparticipants.
| Characteristic | Participants | Nonparticipants (n = 90) | ||||
|---|---|---|---|---|---|---|
| With plasma available (n = 55) | With RNA used in analysis (n = 36) | |||||
| Participants | ||||||
| Age at testing (years) | 28.9 ± 0.5 | 29.0 ± 0.4 | — | |||
| Body mass index (kg/m2) | 25.2 ± 4.5 | 25.0 ± 4.7 | ||||
| College graduate | 83.6 | 91.7 | — | |||
| Currently smoke | 18.2 | 19.4 | — | |||
| Alcohol use > 2 drinks/week | 38.1 | 33.3 | — | |||
| Mother or father with dementia | 3.4 | 0 | — | |||
| Currently taking oral contraceptives | 14.5 | 22.2 | — | |||
| Currently taking medicine other than oral contraceptives | 0 | 0 | — | |||
| Male | 47.2 | 36.8 | 53.3 | |||
| White | 94.5 | 94.7 | 94.4 | |||
| Weeks of gestation | 40.0 ± 1.9 | 39.8 ± 2.3 | 40.0 ± 1.6 | |||
| Birth weight (kg) | 3.4 ± 0.5 | 3.4 ± 0.5 | 3.4 ± 0.5 | |||
| Blood lead concentration (μg/dL) | ||||||
| Cord | 6.3 ± 5.0 | 6.3 ± 5.4 | 7.9 ± 5.4 | |||
| 6 months | 7.8 ± 5.1 | 7.4 ± 5.6 | 9.3 ± 8.0 | |||
| 12 months | 9.8 ± 6.8 | 9.6 ± 7.0 | 9.9 ± 6.4 | |||
| 24 months | 8.1 ± 4.3 | 7.7 ± 4.0 | 8.8 ± 5.1 | |||
| 4 years | 6.6 ± 3.4 | 6.4 ± 3.9 | 6.1 ± 4.0 | |||
| 10 years | 3.0 ± 2.5 | 2.5 ± 2.3 | 2.9 ± 2.4 | |||
| IQ at 4 years of age | 118.4 ± 14.0 | 117.3 ± 14.4 | 114.3 ± 15.4 | |||
| IQ at 10 years of age | 118.7 ± 14.2 | 116.1 ± 16.3 | 114.5 ± 13.8 | |||
| Participants’ mothers | ||||||
| Age at delivery (years) | 30.6 ± 4.0 | 31.4 ± 4.0 | 30.4 ± 4.2 | |||
| College graduate | 60.0 | 72.2 | 62.2 | |||
| Maternal IQ | 124.5 ± 17.8 | 127.6 ± 14.1 | 124.9 ± 14.7 | |||
| Tobacco use during pregnancy | 23.6 | 33.3 | 22.5 | |||
| Alcohol use during pregnancy | 47.2 | 48.4 | 42.7 | |||
| Data are percent yes or mean ± SD. | ||||||
Blood lead concentration. Mean blood lead concentration in the 55 cohort members for whom plasma samples were available was lowest at 10 years of age (3.0 μg/dL) and highest at 12 months of age (9.8 μg/dL; Table 1). Median concentrations followed a similar pattern, except median blood lead concentration was highest at 2 years of age [8.1 μg/dL; see Supplemental Material, Figure 1 (http://dx.doi.org/10.1289/ehp.1104474)]. The highest lead concentrations were seen in infancy and early childhood, possibly reflecting greater lead intake through hand-to-mouth activity and higher exposures in that era.
