Abstract
A genetically heterogeneous population of mice was tested for hearing at 8, 18 and 22 months by auditory brainstem response (ABR), and genotyped at 128 markers to identify loci that modulate late life hearing loss. Half of the test mice were exposed to noise for 2 hr at age 20 months. Polymorphisms affecting hearing at 18 months were noted on chromosomes 2, 3, 7, 10, and 15. Most of these loci had effects only on responses to 48 kHz stimuli, but a subset also influenced the ABR at lower frequencies. Loci on chromosomes 4, 10, 12, and 14 had significant effects on hearing at 22 months in noise-exposed mice, and loci on chromosomes 10 and 11 had effects on mice not exposed to noise. Outer hair cell loss was modulated by polymorphisms on chromosomes 10, 11, 12, 17, and 19. Resistance to age-related hearing loss is thus modulated by a set of genetic effects, some age-specific, some frequency specific, some dependent on prior exposure to noise, and some of which compromise survival of cochlear hair cells.
Keywords: aging, presbycusis, gene mapping, cochlea, mice, animal models
1. Introduction
Age-related hearing impairment (ARHI), also known as presbycusis, affects most people aged 65 and older and represents the most common neurodegenerative disease of aging. The quest to elucidate the causes of ARHI and to find ameliorative treatments is much hampered by the fact that human presbycusis is highly complex (for review see (Van Eyken et al., 2007)), and its progression is modulated both by genetic predisposition and by a variety of insults to the hearing organ. The Framingham Heart Study found significant inherited genetic effects in an analysis of hearing thresholds in sibling or parent/child pairs. The predictive power of family history varied with gender and the type of presbycusis but, overall, about 55% of the variance in ARHI could be ascribed to genetic factors (Gates et al., 1999). This influence is then likely to be modulated by external risk factors that may slowly impair the function and structural integrity of the auditory organ. Risk factors include exposure to noise, environmental chemicals, chemotherapy, or disorders such as diabetes or cardiovascular disease (Cruickshanks et al., 2010). Animal models that can control or account for confounding environmental factors have the potential for clarifying the interaction of genetic and nongenetic factors in the pathobiology of ARHI, and may provide new clues to its physiology and molecular biology.
Noise trauma, as a major contributor to ARHI, poses a serious threat to public health. About 10% of the US population are exposed daily to noise levels that can lead to hearing impairment (Daniel, 2007), and 16% of all disabling hearing loss in adults can be attributed to occupational noise (Nelson et al., 2005), mostly in factory workers and military personnel. However, other population groups such as people professionally or recreationally exposed to loud music are also at risk (Helfer et al., 2010; Phillips et al., 2010). As in most forms of age-related hearing loss, outer hair cells appear most sensitive to damage by noise exposure, although other structures including inner hair cells or the vasculature can eventually be affected as well (Henderson et al., 2008). While the extent of damage is largely determined by the intensity and duration of the exposure, the existence of populations with varying sensitivity to noise-induced hearing loss (Biassoni et al., 2005) suggests the possibility of genetic modulation of susceptibility to noise trauma.
Mice have long been used as models to study the genetics of deafness (Kiernan and Steel, 2000), and are increasingly employed in studies on age-related and noise-induced hearing loss (Ohlemiller, 2006; Sha et al., 2008; Tadros et al., 2008). A number of genetic loci have been identified that modulate the rate of hearing loss or the sensitivity to noise (White et al., 2009) and several that modulate both age and noise effects (Ohlemiller, 2006), suggesting common genetic features underlying each form of pathology. A complication in interpreting most previous genetic studies, however, is their reliance on inbred mouse strains, each of which consists entirely of individuals of a single genotype, and in which all loci have been forced to homozygosity. Human populations, in contrast, include individuals with a wide range of interacting polymorphic loci, and are heterozygous at multiple loci with effects on the phenotypes of interest. In addition, many of the commonly used inbred mouse strains, such as C57BL/6 and BALB/c, harbor the Cdh23Ahl allele, which leads to an accelerated hearing loss within the first quarter of the lifespan, i.e. at stages of the lifespan earlier than those that correspond to the onset of presbycusis in humans, (Cruickshanks et al., 2010).
An animal population featuring a genetically heterogeneous background, late onset of hearing loss and a well defined range of sensitivity to environmental factors might provide a more informative model for human age-related presbycusis and noise sensitivity. Four-way cross mouse populations, originally recommended in 1981 for aging studies by a National Academy advisory panel (Institute of Laboratory Animal Resources, 1981), have previously been used for analyses of the genetics of age-related changes in bone, immune, cataract and endocrine status (Miller et al., 2003; Volkman et al., 2003; Wolf et al., 2004; Hanlon et al., 2006), and in a search for anti-aging pharmaceuticals (Harrison et al., 2009; Strong et al., 2008). In a four-way cross population, each mouse is bred from a mating between two different F1 parents, and thus carries 25% of its genome from each of four distinct inbred grandparental stocks. Thus each of the offspring mice is genetically unique, and heterozygous at many loci, but shares half of its genome with every other mouse in the test population. Four-way cross mice have the advantages of providing robustness, reproducibility, and genetic tractability (Miller et al., 1999).
The UM-HET3 four-way cross mice, used in previous aging studies (Miller et al., 2003; Volkman et al., 2003), are unsuitable for analysis of late-life hearing loss, because three of the four grandparent strains carry the Cdh23753A allele that has been associated with early-onset ARHI (Noben-Trauth et al., 2003). For the current study, therefore, we selected four parental strains, MOLF/Ei, C3H/HeJ, FVB/NJ, and 129/SvImJ, based upon auditory function and genetic criteria. Each strain retains essentially normal hearing until at least 7 months of age, as determined by analysis of auditory brain stem responses (ABR) ((Zheng et al., 1999) and Dolan, unpublished data). In addition, all four strains lack the Cdh23753A allele associated with early onset ARHI (Nichols et al., 1999; Noben-Trauth et al., 2003). Based upon population histories (Beck et al., 2000) and SNP analysis (Wiltshire et al., 2003), these four grandparental strains exhibit relatively high genetic divergence, thus facilitating identification of variants in quantitative trait loci (QTL) associated with late life auditory function. Finally, at least one of the strains (C3H/HeJ) is susceptible to age-related hearing impairment in specific genetic contexts, indicating that QTL influencing hearing loss are present in this strain (Zheng and Johnson, 2001).
We report here a set of gene mapping studies that allowed us to evaluate segregating loci for effects on ABR responses at 8, 18 and 22 months of age, and outer hair cell survival at 22 months. The protocol was designed to permit a search for age-specific and frequency-specific effects, and to discriminate alleles that modulate responses to age, noise, or both age and noise in combination.
2. Materials and methods
2.1 Mice
The tested mice, referred to as the UM-HET4 four-way cross population, were born as the progeny of a cross between female mice of the (MOLF/EiJ × 129S1/SvImJ)F1 stock and males of the (C3H/HeJ × FVB/NJ)F1 stock. Mice of each of the four grandparental inbred stocks were purchased from the Jackson Laboratory, and mated to produce the two F1 hybrid parental stocks, which were then crossed to generate UM-HET4 animals. Each mouse in the population is thus genetically unique, but shares 50% of its genetic alleles with any other mouse in the group; with respect to the nuclear genome, the animals can all be considered as full sibs. The breeding program generated 579 weanling mice, born in roughly equal monthly cohorts between July, 2006, and August, 2008. Only female mice were included in the tests. Weanlings were housed at 4 per cage and given free access to food and water. A tail tip biopsy was taken at 4 months of age to obtain DNA for gene mapping and to create fibroblast cell lines for stress analyses. A second tail skin biopsy was taken at 14 months for further evaluation of cellular traits; the results of these tests have been reported (Miller et al., 2011). Half of the mice were exposed to noise for 2 hr at 20 months of age, as described below. All mice were euthanized at 22.5 – 23 months of age, and cochleae were dissected for histopathological evaluation of hair cell status (see below). Between 526 and 556 mice had technically acceptable genotype and ABR results at 8 months (depending on locus and test frequency). For ABR tests at 18 months, between 476 and 501 mice were available for genotype-ABR association tests. At 22 months of age, data were available for 182–191 noise-exposed mice, and 254–256 mice that had not been deliberately exposed to noise. The colony was tested quarterly for evidence of infection as previously described (Miller et al., 2007); all such tests were negative throughout these experiments, and the mice are thus considered specific-pathogen free. The experimental protocol was approved by the University Committee on the Use and Care of Animals at the University of Michigan.