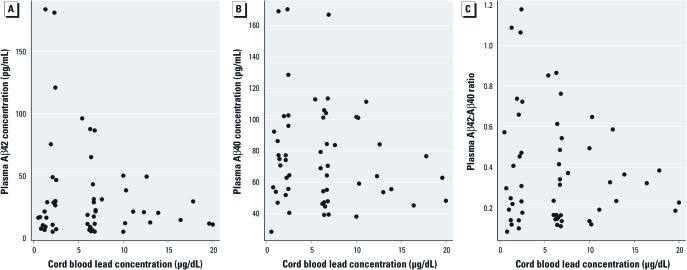
Aβ analysis. Scatterplots of prenatal lead concentration, Aβ42 concentration, and Aβ42:Aβ40 ratio suggest an inverse relationship between prenatal blood lead concentration and plasma Aβ subspecies in adulthood (Figure 1). The mean ± SD Aβ42 concentration among participants with prenatal lead exposure < 10 μg/dL was 36.6 ± 43.1 pM, compared with 22.8 ± 14.7 pM among participants with higher prenatal lead exposure (p = 0.08; Figure 1). The Aβ42:Aβ40 ratio showed a similar pattern between the two groups, with a ratio of 0.40 ± 0.30 for participants with prenatal lead concentration ≥ 10 μg/dL and 0.32 ± 0.17 for participants with prenatal blood lead levels < 10 μg/dL (p = 0.27). Models of untransformed and log-transformed lead concentrations at different time points did not indicate clear or consistent associations with plasma Aβ42 or the Aβ42:Aβ40 ratio. Visual inspection of the scatterplots among participants with high prenatal lead exposure reveals an apparent inverse relationship between umbilical cord lead concentration and plasma Aβ42 concentration as well as plasma Aβ42:Aβ40 ratio (Figure 1).
Figure 1.
Scatterplots of prenatal (umbilical cord) lead concentrations versus plasma Aβ subspecies concentrations (n = 55 plasma samples). Aβ42 (A) and Aβ40 (B) at 28 years of age and Aβ42:Aβ40 ratio at 29 years of age (C).
AD-related gene expression. Forty-nine samples were available for RNA analysis, and 36 samples that met standard quality control criteria were included in analyses. All of the 36 samples used in the analyses were processed at the IDDRC. The expression of 196 genes was significantly associated (5% FDR) with increasing cord blood lead level. Because of the small sample size, an additional filter was applied, retaining only genes with a correlation of |r| ≥ 0.9 with an increasing (or decreasing) trend between cord blood levels of 10–20 μg/dL. The 39 retained genes [listed in Supplemental Material, Table 1 (http://dx.doi.org/10.1289/ehp.1104474)] exhibited a much higher level of variation in gene expression in relation to cord blood level concentrations < 10 μg/dL (n = 28) compared with concentrations > 10 μg/dL (n = 10; data not shown).
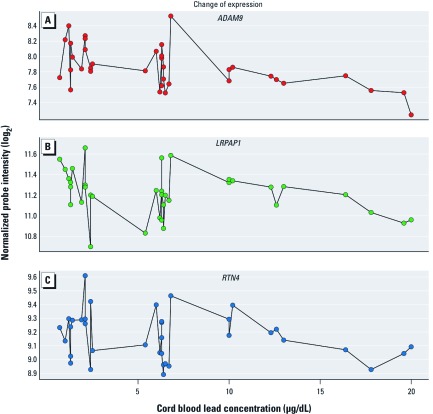
Three of the 39 identified genes encode proteins that have been reported to affect Aβ production and deposition in the brain; ADAM metallopeptidase domain 9 (ADAM9), low-density lipoprotein receptor-related protein associated protein 1 (LRPAP1) and reticulon 4 (RTN4). Figure 2 shows normalized probe intensities (measures of relative gene expression) of these genes in relation to prenatal blood lead concentrations for individual participants.
Figure 2.
Gene expression levels for selected AD-related genes: normalized probe intensities for ADAM9 (A), RTN4 (B), and LRPAP1 (C) at 29 years of age in relation to prenatal (umbilical cord) blood lead concentrations. Data points from samples with cord blood lead level concentrations < 10 μg/dL (n = 26) were not used to identify genes whose expression was significantly associated with blood lead levels > 10 μg/dL (n = 10) and are plotted for reference only.
Genes whose expression showed an increasing or decreasing trend in relation to prenatal lead level were found to interact directly or indirectly in physical interaction and coexpression networks [see Supplemental Material, Figure 2 (http://dx.doi.org/10.1289/ehp.1104474)]. Although the network analysis did not exhibit a direct involvement with amyloid pathways, it identified enrichment of genes associated with the (negative) regulation of neurogenesis, nerve growth, and general cell development (Table 2).