2.2 Acoustic brainstem responses (ABR)
Animals were anesthetized (ketamine 65 mg/kg, xylazine 3.5 mg/kg, and acepromazine 2 mg/kg) and placed on a hot water heating pad so that normal rectal temperature could be maintained. Additional anesthetic (1/2 dose ketamine and xylazine) was administered if needed to maintain anesthesia depth sufficient to insure immobilization and relaxation. ABRs were recorded in an electrically and acoustically shielded chamber (CA Tegner, Sweden). Needle electrodes (Grass F-E2M-48) were placed subcutaneously at vertex (active) and the base of the test ear's pinna (reference) and at the base of the contralateral pinna (ground). Tucker Davis Technologies (TDT) System III hardware and SigGen/BioSig software (TDT, Alachua, FL USA) were used to present the stimulus and record responses. Tones were delivered monaurally (left ear) to an EC1 earphone mated to a customized vinyl speculum tube inserted into the ear canal. Sound calibration was performed with a 1/8 in. condenser microphone in a volume approximating the external ear canal. Threshold is expressed as dB SPL. Threshold was determined by following the largest wave, typically wave III. Thresholds were visually interpolated between the lowest stimulus level where a response was observed, and 5 dB lower, where no response was observed. Stimulus presentation was 15 ms tone bursts, with a 1 ms cosine-shaped rise-fall time, presented 10 per second. The voltage from the electrodes was filtered (0.3–3 kHz), and amplified (× 100 K). The ABR response was measured at frequencies of 4, 12, 24, and 48 kHz. Up to 1024 responses were averaged for each stimulus level. Responses were collected for stimulus levels in 10 dB steps at higher stimulus levels, with additional 5 dB steps near threshold.
2.3 Noise exposure
Half of the mice were noise exposed at 20 months of age, two months before the 22 month ABR acquisition point. The system was initially calibrated using a white noise source (General Raio 1381) emitting sound between 62.5 Hz and 22 kHz, as measured with a 1/2-inch microphone (B&K 4134) and fast Fourier transform (FFT) spectrum analyzer (Stanford Research SR760). The spectrum data were then used to create an equalizing FFT filter in an audio file editor (Adobe Audition), which was in turn used to create an audio CD for the noise-exposure treatment. Mice to be treated were placed, while awake, into a ventilated chamber in individual wire cages. The noise (2 – 20 kHz, 110 dB SPL for 2 hr) was then presented though a loudspeaker mounted on the top of the chamber. The level of the noise was measured at the cage position using a B&K Sound level meter (model 2231, with type 4155 1/2” microphone, and type 1625 octave band filter).
2.4 Cytocochleogram (CCG) analysis
The left cochlea, from all mice, were rapidly removed following termination and cochleae received intrascalar fixation with 4% paraformaldehyde in phosphate buffer. Following a partial decalcification for 24 h in 5% EDTA the boney shell of the otic capsule was removed. Cochleae were then placed in 1% phalloidin with an Alexa 305 label for 1 hour followed by rinse. The cochlear spiral was then removed as segments and placed on glass slides for assessment under epifluorescent optics. A quantitative assessment of the three rows of outer hair cells and the inner hairs was then carried out using a 50× objective and a 0.19 mm reticule in the microscope eyepiece. The assessment started at the apex of the cochlear spiral and continued all along the length of the cochlea (average length was 5.93 mm) moving to different segments as necessary. The number of hair cells present or absent within the reticule was noted, the reticule was moved towards the basal pole, and the assessment repeated until the entire length of the cochlea had been examined. Data were then summed to provide five indices for each mouse: the number of missing inner hair cells (IHC), and the number of missing outer hair cells (OHC) in each of four equally sized regions, i.e. apex, mid-1, mid-2, and base. The missing OHC estimates were derived as an average over each of the three OHC rows in each of the four regional segments.
2.5 Genotyping and gene mapping
Genomic DNAs were prepared from 1-cm sections of tail obtained at 3 months of age. Genomic DNA for each animal was genotyped at single-nucleotide polymorphisms known to exist between the grandparental inbred strains using fluorescent-labeled ligation-detection reactions, followed by capillary gel electrophoresis. In total, 128 biallelic informative loci were examined across the genome, with 71 loci informative for the maternal-derived chromosomes and 53 loci informative for paternal-derived chromosomes. Each maternal- or paternal-derived chromosome was assessed for a minimum of two SNP polymorphic loci. A single-point, genome-wide search was performed for each trait to detect loci associated with the trait. One-way ANOVA models, with trait as the dependent variable and biallelic marker as the factor with two levels,were used to perform genome-wide searches for all biallelic markers. The strength of associations between genetic markers and each trait was evaluated using a permutation-based test of statistical significance. This test generates “experiment-wise” p-values to compensate for multiple locus-by-trait comparisons. The phenotype and genotype data were permuted 1000 times to generate a null distribution, and the resulting distribution compared to the actual, observed F statistic to provide the experimentwise p-value. An experimentwise p ≤ 0.05 was the chosen criterion for inferring that a specific marker has a statistically significant linkage to an effector locus of interest. The percent of variance in each trait that can be explained by genetic effects was estimated in a standard way from corresponding regression models. All tests were performed using SAS software (v9-1, SAS Institute Inc., Cary, NC).
3. Results
3.1 QTL for hearing tested at 8 months of age
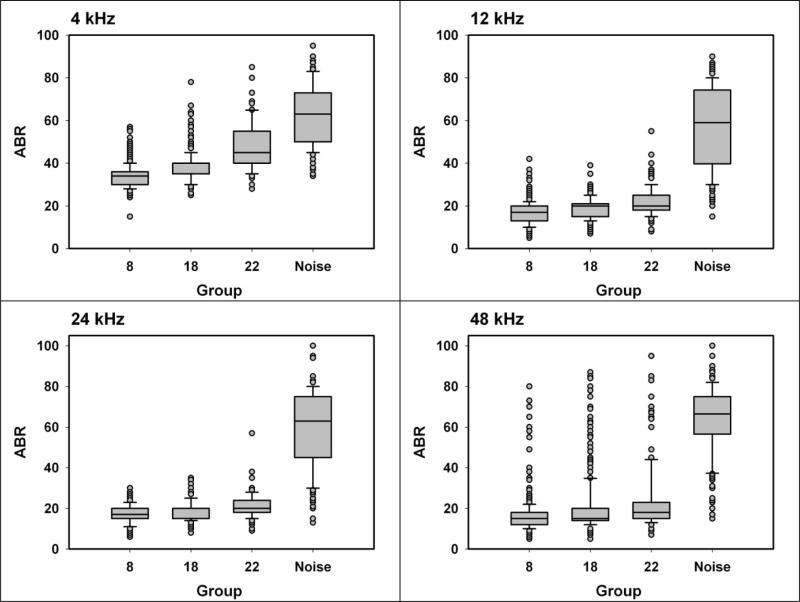
A population of genetically heterogeneous mice was produced using a cross between (MOLF/Ei × 129S1/SvImJ)F1 females and (C3H/HeJ × FVB/NJ)F1 males. All progeny in the population can be considered full sibs of one another, each sharing a random 50% of its genome. Each of the four grandparental stocks was selected because it does not carry the Cdh23Ahl allele, which leads to an accelerated hearing loss within the first quarter of the lifespan, i.e. at stages of the lifespan earlier than those which are typical for onset of presbycusis in humans. Mice were genotyped at loci that discriminate among the two maternal alleles (MOLF and 129) or between the two paternal alleles (C3H and FVB), and the genotypes compared to results of ABR tests conducted at 8, 18, or 22 months of age. Half of the mice were exposed, at 20 months, to 2 hr of noise at 110 dB SPL. >Figure 1 presents box plots showing the distribution of ABR test results at each combination of age, frequency, and noise-exposure for the mice in this study.
1.
Distribution of ABR test results at ages 8, 18, and 22 months, and in 22 month old noise-exposed UM-HET4 mice, evaluated at frequencies of 4, 12, 24, and 48 kHz. Box boundaries indicate 25th and 75th percentiles, with the median indicated by the line in the box center. Whiskers indicate 10th and 90th percentiles, and circles indicate individual mice whose scores were outside the 10 – 90th percentile range.
Table 1 (top) lists polymorphisms that achieved experiment-wise statistical significance for ABR values measured at 8 months of age, i.e. in young adult mice. Chromosome 3 carries at least two QTL, one near the proximal end for which the MOLF allele is associated with higher ABR values, and a more distal QTL in which higher ABR values are associated with the 129 allele. Chromosome 13 carries a maternal 129 allele associated with high ABR at 48 kHz, and a paternal FVB allele associated with high ABR at 12 kHz. We cannot exclude the possibility that these two markers cosegregate with the same effector gene, potentially polymorphic in each of the two parental F1s. Each allele accounts from 2% – 4% of the total phenotypic variance.
Table 1.