Table 2.
Network analysis representing functional association of 39 genes with altered expression at prenatal lead levels > 10 μg/dL.
| Gene Ontology biological process | Q-value | Count [annotated (overall)] | ||
|---|---|---|---|---|
| Neuron development | ||||
| Negative regulation of axonogenesis | 0.00080 | 4 (13) | ||
| Negative regulation of cell projection organization | 0.00328 | 4 (21) | ||
| Negative regulation of neurogenesis | 0.00634 | 4 (27) | ||
| Nerve growth factor receptor signaling pathway | 0.00990 | 7 (215) | ||
| Regulation of axonogenesis | 0.00990 | 4 (34) | ||
| Regulation of neuron projection development | 0.08320 | 4 (63) | ||
| Cell development | ||||
| Negative regulation of cell development | 0.01040 | 4 (36) | ||
| Negative regulation of cell differentiation | 0.10500 | 5 (148) | ||
| Immune response | ||||
| Response to abiotic stimulus | 0.08320 | 6 (218) | ||
| T-cell receptor signaling pathway | 0.08490 | 4 (67) | ||
| Data are Gene Ontology categories (http://www.geneontology.org/) and Q-values (minimum FDR at which the test is considered significant) from an FDR-corrected hypergeometric test for enrichment. Q-Values were estimated using the Benjamini–Hochberg procedure. Counts reflect genes with this annotation in the query set (39 genes of interest identified by differential monotonous change of expression in response to lead exposure) compared with the overall number of genes in the GeneMANIA background set with this annotation and at least one interaction (numbers in parentheses). | ||||
Discussion
Two observations from this study support the hypothesis that early-life (prenatal) lead exposure influences the biological pathways believed to be involved in AD. First, participants with higher prenatal lead exposure (umbilical cord lead concentration > 10 μg/dL) had lower mean plasma Aβ42 concentrations than did participants with low and moderate prenatal lead levels. Data from participants with higher prenatal lead levels showed an apparent inverse relationship between umbilical cord lead concentration and both plasma Aβ42 concentration and plasma Aβ42:Aβ40 ratios.
The second finding is that among the 10 participants with higher prenatal lead exposure (> 10 μg/dL), there were inverse relationships between umbilical cord lead concentrations and expression of ADAM9, RTN4, and LRPAP1, genes whose products are believed to affect Aβ production and deposition in the brain (Allinson et al. 2003; Cong and Jia 2011; He et al. 2004; Hyman et al. 2000; Murayama et al. 2006). The expression of 39 genes appeared to be associated with prenatal blood lead levels within this subgroup, but these three genes have been previously found relevant to AD pathogenesis.
The product of ADAM9 is a member of the ADAM (a disintegrin and metalloprotease domain) family. Members of this family are membrane-anchored proteins that have been implicated in a variety of biological processes, including neurogenesis. The product of ADAM9 is believed to act as an α-secretase and cause nonamyloidogenic cleavage of APP (Allinson et al. 2003; Cong and Jia 2011; Deuss et al. 2008).
The product of RTN4 is a potent neurite outgrowth inhibitor that may also help block the regeneration of the central nervous system in higher vertebrates. RTN4 is also implicated in the amyloid cascade. Animal studies have suggested that the product of RTN4 decreases cleavage of APP to its amyloidogenic products (He et al. 2004; Murayama et al. 2006).
Low-density lipoprotein receptor-related protein (LRP) is thought to play an important role in determining the balance between Aβ synthesis and clearance mechanisms (Hyman et al. 2000). The product of the LRPAP1 gene blocks ligand-binding sites of LRP and interferes with Aβ clearance. In accordance with its role in the amyloidogenic pathway, variation in LRPAP1 has been associated with the risk of developing AD (Sanchez et al. 2001).
Although gene network analysis did not suggest a direct involvement with amyloid pathways, several genes that exhibited decreased expression in relation to prenatal lead concentration are involved in the (negative) regulation of neurogenesis, nerve growth, and general cell development. This provides additional support to the hypothesis that developmental exposure to lead can cause changes in expression for genes important in neurological development and disease. Our findings were similar to those seen in zebrafish embryos (Peterson et al. 2011) after researchers exposed them to a sublethal dose of lead and analyzed global transcriptional alterations.
The known association between AD and both limited education and occupational attainment (Cobb et al. 1995; Snowdon et al. 2000; Stern et al. 1994), often referred to as the cognitive reserve hypothesis, suggests that risk factors for AD may be established early in life. The cognitive reserve hypothesis posits that higher levels of education, or other measures of cognitive ability early in life, provide protection against cognitive decline, suggesting that early education leads to the development of efficient and/or flexible neural networks more capable of coping with the disruption imposed by brain pathology (Stern 2009). An alternative explanation is that the same environmental influences that result in poor cognition in childhood and early adulthood have persistent and/or latent effects that contribute to the development of dementia during old age.
Our study used prospectively collected lead exposure information from a cohort study started > 30 years ago. The long latent period between exposure and outcome poses significant challenges to the study of the FeBAD hypothesis, and the imprecision of exposure classification based on ecological data or recall is one of the biggest criticisms of this work (Joseph and Kramer 1996). Our study minimized the uncertainty around exposure assessment by using samples that were collected and analyzed in real time.
Our study has a number of limitations, the most important of which is a small number of participants. Specifically, we had only 13 participants with prenatal blood lead levels > 10 μg/dL (including 10 with gene expression data), which turned out to be the exposure level of greatest interest. The Boston cohort was specifically assembled to investigate the cognitive effects of what was then considered to be low-level lead exposure. Repeating this analysis among participants from cohorts with higher prenatal lead levels may provide more data around the exposure levels of interest, therefore providing more power and more precise estimates of association.
Another important limitation is the use of plasma measurements of Aβ instead of cerebrospinal fluid measurements or neuropathological changes (as would be seen at autopsy) to define the outcome. The published data on plasma Aβ levels in AD are contradictory, with plasma Aβ levels reported to be higher (Kosaka et al. 1997; Sobow et al. 2005), lower (Buerger et al. 2009; Cosentino et al. 2010), or unchanged (Abdullah et al. 2007; Fukumoto et al. 2003; Giedraitis et al. 2007; Tamaoka et al. 1996) in cases with AD compared with controls. A recent meta-analysis found that in cross-sectional studies, AD patients in their 60s and 70s had lower Aβ42 plasma concentrations compared with cognitively normal individuals (Song et al. 2011), although this pattern was not seen in longitudinal studies. A proposed mechanism for the discrepancy between high Aβ levels seen in the brains of AD cases and low Aβ subspecies levels in plasma is that there is compartmentalization of Aβ peptides in the brain among patients with AD (Schupf et al. 2008). There are no reports regarding the association between plasma Aβ among young adults and the development of AD.
An additional limitation of our study is that we did not measure blood lead levels at the same time that we measured plasma Aβ and gene expression levels. In a recent pooled analysis, current blood lead levels and average lifetime estimates were stronger predictors of intellectual deficits in late childhood than was early childhood lead concentration (Lanphear et al. 2005). In older adults, however, recent studies found that current blood lead levels were not associated with cognitive function in adults, suggesting that early-life or accumulated exposures may be etiologically more important (van Wijngaarden et al. 2011). Future studies would be strengthened if associations are estimated between outcome and current blood lead levels, as well as early blood lead levels.
An additional concern is the use of multiple testing in microarray experiments. Given thousands of genes, it is possible that the ones we found to be associated with prenatal lead exposure were so associated by chance (Ioannidis 2005). We corrected for this limitation by looking at genes with continuous changes at higher prenatal levels (following the trend seen in scatterplots) and also by comparing these with genes implicated in AD pathogenesis. However, our findings should be considered preliminary.