QTL with significant effects on ABR values at 8 or at 18 months.
| Age (months) | Frequency kHz | Positiona | Pct Expb | Higher ABR in: | p(e)c |
|---|---|---|---|---|---|
| 8 | 48 | 3·55 | 3.5 | MOLF | 0.000 |
| 8 | 48 | 3·61 | 2.3 | 129 | 0.034 |
| 8 | 48 | 3·75 | 3.4 | 129 | 0.000 |
| 8 | 48 | 3·115 | 2.2 | 129 | 0.027 |
| 8 | 48 | 3·122 | 2.5 | 129 | 0.007 |
| 8 | 48 | 13·52 | 3.0 | 129 | 0.002 |
| 8 | 12 | 13·84 | 2.3 | FVB | 0.043 |
| 18 | 48 | 2·166 | 2.7 | MOLF | 0.016 |
| 18 | 48 | 3·55 | 4.0 | MOLF | 0.000 |
| 18 | 48 | 3·75 | 3.2 | 129 | 0.005 |
| 18 | 24 | 7·36 | 3.0 | 129 | 0.014 |
| 18 | 48 | 10·44 | 6.5 | 129 | 0.000 |
| 18 | 48 | 10·93 | 2.6 | 129 | 0.026 |
| 18 | 48 | 15·25 | 2.5 | FVB | 0.039 |
| 18 | 48 | 15·36 | 2.7 | 129 | 0.016 |
| 18 | 48 | 15·78 | 2.4 | 129 | 0.047 |
Position is given as [chromosome] • [position in megabase pairs]
“Pct Exp” is the percentage of phenotypic variance attributable to the indicated allele.
p(e) is experiment-wise p-value
Numbers of mice: 526 – 556 for tests at 8 months; 476 – 501 for tests at 18 months Colored bands indicate the most parsimonious genetic model for ABR results at the age indicated, based on the distinctions of chromosome and grandparental allele.
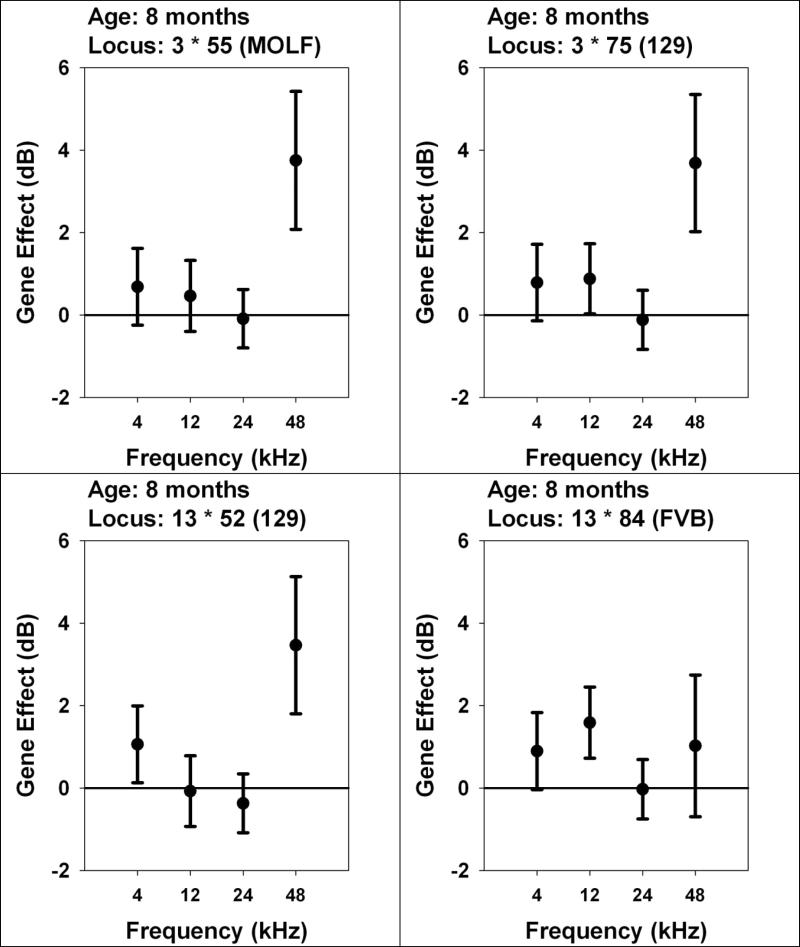
Using post-hoc analysis, we tested for possible genetic effects on ABR across the whole spectrum of measured frequencies in mice discriminated on the basis of a significant QTL at a specific frequency (Figure 2). Each of the two loci on chromosome 3 appears to affect high frequency hearing preferentially, although the 129 allele at 3•75 is associated with significantly poorer hearing at 12 kHz as well as at 48 kHz. The 129 allele at 13•52 has significant effects on hearing at both 48 kHz and 4 kHz, with larger effects at the higher frequency. The FVB allele at 13•84 has a significant effect only at 12 kHz, although there are suggestions of similar effects at both higher and lower test frequencies.
2.
Are QTL effects frequency-specific? Effects of QTL, detected at 8 months, on ABR thresholds at same age tested at multiple frequencies. Each panel shows one locus selected because of a significant effect on ABR tested at 8 months of age. Each symbol shows an effect size for the indicated frequency (4, 12, 24, or 48 kHz), calculated as mean ABR in mice with the indicated allele (MOLF, 129, or FVB) minus mean ABR for mice with the opposite allele, in dB. The error bars show the 95% confidence interval for the estimated effect. Entries that do not cross the horizontal line at zero are significant, at two-tailed p < 0.05, without correction for multiple comparisons.
3.2 QTL for hearing tested at 18 months of age
We identified several alleles that attained experiment-wise significance for mice tested at 18 months of age (bottom portion of Table 1). Hearing acuity is modulated, in this stock at this age, by loci on maternal chromosomes 2, 3, 7, 10, 15, and on paternal chromosome 15. Maternal chromosome 3 appears to contain at least two relevant polymorphisms, one in which the MOLF allele is associated with high ABR, and one in which the 129 allele co-segregates with high ABR values. Each of the detected alleles accounts for between 2% and 7% of the phenotypic variance.
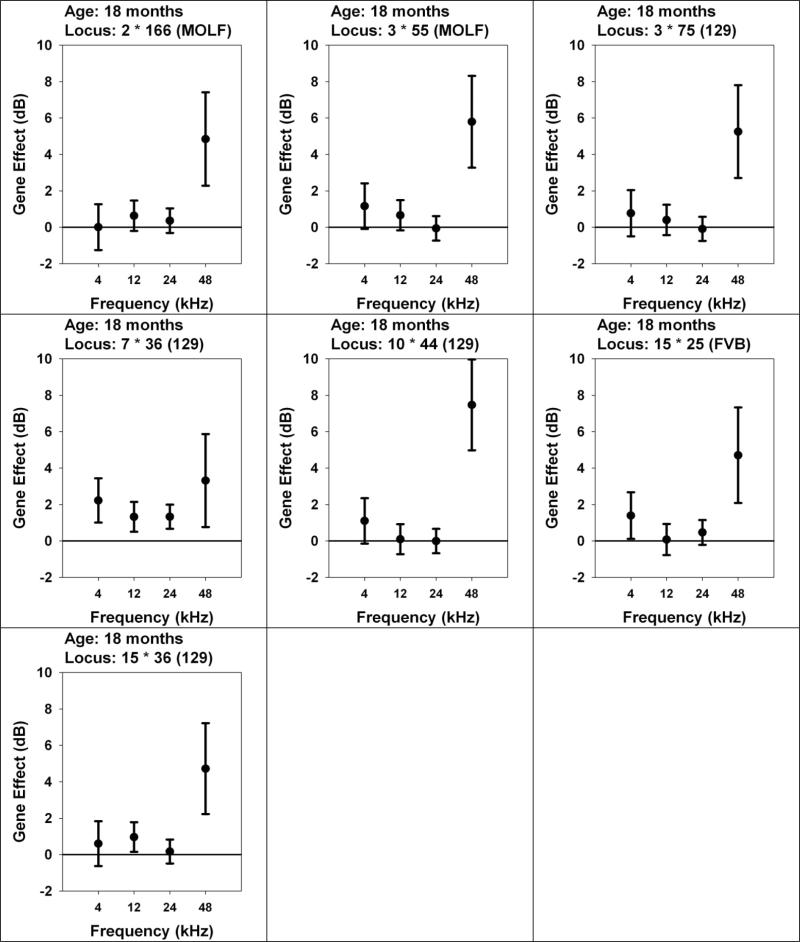
Post-hoc analyses indicated that several of the alleles (at 2•166, 3•55, 3•75, and 10•44) appear to affect hearing only at the 48 kHz test frequency (Figure 3). In contrast, the 129 allele at 7•36, detected by significance of its association with hearing at 24 kHz, also discriminates among mice for hearing tested at 4 kHz, 12 kHz and 48 kHz. This appears to be a polymorphism whose effect on age-dependent hearing loss is thus apparently not frequency-specific. Each of the alleles on chromosome 15 (FVB paternal allele at 15•25 and 129 maternal allele at 15•36) has its strongest effect on hearing at 48 kHz, but also a significant effect on ABR at either 4 kHz or 12 kHz.
3.
Are QTL effects frequency-specific? Effects of QTL, detected at18 months, on ABR thresholds at same age tested at multiple frequencies. Each panel shows a locus selected because of a significant effect on ABR tested at 18 months of age. Each symbol shows an effect size for the indicated frequency (4, 12, 24, or 48 kHz), calculated as mean ABR in mice with the indicated allele (MOLF, 129, or FVB) minus mean ABR for mice with the opposite allele, in dB. The error bars show the 95% confidence interval for the estimated effect. Entries that do not cross the horizontal line at zero are significant, at two-tailed p < 0.05, without correction for multiple comparisons.
3.3 QTL for hearing tested at 22 months of age, with or without noise exposure
We identified several QTL with significant association with hearing function at 22 months of age (Table 2). QTL were evaluated in three subsets of the mice: (a) those not exposed to noise, (b) those exposed to noise at 20 months of age, and (c) the pooled population, with noise exposure used as a covariate in the calculation. In two cases (loci at 10•44 and 10•93) we noted significant associations both in the combined population and in one or two of the subsets, and Table 2 indicates these relationships separately. Hearing in the absence of deliberate noise exposure is influenced by at least two QTL, on chromosomes 10 and 11. Hearing in mice exposed to prior noise is influenced by at least 3 QTL, on chromosomes 9, 10, and 12.
Table 2.
QTL with significant effects on ABR tested at 22 months
| Test Groupa | Frequency kHz | Positionb | Pct Expc | Higher ABR in | p (e)d |
|---|---|---|---|---|---|
| Noise-Rx | 4 | 9·60 | 7.0 | MOLF | 0.038 |
| Combined | 48 | 10·44 | 3.7 | 129 | 0.000 |
| No Noise | 48 | 10·44 | 8.4 | 129 | 0.001 |
| Noise-Rx | 48 | 10·44 | 9.1 | 129 | 0.004 |
| Combined | 12 | 10·93 | 1.3 | 129 | 0.010 |
| Noise-Rx | 12 | 10·93 | 7.7 | 129 | 0.011 |
| No Noise | 12 | 11·107 | 4.9 | 129 | 0.031 |
| Noise-Rx | 4 | 12·17 | 7.3 | 129 | 0.031 |
| Noise-Rx | 4 | 12·40 | 7.3 | 129 | 0.030 |
Test group: Noise-Rx - mice exposed to 2 hr noise at 20 months of age. No Noise: mice not exposed to Noise stress at 20 months of age. Combined: both groups pooled, with noise exposure treated as a covariate in estimation of QTL effect
Position is given as [chromosome] • [position in megabase pairs]
“Pct Exp” is the percentage of phenotypic variance attributable to the indicated allele.
p(e) is experiment-wise p-value
Numbers of mice: 182 – 191 for Noise-Rx; 254 – 256 for No Noise; 438 – 447 for Combined groups.
Colored bands indicate the most parsimonious genetic model for ABR results at 22 months, based on the distinctions of chromosome and grandparental allele. Separate rows are included for Noise-Rx and No Noise groups only when p(e) < 0.05 for the analysis in the indicated subset of mice.
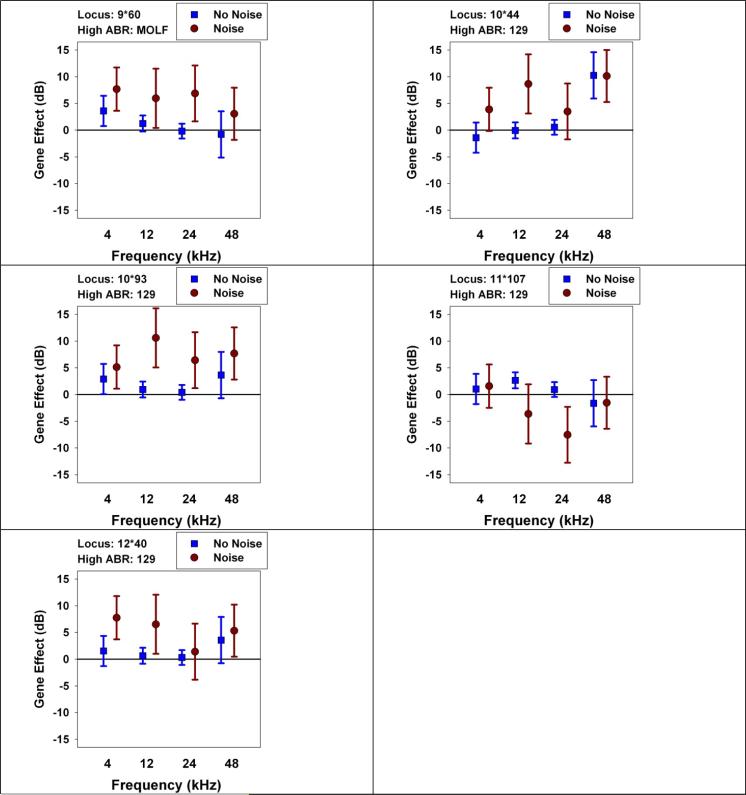
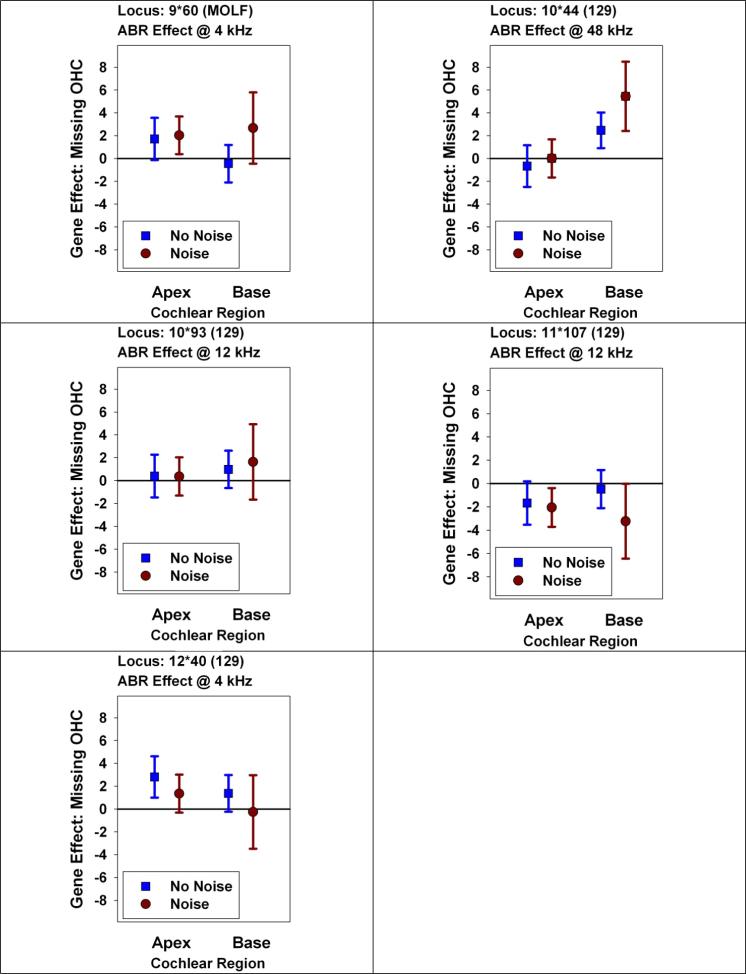
To dissect this set of gene-trait associations further, we looked at the ability of each of these QTL to predict hearing function, at each frequency, in 22-month old mice with or without prior noise exposure (Figure 4). Three of the polymorphisms (MOLF allele at 9•60, 129 allele at 10•93, and 129 allele at 12•40) have similar patterns of effects: they are associated with significant hearing loss in the noise-exposed mice at three or four of the test frequencies, but smaller (or absent) effects on hearing in mice not exposed to noise. There is a significant association of the MOLF allele at 9•60 with poor hearing at 4 kHz in the “no noise” group, but the effect is smaller than that seen for noise-exposed mice. The pattern for the 129 allele at 10•44 is similar to that seen for 10•93, and these markers may reflect the same (unknown) effector locus, although the association of 10•44 with hearing loss at 48 kHz is strong enough to reach statistical significance even in mice not exposed to noise.
4.