If early-life lead exposure influences biological processes involved in AD pathogenesis, the implications for public health practice are substantial. Lead exposure in the United States reached its peak in the 1970s, and the children exposed at that time, now adults, may be at increased risk for neurodegenerative disease. If similar results are achieved in a larger cohort, our study could provide support for screening strategies that identify adults who may be at higher risk for neurodegenerative disease because of childhood exposures, as well as identify an opportunity for disease prevention.
Conclusions
Data from our study suggest that early-life lead exposure may influence the biological pathways that have been implicated in AD in old age. Gene network analysis identified further candidates for the study of the mechanisms of developmental lead neurotoxicity in humans.
Supplemental Material
Acknowledgments
We thank K. Abanilla and G. Dieudonne, who coordinated participant recruitment. We thank D.-L. Destouche, B. Deutsch, and J. Medeiros, technicians in the General Clinical Research Center (GCRC) Laboratory at Children’s Hospital Boston who drew samples and isolated plasma, as well as R. Grand, F. Bonilla, and K. Jordan of the GCRC staff. We also thank D. Christiani and D. Selkoe. For RNA extraction and microarray, we thank E. Fox of Dana Farber Cancer Institute and H. Schneider of the Children’s Hospital Boston Molecular Genetics Core Facility, supported by the Intellectual and Developmental Disabilities Center and the Sir Edwin Manton Center for Orphan Disease Research.
Footnotes
M.M. was supported by K23 ES017437 from the National Institute of Environmental Health Sciences (NIEHS). Additional support was provided by pilot project funding from the Harvard School of Public Health–NIEHS Center for Environmental Health (ES000002), the Annie E. Casey Foundation, the Heinz Foundation, and the Harvard Clinical and Translational Science Center (National Institutes of Health award UL1 RR024758 and financial contributions from Harvard University and its affiliated academic health care centers).
The content is solely the responsibility of the authors and does not necessarily represent the official views of Harvard University, and its affiliated academic health care centers, or the National Institutes of Health.
The authors declare they have no actual or potential competing financial interests.
References
- Abdullah L, Paris D, Luis C, Quadros A, Parrish J, Valdes L, et al. The influence of diagnosis, intra- and inter-person variability on serum and plasma Abeta levels. Neurosci Lett. 2007;428:53–58. doi: 10.1016/j.neulet.2007.09.058. [DOI] [PubMed] [Google Scholar]
- Allinson TM, Parkin ET, Turner AJ, Hooper NM. ADAMs family members as amyloid precursor protein α-secretases. J Neurosci Res. 2003;74:342–352. doi: 10.1002/jnr.10737. [DOI] [PubMed] [Google Scholar]
- Barker DJ. Fetal origins of coronary heart disease. BMJ. 1995;311:171–174. doi: 10.1136/bmj.311.6998.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker DJ, Osmond C. Infant mortality, childhood nutrition, and ischaemic heart disease in England and Wales. Lancet. 1986;1:1077–1081. doi: 10.1016/s0140-6736(86)91340-1. [DOI] [PubMed] [Google Scholar]
- Basha MR, Wei W, Bakheet SA, Benitez N, Siddiqi HK, Ge YW, et al. The fetal basis of amyloidogenesis: exposure to lead and latent overexpression of amyloid precursor protein and β-amyloid in the aging brain. J Neurosci. 2005;25:823–829. doi: 10.1523/JNEUROSCI.4335-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellinger DC. Lead. Pediatrics. 2004;113:1016–1022. [PubMed] [Google Scholar]
- Bellinger D, Leviton A, Waternaux C, Needleman H, Rabinowitz M. Longitudinal analyses of prenatal and postnatal lead exposure and early cognitive development. N Engl J Med. 1987;316:1037–1043. doi: 10.1056/NEJM198704233161701. [DOI] [PubMed] [Google Scholar]
- Bellinger D, Sloman J, Leviton A, Rabinowitz M, Needleman HL, Waternaux C. Low-level lead exposure and children’s cognitive function in the preschool years. Pediatrics. 1991;87:219–227. [PubMed] [Google Scholar]
- Bellinger DC, Stiles KM, Needleman HL. Low-level lead exposure, intelligence and academic achievement: a long-term follow-up study. Pediatrics. 1992;90:855–861. [PubMed] [Google Scholar]
- Buerger K, Frisoni G, Uspenskaya O, Ewers M, Zetterberg H, Geroldi C, et al. Validation of Alzheimer’s disease CSF and plasma biological markers: the multicentre reliability study of the pilot European Alzheimer’s Disease Neuroimaging Initiative (E-ADNI). Exp Gerontol. 2009;44:579–585. doi: 10.1016/j.exger.2009.06.003. [DOI] [PubMed] [Google Scholar]
- Cobb JL, Wolf PA, Au R, White R, D’Agostino RB. The effect of education on the incidence of dementia and Alzheimer’s disease in the Framingham Study. Neurology. 1995;45:1707–1712. doi: 10.1212/wnl.45.9.1707. [DOI] [PubMed] [Google Scholar]
- Cong L, Jia J. Promoter polymorphisms which regulate ADAM9 transcription are protective against sporadic Alzheimer’s disease. Neurobiol Aging. 2011;32:54–62. doi: 10.1016/j.neurobiolaging.2009.01.001. [DOI] [PubMed] [Google Scholar]
- Cosentino SA, Stern Y, Sokolov E, Scarmeas N, Manly JJ, Tang MX, et al. Plasma β-amyloid and cognitive decline. Arch Neurol. 2010;67:1485–1490. doi: 10.1001/archneurol.2010.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuss M, Reiss K, Hartmann D. Part-time α-secretases: the functional biology of ADAM 9, 10 and 17. Curr Alzheimer Res. 2008;5:187–201. doi: 10.2174/156720508783954686. [DOI] [PubMed] [Google Scholar]
- Fukumoto H, Tennis M, Locascio JJ, Hyman BT, Growdon JH, Irizarry MC. Age but not diagnosis is the main predictor of plasma amyloid β-protein levels. Arch Neurol. 2003;60:958–964. doi: 10.1001/archneur.60.7.958. [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. 2004Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5R80; doi: 10.1186/gb-2004-5-10-r80[Online 15 September 2004] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giedraitis V, Sundelof J, Irizarry MC, Garevik N, Hyman BT, Wahlund LO, et al. The normal equilibrium between CSF and plasma amyloid beta levels is disrupted in Alzheimer’s disease. Neurosci Lett. 2007;427:127–131. doi: 10.1016/j.neulet.2007.09.023. [DOI] [PubMed] [Google Scholar]
- Gu H, Wei X, Monnot AD, Fontanilla CV, Behl M, Farlow MR, et al. Lead exposure increases levels of β-amyloid in the brain and CSF and inhibits LRP1 expression in APP transgenic mice. Neurosci Lett. 2011;490:16–20. doi: 10.1016/j.neulet.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- He W, Lu Y, Qahwash I, Hu XY, Chang A, Yan R. Reticulon family members modulate BACE1 activity and amyloid-β peptide generation. Nat Med. 2004;10:959–965. doi: 10.1038/nm1088. [DOI] [PubMed] [Google Scholar]
- Hess K, Straub PW. Chronische Bleivergiftung (Chronic lead poisoning) Schweiz Rundsch Med Prax. 1974;63:177–183. [in German] [PubMed] [Google Scholar]
- Huang H, Bihaqi SW, Cui L, Zawia NH. In vitro Pb exposure disturbs the balance between Aβ production and elimination: the role of AßPP and neprilysin. Neurotoxicology. 2011;32:300–306. doi: 10.1016/j.neuro.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman BT, Strickland D, Rebeck GW. Role of the low-density lipoprotein receptor–related protein in β-amyloid metabolism and Alzheimer disease. Arch Neurol. 2000;57:646–650. doi: 10.1001/archneur.57.5.646. [DOI] [PubMed] [Google Scholar]