Do QTLs with effects on hearing at 22 months affect mice with and without noise exposure? Each panel shows a locus detected by a significant effect on ABR tested at 22 months of age (see Table 2), either in noise-exposed mice, mice not exposed to noise, or in both groups pooled. Each symbol shows the effect size, in dB, at 22 months of age, at the indicated frequency, either in the No Noise group or in the Noise Rx group. The allele shown (MOLF or 129) indicates the allele associated with higher ABR values at the frequency that achieved experiment-wise significance (see Table 2). The error bars show the 95% confidence interval for the estimated effect. Entries that do not cross the horizontal line at zero are significant, at two-tailed p < 0.05, without correction for multiple comparisons. Evaluation of the QTL at 12•17 (not shown) closely resembles that shown for 12•40.
The 129 allele at 11•107 has more complex effects. This polymorphism was detected because it led to a small, but significant, increase in ABR values at 12 kHz in mice not exposed to noise. The post-hoc evaluation showed, unexpectedly, that this allele is associated with significantly lower ABR values, at 24 kHz, in noise-exposed mice. The latter association was not detected using experiment-wise criteria, and may represent a chance fluctuation.
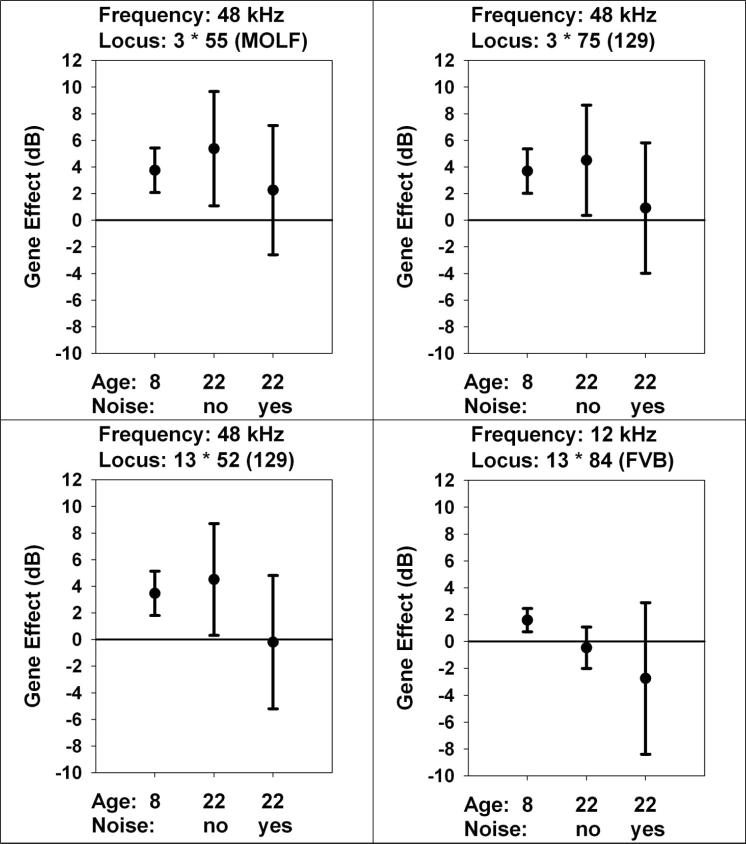
We evaluated alleles that were initially associated with effects on hearing loss at 8 months of age for similar effects at 22 months of age, either with or without prior exposure to 2 hr of noise at 20 months of age (Figure 5). Three of the four loci have effects, in the 22 month old mice that had not been exposed to noise, that are statistically significant and as large at 22 months as they were when tested at 8 months of age. None of these three QTL has a statistically significant effect on hearing in 22 month old mice of the noise-exposed group. Hearing in the noise-exposed mice is worse (higher ABR values) in mice carrying the FVB allele at 13•84, but this effect is not statistically significant, and there is no apparent effect of this allele in mice not exposed to noise.
5.
Are QTL effects seen at 8 months also present in 22 month old mice? Each panel shows a locus selected because of a significant effect on ABR tested at 8 months of age. Each symbol shows the effect size, in dB, either at 8 months (repeated from Figure 2), or at 22 months either in mice not exposed to noise, or in mice exposed to noise for 2 hr at age 20 month. The allele (MOLF, 129, or FVB) reflects the variant associated with higher ABR responses at 8 months of age. The frequency for ABR testing is also indicated in each panel. The error bars show the 95% confidence interval for the estimated effect. Entries that do not cross the horizontal line at zero are significant, at two-tailed p < 0.05, without correction for multiple comparisons.
3.4 QTL that influence survival of outer hair cells
At euthanasia, one cochlea from each mouse was evaluated histologically to count the number of missing IHC and OHC in each of four equally sized regions: apex, upper-middle, lower-middle, and base. The numbers of missing IHC were low at this age, and the analysis thus focused on OHC count. The genetic influence on OHC loss was evaluated in two ways. First, QTL analyses were conducted using OHC in each of the cochlear regions as the outcome variables. These analyses were conducted on three subsets of the mice: once in mice exposed to noise, once in mice not exposed to noise, and once in the combined population using noise exposure as a covariate. Results are shown in Table 3. Using an experiment-wise significance criterion (p < 0.05), we noted QTL with an effect on basal OHC on chromosome 10, and QTL with effects on apical OHC loss on chromosomes 11, 12, 17, and 19. The QTL on chromosomes 10 and 19 reached significance in the noise-exposed group, and each listed QTL was significant in the combined population, but no QTL was significant in the mice not exposed to noise. Each of these loci accounted for 5% to 7% of the OHC variance in the combined population, and for up to 15% of the variance in the noise-exposed group. A post-hoc calculation (not shown) was performed to see if variation at any of these loci affected ABR results at 22 months of age. The 129 allele at 10•44 was associated with a 10 dB hearing decrement at 48 kHz, both in noise-exposed and no-noise mice, significant in each case by t-test at p < 0.001. It thus seems plausible that the high frequency hearing loss associated with this allele may reflect age- and noise-related loss of OHC. The 129 allele at 12•117 was associated with hearing deficits (2 to 6 dB) at each test frequency, but only in noise-exposed mice; only the 6 dB loss at 12 kHz reached statistical significance (p = 0.04).
Table 3.
QTL with effects on the number of missing outer hair cells (OHC) tested at 22 months of age
| Test Groupa | Region | Positionb | Pct Expc | Cells Missing in: | p(e) |
|---|---|---|---|---|---|
| Combined | Base | 10*3 | 6.7 | 129 | 0.000 |
| Noise Rx | Base | 10*3 | 14.5 | 129 | 0.005 |
| Combined | Base | 10*44 | 6.2 | 129 | 0.000 |
| Combined | Apex | 11*72 | 6.4 | MOLF | 0.000 |
| Combined | Apex | 12*117 | 6.7 | 129 | 0.000 |
| Combined | Apex | 17*42 | 5.6 | 129 | 0.005 |
| Combined | Apex | 19*12 | 5.6 | 129 | 0.002 |
| Noise Rx | Apex | 19*12 | 9.2 | 129 | 0.012 |
| Combined | Apex | 19*17 | 4.8 | 129 | 0.013 |
| Noise Rx | Apex | 19*17 | 10.8 | 129 | 0.004 |
Test group: Noise-Rx - mice exposed to 2 hr noise at 20 months of age. Combined: both groups pooled, with noise exposure treated as a covariate in estimation of QTL effect.
Position is given as [chromosome] • [position in megabase pairs]
“Pct Exp” is the percentage of phenotypic variance attributable to the indicated allele.
dp(e) is experiment-wise p-value Numbers of mice: 122 – 164 for Noise-Rx; 253 – 322 for Combined groups.
Colored bands indicate the most parsimonious genetic model for OHC pathology at 22 months, based on the distinctions of chromosome and grandparental allele. Separate rows are included for Noise-Rx mice only when p(e) < 0.05 for the analysis in that subset of mice.
The second analysis took the converse approach: we evaluated the loci detected by their effects on ABR at 22 months of age (illustrated in Table 2 and Figure 5), to see if the corresponding alleles were associated with differences in OHC loss. The results are shown in Figure 6.
6.
6. Do loci that affect hearing at 22 months also have an effect on OHC loss? Each panel shows a locus detected by a significant effect on ABR tested at 22 months of age (see Table 2), either in noise-exposed mice, mice not exposed to noise, or both groups pooled. Each symbol shows differences in missing OHC count, at cochlear apex or base, associated with the “high ABR” allele, with mice in the Noise treatment a “no noise” groups evaluated separately. The error bars show the 95% confidence interval for the estimated effect. Entries that do not cross the horizontal line at zero are significant, at two-tailed p < 0.05, without correction for multiple comparisons. Numbers of mice: 130–153 for No Noise group, and 118 – 167 for Noise group. The allele and test frequency named in each panel refer to experiment-wise significance as shown in Table 2. The arrangement of loci is the same as that used in Figure 5.