- Ioannidis JP.2005Why most published research findings are false. PLoS Med 2e124; doi: 10.1371/journal.pmed.0020124[Online 30 August 2005] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph KS, Kramer MS. Review of the evidence on fetal and early childhood antecedents of adult chronic disease. Epidemiol Rev. 1996;18:158–174. doi: 10.1093/oxfordjournals.epirev.a017923. [DOI] [PubMed] [Google Scholar]
- Kauffmann A, Gentleman R, Huber W. arrayQualityMetrics—a bioconductor package for quality assessment of microarray data. Bioinformatics. 2009;25:415–416. doi: 10.1093/bioinformatics/btn647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaka T, Imagawa M, Seki K, Arai H, Sasaki H, Tsuji S, et al. The beta APP717 Alzheimer mutation increases the percentage of plasma amyloid-beta protein ending at A beta 42(43). Neurology. 1997;48:741–745. doi: 10.1212/wnl.48.3.741. [DOI] [PubMed] [Google Scholar]
- Lanphear BP, Hornung R, Khoury J, Yolton K, Baghurst P, Bellinger DC, et al. Low-level environmental lead exposure and children’s intellectual function: an international pooled analysis. Environ Health Perspect. 2005;113:894–899. doi: 10.1289/ehp.7688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D, Shkedy Z, Yekutieli D, Burzykowski T, Gohlmann HW, De Bondt A, et al. Testing for trends in dose-response microarray experiments: a comparison of several testing procedures, multiplicity and resampling-based inference. Stat Appl Genet Mol Biol. 2007;6:1–26. doi: 10.2202/1544-6115.1283. [DOI] [PubMed] [Google Scholar]
- MacArthur BD, Lachmann A, Lemischka IR, Ma’ayan A. GATE: software for the analysis and visualization of high-dimensional time series expression data. Bioinformatics. 2010;26:143–144. doi: 10.1093/bioinformatics/btp628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumdar M, Bellinger DC, Gregas M, Abanilla K, Bacic J, Needleman HL.2011Low-level environmental lead exposure in childhood and adult intellectual function: a follow-up study. Environ Health 1024; doi: 10.1186/1476-069X-10-24[Online 30 March 2011] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama KS, Kametani F, Saito S, Kume H, Akiyama H, Araki W. Reticulons RTN3 and RTN4-B/C interact with BACE1 and inhibit its ability to produce amyloid β-protein. Eur J Neurosci. 2006;24:1237–1244. doi: 10.1111/j.1460-9568.2006.05005.x. [DOI] [PubMed] [Google Scholar]
- Needleman H. Lead poisoning. Annu Rev Med. 2004;55:209–222. doi: 10.1146/annurev.med.55.091902.103653. [DOI] [PubMed] [Google Scholar]
- Niklowitz WJ. Neurofibrillary changes after acute experimental lead poisoning. Neurology. 1975;25:927–934. doi: 10.1212/wnl.25.10.927. [DOI] [PubMed] [Google Scholar]
- Peterson SM, Zhang J, Weber G, Freeman JL. Global gene expression analysis reveals dynamic and developmental stage-dependent enrichment of lead-induced neurological gene alterations. Environ Health Perspect. 2011;119:615–621. doi: 10.1289/ehp.1002590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez L, Alvarez V, Gonzalez P, Gonzalez I, Alvarez R, Coto E. Variation in the LRP-associated protein gene (LRPAP1) is associated with late-onset Alzheimer disease. Am J Med Genet. 2001;105:76–78. [PubMed] [Google Scholar]
- Schupf N, Tang MX, Fukuyama H, Manly J, Andrews H, Mehta P, et al. Peripheral Abeta subspecies as risk biomarkers of Alzheimer’s disease. Proc Natl Acad Sci USA. 2008;105:14052–14057. doi: 10.1073/pnas.0805902105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selkoe DJ. Biochemistry and molecular biology of amyloid beta-protein and the mechanism of Alzheimer’s disease. Handb Clin Neurol. 2008;89:245–260. doi: 10.1016/S0072-9752(07)01223-7. [DOI] [PubMed] [Google Scholar]
- Snowdon DA, Greiner LH, Markesbery WR. Linguistic ability in early life and the neuropathology of Alzheimer’s disease and cerebrovascular disease. Findings from the Nun Study. Ann N Y Acad Sci. 2000;903:34–38. doi: 10.1111/j.1749-6632.2000.tb06347.x. [DOI] [PubMed] [Google Scholar]
- Sobow T, Flirski M, Kloszewska I, Liberski PP. Plasma levels of Aβ peptides are altered in amnestic mild cognitive impairment but not in sporadic Alzheimer’s disease. Acta Neurobiol Exp (Wars) 2005;65:117–124. doi: 10.55782/ane-2005-1544. [DOI] [PubMed] [Google Scholar]
- Song F, Poljak A, Valenzuela M, Mayeux R, Smythe GA, Sachdev PS. Meta-analysis of plasma amyloid-β levels in Alzheimer’s disease. J Alzheimers Dis. 2011;26(2):365–375. doi: 10.3233/JAD-2011-101977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern Y. Cognitive reserve. Neuropsychologia. 2009;47:2015–2028. doi: 10.1016/j.neuropsychologia.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern Y, Gurland B, Tatemichi TK, Tang MX, Wilder D, Mayeux R. Influence of education and occupation on the incidence of Alzheimer’s disease. JAMA. 1994;271:1004–1010. [PubMed] [Google Scholar]
- Tamaoka A, Fukushima T, Sawamura N, Ishikawa K, Oguni E, Komatsuzaki Y, et al. Amyloid β protein in plasma from patients with sporadic Alzheimer’s disease. J Neurol Sci. 1996;141:65–68. doi: 10.1016/0022-510x(96)00143-8. [DOI] [PubMed] [Google Scholar]
- van Wijngaarden E, Campbell JR, Cory-Slechta DA. Bone lead levels are associated with measures of memory impairment in older adults. Neurotoxicology. 2009;30:572–580. doi: 10.1016/j.neuro.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wijngaarden E, Winters PC, Cory-Slechta DA. Blood lead levels in relation to cognitive function in older U.S. adults. Neurotoxicology. 2011;32:110–115. doi: 10.1016/j.neuro.2010.11.002. [DOI] [PubMed] [Google Scholar]
- Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–W220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisskopf MG, Proctor SP, Wright RO, Schwartz J, Spiro A, III, Sparrow D, et al. Cumulative lead exposure and cognitive performance among elderly men. Epidemiology. 2007;18:59–66. doi: 10.1097/01.ede.0000248237.35363.29. [DOI] [PubMed] [Google Scholar]
- Weisskopf MG, Wright RO, Schwartz J, Spiro A, III, Sparrow D, Aro A, et al. Cumulative lead exposure and prospective change in cognition among elderly men: the VA Normative Aging Study. Am J Epidemiol. 2004;160:1184–1193. doi: 10.1093/aje/kwh333. [DOI] [PubMed] [Google Scholar]
- Weuve J, Korrick SA, Weisskopf MG, Ryan LM, Schwartz J, Nie H, et al. Cumulative exposure to lead in relation to cognitive function in older women. Environ Health Perspect. 2009;117:574–580. doi: 10.1289/ehp.11846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski K, Jervis GA, Moretz RC, Wisniewski HM. Alzheimer neurofibrillary tangles in diseases other than senile and presenile dementia. Ann Neurol. 1979;5:288–294. doi: 10.1002/ana.410050311. [DOI] [PubMed] [Google Scholar]
- Wright RO, Tsaih SW, Schwartz J, Spiro A, III, McDonald K, Weiss ST, et al. Lead exposure biomarkers and mini-mental status exam scores in older men. Epidemiology. 2003;14:713–718. doi: 10.1097/01.EDE.0000081988.85964.db. [DOI] [PubMed] [Google Scholar]
- Wu J, Basha MR, Brock B, Cox DP, Cardozo-Pelaez F, McPherson CA, et al. Alzheimer’s disease (AD)-like pathology in aged monkeys after infantile exposure to environmental metal lead (Pb): evidence for a developmental origin and environmental link for AD. J Neurosci. 2008;28:3–9. doi: 10.1523/JNEUROSCI.4405-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.