The 129 allele at 10•44, detected through its significant effect on ABR at 48 kHz, was associated with higher numbers of missing basal OHC in both noise-exposed and unexposed mice. Interestingly, there was no apparent effect on apical OHC. This allele is also associated with poor hearing at 18 months of age, i.e. prior to any deliberate noise exposure.
Two alleles, i.e. the MOLF allele at 9•60, and the 129 allele at 12•40, were each detected because of effects on hearing at 4 kHz. Each of these is associated with loss of apical OHC, though the effects are significant for 9•60 only in noise-exposed mice and for 12•40 only for mice not exposed to noise. Effects on OHC loss at the base of the cochlea are not significant, but consistent with the hypothesis that 9•60 may have preferential effects after noise exposure.
The 129 allele at 10•93 and the 129 allele at 11•107 were each detected through effects at 12 kHz. The former allele does not produce any significant effects on basal or apical OHC counts. Unexpectedly, the allele at 11•107 is associated with fewer missing OHCs, in both apical and basal regions of the cochlea, and regardless of noise exposure; the effects are significant only in mice exposed to noise.
Table 4 provides a summary showing each of the 16 distinct QTL alleles detected by the work described above. This list represents the mimimum number of polymorphic alleles needed to account for the gene/trait associations seen in our study. It is possible that one or more of the marker loci are linked to multiple loci with effects on hearing or OHC survival; further work using additional animals and a higher density set of markers would be needed to evaluate this idea. Conversely, it is possible that pairs of alleles linked to markers segregating separately in maternal and paternal meioses might reflect a single underlying effector locus; the 129 allele at 13 • 52 and the FVB allele at 13 • 84, for example, may each be linked to a locus with effects on hearing at 8 months of age, and similar considerations apply to markers on chromosomes 3, 11 and 15.
Table 4.
Summary of QTL described in this paper
| Index | Marker Loci | Allele | 8 months | 18 months | 22 months | 22 months, noise | Missing OHC |
|---|---|---|---|---|---|---|---|
| 1 | 3 • 55 | MOLF | 48 | 48 | 48 | ||
| 2 | 3 • 61, 75, 115, 122 | 129 | 12, 48 | 48 | 48 | ||
| 3 | 13 • 52 | 129 | 4, 48 | 48 | |||
| 4 | 13 • 84 | FVB | 12 | ||||
| 5 | 2 • 166 | MOLF | 48 | ||||
| 6 | 7 • 36 | 129 | 4, 24, 24, 48 | ||||
| 7 | 10 • 44, 93 | 129 | 48 | 4, 48 | 4,12, 24, 48 | Base (10 • 3, 44) | |
| 8 | 15 • 25 | FVB | 4, 48 | ||||
| 9 | 15 • 36, 78 | 129 | 12, 48 | ||||
| 10 | 9 • 60 | MOLF | 4 | 4, 12, 24 | |||
| 11 | 11 • 107 | 129 | 12 | 24 (opposite direction) | |||
| 12 | 12 • 17, 40 | 129 | 4, 12, 48 | ||||
| 13 | 11 • 72 | MOLF | Apex | ||||
| 14 | 12 • 117 | 129 | Apex | ||||
| 15 | 17 • 42 | 129 | Apex | ||||
| 16 | 19 • 12, 17 | 129 | Apex |
Boldface denotes experiment-wise significance at p < 0.05. Order of marker loci reflects the order in which they appear in Tables 1 – 3. Entries shown without boldface reflect significance in post-hoc secondary analyses, based on data shown in Figures 2 – 5. The Allele column shows the allele associated with high ABR thresholds or with increased number of missing OHC. When two marker loci are listed on the same row, separated by commas, this indicates that there is insufficient evidence to rule out the hypothesis that the chromosomal region contains more than a single locus with effect on the traits evaluated.
3.5 Additive effects of QTL in combination
There are several cases in which hearing at a specific age and frequency is influenced by alleles at three or more loci, as listed in Tables 1 and 2. To see how large a difference in ABR thresholds could be produced by effects of multiple alleles in combination, we calculated the mean ABR thresholds in subsets of mice that had inherited the “high ABR” alleles at all 3, 4 or 5 relevant loci, and compared this to the ABR threshold in the subset of mice that had inherited the opposite allele at each locus. The results are shown in Table 5. When tested at 8 months of age for hearing at 48 kHz, mice with three “high ABR” alleles had an impairment of 8 dB compared to sibs with the opposite allele combination. A combination of 5 alleles with effects at 18 months of age can produce a 22 dB deficit, and combinations of 3 or 4 alleles can lead to deficits of 15 – 23 dB in noise-exposed mice tested at various frequencies at 22 months of age.
Table 5.
Effect on ABR of allele combinations.
| Age | Noise? | Frequency | Loci (Alleles)a | Combined effectb | Number of micec |
|---|---|---|---|---|---|
| 8 | N/A | 48 | 3·55 (MOLF), 3·75 (129), 13·52 (129) | 8 dB | 112, 111 |
| 18 | N/A | 48 | 2·166 (MOLF), 3·55 (MOLF), 3·75 (129), 10·44 (129), 10·93 (129) | 22 dB | 38, 24 |
| 22 | yes | 4 | 9·60 (MOLF), 10·93 (129), 12·40 (129) | 23 dB | 22, 24 |
| 22 | yes | 12 | 9·60 (MOLF), 10·44 (129), 10·93 (129), 12·40 (129) | 22 dB | 16, 20 |
| 22 | yes | 48 | 10·44 (129), 10·93 (129), 12·40 (129) | 15 dB | 35, 41 |
Loci (with alleles in parentheses) that reach experiment-wise significance in the QTL analysis for ABR results at the indicated age and test frequency. The alleles shown are those which are associated with higher ABR thresholds.
Difference, in dB, between mice with the indicated combination of alleles, and mice with the opposite alleles at all of the loci listed.
The number of mice with the indicated allele combination, followed by the number of mice with the corresponding set of opposite alleles.
4. Discussion
A new model for age-related hearing loss in mice
The UM-HET4 four-way cross mice that are the topic of this report present a promising model for analysis of the interaction between genetic and non-genetic factors that predispose to age-related and noise-induced hearing loss. In contrast to other frequently used models, e.g. C57BL/6, auditory threshold in these animals remain robust throughout the first half of the lifespan, and loss of hearing occurs late in life, as it typically does in humans. In this respect UM-HET4 mice resemble CBA/J (Sha et al., 2008), but provide genetic heterogeneity. The effects of noise exposure, likewise, show a variability that appears to reflect a range of sensitivities similar to those observed in a human population.
Auditory thresholds among UM-HET4 mice are stable until at least 8 months of age. There are small, but significant declines in mean ABR results at each frequency by 18 months. At 22 months, mean threshold shifts are still relatively small, averaging less than 10 dB at 12, 24 and 48 kHz, and slightly over 10 dB at 4 kHz, but there is a great deal of potentially informative variation among the mice, with some individuals showing no effects of age, and others with clear manifestations of hearing loss. Such a distribution is similar to that seen in human populations, in which some individuals with excellent hearing can be found among the aging population, particularly among people who have not been exposed to chronic noise. In the UM-HET4 mice, a single episode of moderate noise exposure at 20 months induces a significant elevation in thresholds at all frequencies at 22 months and also induces a wider spread of individual thresholds than seen in the non-exposed cohort. The wide spread in ABR performance among the noise-exposed animals, some of which show essentially no noise effect, with others showing severe hearing loss of 80 dB or more compared to the mean of non-exposed, age-matched mice, again resembles the human conditions of “tough” and “tender” ears (Cody and Robertson, 1983; Maison and Liberman, 2000). This variation in ABR responses, with or without noise exposure, provided an opportunity to evaluate genetic influences on age-dependent hearing loss and on the interaction of age and noise trauma. Differences among the effects of the individual QTL suggest that several types of presbycusis emerge among the mice. For example, there is a preponderance of both low-frequency and high-frequency hearing loss which is well correlated in most but not all animals thus potentially representing at least three forms (low-frequency, high-frequency and combined low- and high-frequency) of hearing loss. While a relatively robust correlation in a large segment of animals points to loss of hair cells as a cause of threshold shifts, another subgroup lacks such a direct link, further extending the complexity of the observed presbycusis. Interestingly, there is very little change at the mid-frequencies of 12 and 24 kHz. This suggests a virtual absence of `metabolic' or `strial' presbycusis which would lead to a hearing loss affecting all frequencies.
Polymorphisms in UM-HET4 mice with effects on adult and late-life hearing
The genetic results presented here, as summarized in Table 4, provide evidence for alleles – and therefore pathophysiological pathways – that can affect specific frequencies, or mice of specific ages, or mice with specific histories of noise-induced injury. Even in young adult mice, i.e. mice 8 months of age, roughly equivalent to one third of the expected median lifespan, hearing at high frequency is modulated by at least four segregating alleles, two on chromosome 3 and two others on chromosome 13 (Figure 2, Table 4). Two of these markers also have significant associations with hearing at 4 or 12 kHz, although the current data cannot rule out the possibility that the marker loci may be linked to distinct frequency-specific effector loci. The loci on chromosomes 3 and 13 that modulate the response to high frequency stimulation at 8 months of age have similar effects at 22 months of age in those mice not exposed to noise (Figure 5), although there is no apparent effect in mice that had been exposed to noise trauma at age 20 months, suggesting that the pathways modulated by these three loci may be age-sensitive but not noise-sensitive. The allele at 13•84, with a significant effect on responses at 12 kHz in young mice (Figure 2), does not have a significant association with hearing at 22 months of age, regardless of prior noise exposure (Figure 5).
Evaluation of mice at 18 months of age, about 75% of the expected median lifespan, revealed significant effects of at least 7 distinct alleles, on chromosomes 2, 3, 7, 10 and 15 (see Table 4), of which only the alleles on chromosome 3 had been detected in tests of 8 month old mice. The current data do not allow an inference as to whether the two associations with alleles on chromosome 15, one from FVB and the other from 129, reflect variation at the same effector locus. Each of these seven alleles has a greater effect on ABR at 48 kHz than on responses to lower frequencies. Three of the alleles have significant post-hoc associations with hearing at lower frequencies as well, but in general genetic variation in this population seems to have a preferential effect on high frequency responses in these older mice.
Because half of the mice were exposed to noise at 20 months of age, and because 8% of the mice died between 18 and 22 months, QTL scans done at 22 months involved smaller numbers of tested mice, and these tests therefore lose a good deal of statistical power. Alleles on chromosomes 10 and 11 were found to have significant effects in mice not exposed to noise (Table 2), and alleles on chromosomes 9, 10, and 12 were found to have effects in mice that had been noise exposed. Post-hoc analyses (Fig. 4) showed several different patterns of effect in this group of five QTL. Locus 9•60 seems to modulate responses at 4 kHz regardless of deliberate noise exposure, and similarly locus 10•44 has parallel, significant effects on hearing at 48 kHz in both aged subpopulations. The locus linked to 10•93 has effects on hearing at all test frequencies, but only in noise-damaged mice (there is a smaller, but significant, effect at 4 kHz in the mice not exposed to noise.) A similar pattern, with stronger effects after noise exposure, is also seen for 12•40. The QTL linked to 11•107, detected by its effect at 12 kHz in mice not exposed to noise, is also associated, unexpectedly, with a significant improvement in hearing at 24 kHz in those mice that had been noise-exposed. A subset of UM-HET4 mice also exhibited threshold increases at 22 months of age that are associated with 129 alleles linked to markers at 10•44 and 10• 93. These associations were observed in mice that were exposed to noise at 20 months of age and in unexposed mice, suggesting QTLs on chromosome 10 segregating in the UM-HET4 cross that affect pathways important for general maintenance of hearing in late life and also responses to noise trauma.
Interactions among loci
Several types of genetic interactions have been described between variants associated with age related hearing loss in the mouse. These include digenic influences that affect the extent or timing of hearing loss such as those observed in crosses involving NOD/LtJ strain mice, which are susceptible to early onset hearing loss that progresses with age (Johnson and Zheng, 2002). Threshold elevations were found to depend upon inheritance of susceptibility alleles at the Cdh23 locus, while the early onset nature of the shifts required co-inheritance of homozygous NOD strain-derived alleles at the Ahl2 locus (Johnson and Zheng, 2002). The Ahl2 locus alone exhibited no significant effect on hearing function. In contrast, the effects on later life hearing demonstrated in UM-HET4 mice that carry multiple QTL variant alleles (see Table 5) resemble the additive effects found in crosses involving BUB/BnJ strain mice, which also exhibit early onset hearing loss (Zheng et al., 1999). Hearing function in this strain is influenced strongly by variants at two loci, each of which confer significant individual effects without evidence of epistasis (Johnson et al., 2005). Early hearing loss is linked to variants at a QTL in the Gpr98 gene, while the Cdh23Ahl allele confers additional susceptibility to hearing loss in later life.
Genetic modulation of OHC loss
Some of the loci that lead to functional deficits may be working through pathways that regulate survival of cochlear hair cells. Loss of hair cells is a consistent feature in sensorineural presbycusis and noise trauma. Outer hair cells are considered more sensitive to trauma than inner hair cells (Henderson et al., 2008), and the findings presented here largely follow this pattern. The loss of sensory cells is generally held responsible for the accompanying threshold shifts since the two parameters broadly correlate in most species. Spongr et al. (Spongr et al., 1997) claimed a quantitative correlation between patterns of hair cell loss and threshold shifts in C57BL/6 and CBA mice. Other studies, however, find discrepancies between the extent of the anatomical and physiological changes (Chen et al., 2009). Poor performance in ABR was generally well correlated with hair cell loss in our study but not completely so. While loss of hair cells should result in hearing loss, a small outer hair cell loss may not influence hearing threshold and a loss of hair cells in regions where the auditory brain response is not being measured would not be detected in the hearing test. Hearing loss can also result from other factors than hair cell loss, including loss of auditory nerve – inner hair cell connections and damaged spiral ganglion neurons that can occur independently of hair cell loss. As noted above, we found several QTL that were associated with OHC survival at apex or base of the cochlea and whose associated ABR differences were seen at low or at high frequencies respectively. Some loci (e.g. 10•93, see Figure 6) influence ABR responses but do not have any effect on OHC (or IHC) survival; these may be acting through pathways unrelated to hair cell death. Other QTL (see 17•42 and 19•12 in Table 3) have effects on hair cell survival, but did not lead to significant alterations in ABR responses in the aged mice. It is possible that this disparity reflects damage to structures other than hair cells, but possibly also low statistical power for the ABR tests.
Comparison of ABR and OHC effects
In most of the aging four-way cross mice hearing (or lack thereof) is also well correlated between low and high frequencies, but a small number of polymorphic alleles mediate preferential loss of either high or low frequencies. An important result in this context is that different loci are associated with age-related loss of hair cells in the base (QTL on chromosome 10) versus the apex (QTL on chromosomes 11,12, 17 and 19). This suggests that some of the mechanisms underlying age-related hair cell degeneration are different for the basal and apical cochlea. This is consistent with the different patterns of age-related hearing loss and hair cell loss in different species and even strains within species (for reviews, see (Frisina, 2009; Ohlemiller, 2009; Ohlemiller, 2004).) For example, aging gerbils characteristically show apical hair cell loss and low frequency hearing loss (e.g. (Boettcher et al., 1995), while in contrast aging humans more often show basal hair cell loss and declines in high frequency hearing with aging (Gates and Mills, 2005; Ohlemiller, 2004). Therapeutic approaches to prevent human ARHL may need to be tested in an animal model with comparable underlying mechanisms.
It is interesting that the 129 allele at 10•44 is associated with increased hair cell loss in the base induced both by aging and noise. This allele may confer increased susceptibility to multiple stresses to the apical cochlea, which is normally more resistant to ototoxins. In contrast, some loci (e.g. 129 allele at 12•40) are associated with increased susceptibility to noise-induced hearing loss, but do not lead to decline in aged mice not exposed to noise, implying that underlying mechanisms may also differ.
Comparison to previous reports of mouse QTL
Most previous QTL studies in mouse have evaluated associations in crosses using two inbred strains, including backcrosses and recombinant inbred lines derived from intercrosses, and have uncovered several loci that are associated with susceptibility to age-related hearing loss (for review, see (Noben-Trauth and Johnson, 2009). Analysis of two-strain crosses has typically revealed single QTL with relatively large effect sizes, in some cases approaching Mendelian segregation patterns. Analysis of an outbred strain, Black Swiss, has also been performed and provided evidence for two apparently unique loci (Drayton and Noben-Trauth, 2006). These previously defined QTL were associated with hearing loss that initiates before 8 months of age and were often localized using ABR thresholds at one frequency measured at a single time point. Overall, the large number of QTL, each with relatively small individual effects, seen in our study of UM-HET4 mice is consistent with previous quantitative trait studies in many species, including heterogeneous mouse stocks (Valdar et al., 2006), and may better reflect the complex genetic architecture controlling hearing function across the lifespan. The QTL detected in 18 month old mice, those listed in Table 1 on chromosomes 2, 7, 10, and 15, are, we believe, the first mouse genetic variants whose effects on hearing become detectable only in late life, i.e. at ages greater than one year.
The 8-month QTL locations on chromosomes 3 and 13 identified in the current cross do not overlap with those of previously defined QTL associated with age-related hearing loss, and thus represent novel loci that modulate hearing in young adult UM-HET4 mice. Similarly, the majority of the QTL associated with hearing loss at 18 months in UM-HET4 mice, like those that affect hearing in 8 month old mice, have not previously been associated with age-related loss in mice. The potential exception is the QTL located on chromosome 10 and associated with effects on hearing at 48 kHz. The original Cdh23 locus on central chromosome 10 is unlikely to be responsible for the effects noted in our study, because all grandparental strains in the present cross carry the Cdh23753G variant associated with better hearing. Four other auditory QTL have also previously been localized to chromosome 10. Ahl4 on the distal region of the chromosome was identified in mapping studies with the A/J mouse strain (Zheng et al., 2009). A/J-derived variants at this locus were associated with threshold increases by 6 months of age. Ahl5, detected in crosses involving Black Swiss outbred mice (Drayton and Noben-Trauth, 2006), Phl2, detected in strain 101/H crosses (Mashimo et al., 2006), and Snhl1, detected in recombinant inbred lines derived from a multigenerational cross of several laboratory strains (Noben-Trauth et al., 2010), also map to central chromosome 10. Ahl5, Phl2, and Snhl1 appear to be distinct from the original Ahl allele at Cdh23 based upon haplotype studies of the Cdh23 genomic region in these strains. Similar to Ahl4, variants at these other chromosome 10 QTL are associated with onset of hearing loss before 1 year of age.
Comparison to human loci that affect late life hearing
In contrast to precise genetic information on both syndromic and non-syndromic forms of deafness, the genetic factors contributing to human ARHI remain to be established. Several studies have found suggestive linkages, but a recent multi-center European study found only one gene significantly associated with age-related hearing loss (Van Laer et al., 2008), a variant allele in the gene GRHL2 on chromosome 8 (Van Laer et al., 2008). Interestingly, the current study provided evidence in UM-HET4 mice of a QTL with an influence on hearing at 48 kHz, located on chromosome 15 in a location syntenic with the region of human chromosome 8 that includes GRHL2. A protein truncating mutation in this gene had been previously identified in individuals segregating a progressive hearing loss trait with dominant Mendelian inheritance at the DFNA28 locus (Peters et al., 2002). More recently, a genome-wide association study which tested 169,154 single-nucleotide polymorphisms found no variants with significance for hearing impairment (Huyghe et al., 2008), while another identified a significant association of variant allele in the gene GRM7 with hearing loss (Friedman et al., 2009).
Acknowledgements
We are grateful for the technical support of Lisa Burmeister, Sabrina Friedline, Lisa Kabara, Ariane Kanicki, Catherine Martin. We thank Margaret Lomax for advice. This work was supported by P01-AG025164, P30-DC05188 and P30-AG024824.
Abbreviations
- ABR
auditory brainstem response
- AHRI
age-related hearing impairment
- CCG
cytocochleogram
- dB
decibel
- IHC
inner hair cell
- kHz
kilohertz
- OHC
outer hair cell
- SOD
superoxide dismutase.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure statement: None of the authors have conflicts of interest relevant to the work presented in this manuscript. All procedures using mice were carried out after review and approval of the University of Michigan's Committee on Use and Care of Animals.
Reference List
- Beck JA, Lloyd S, Hafezparast M, Lennon-Pierce M, Eppig JT, Festing MF, Fisher EM. Genealogies of mouse inbred strains. Nat. Genet. 2000;24:23–25. doi: 10.1038/71641. [DOI] [PubMed] [Google Scholar]
- Biassoni EC, Serra MR, Richtert U, Joekes S, Yacci MR, Carignani JA, Abraham S, Minoldo G, Franco G. Recreational noise exposure and its effects on the hearing of adolescents. Part II: development of hearing disorders. Int. J. Audiol. 2005;44:74–85. doi: 10.1080/14992020500031728. [DOI] [PubMed] [Google Scholar]
- Boettcher FA, White DR, Mills JH, Schmiedt BN. Age-related changes in auditory evoked potentials of gerbils. III. Low-frequency responses and repetition rate effects. Hear. Res. 1995;87:208–219. doi: 10.1016/0378-5955(95)00091-h. [DOI] [PubMed] [Google Scholar]
- Chen GD, Li M, Tanaka C, Bielefeld EC, Hu BH, Kermany MH, Salvi R, Henderson D. Aging outer hair cells (OHCs) in the Fischer 344 rat cochlea: function and morphology. Hear. Res. 2009;248:39–47. doi: 10.1016/j.heares.2008.11.010. [DOI] [PubMed] [Google Scholar]
- Cody AR, Robertson D. Variability of noise-induced damage in the guinea pig cochlea: electrophysiological and morphological correlates after strictly controlled exposures. Hear. Res. 1983;9:55–70. doi: 10.1016/0378-5955(83)90134-x. [DOI] [PubMed] [Google Scholar]
- Cruickshanks KJ, Zhan W, Zhong W. In: The Aging Auditory System. Gordon-Salant S, Frisina DR, Popper AN, Fay RR, editors. Springer; New York: 2010. pp. 259–274. [Google Scholar]
- Daniel E. Noise and hearing loss: a review. J. Sch. Health. 2007;77:225–231. doi: 10.1111/j.1746-1561.2007.00197.x. [DOI] [PubMed] [Google Scholar]
- Drayton M, Noben-Trauth K. Mapping quantitative trait loci for hearing loss in Black Swiss mice. Hear. Res. 2006;212:128–139. doi: 10.1016/j.heares.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Friedman RA, Van Laer L, Huentelman MJ, Sheth SS, Van Eyken E, Corneveaux JJ, Tembe WD, Halperin RF, Thorburn AQ, Thys S, Bonneux S, Fransen E, Huyghe J, Pyykko I, Cremers CW, Kremer H, Dhooge I, Stephens D, Orzan E, Pfister M, Bille M, Parving A, Sorri M, Van de Heyning PH, Makmura L, Ohmen JD, Linthicum FHJ, Fayad JN, Pearson JV, Craig DW, Stephan DA, Van Camp G. GRM7 variants confer susceptibility to age-related hearing impairment. Hum. Mol. Genet. 2009;18:785–796. doi: 10.1093/hmg/ddn402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisina RD. Age-related hearing loss: ear and brain mechanisms. Ann. N. Y. Acad. Sci. 2009;1170:708–717. doi: 10.1111/j.1749-6632.2009.03931.x. [DOI] [PubMed] [Google Scholar]
- Gates GA, Couropmitree NN, Myers RH. Genetic associations in age-related hearing thresholds. Arch. Otolaryngol. Head Neck Surg. 1999;125:654–659. doi: 10.1001/archotol.125.6.654. [DOI] [PubMed] [Google Scholar]
- Gates GA, Mills JH. Presbycusis. Lancet. 2005;366:1111–1120. doi: 10.1016/S0140-6736(05)67423-5. [DOI] [PubMed] [Google Scholar]
- Hanlon P, Lorenz WA, Shao Z, Harper JM, Galecki AT, Miller RA, Burke DT. Three-locus and four-locus QTL interactions influence mouse insulin-like growth factor-I. Physiol. Genomics. 2006;26:46–54. doi: 10.1152/physiolgenomics.00247.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, Pahor M, Javors MA, Fernandez E, Miller RA. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfer TM, Canham-Chervak M, Canada S, Mitchener TA. Epidemiology of hearing impairment and noise-induced hearing injury among U.S. military personnel, 2003-2005. Am. J. Prev. Med. 2010;38:S71–S77. doi: 10.1016/j.amepre.2009.10.025. [DOI] [PubMed] [Google Scholar]
- Henderson D, Hu B, Bielefeld EC. Patterns and mechanisms of noise-induced cochlear pathology. In: Schacht J, Popper AN, Fay RR, editors. Springer Handbook of Auditory Research. Springer; US: 2008. pp. 195–217. [Google Scholar]
- Huyghe JR, Van Laer L, Hendrickx JJ, Fransen E, Demeester K, Topsakal V, Kunst S, Manninen M, Jensen M, Bonaconsa A, Mazzoli M, Baur M, Hannula S, Maki-Torkko E, Espeso A, Van Eyken E, Flaquer A, Becker C, Stephens D, Sorri M, Orzan E, Bille M, Parving A, Pyykko I, Cremers CW, Kremer H, Van de Heyning PH, Wienker TF, Nurnberg P, Pfister M, Van Camp G. Genome-wide SNP-based linkage scan identifies a locus on 8q24 for an age-related hearing impairment trait. Am. J. Hum. Genet. 2008;83:401–407. doi: 10.1016/j.ajhg.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KR, Zheng QY. Ahl2, a second locus affecting age-related hearing loss in mice. Genomics. 2002;80:461–464. [PMC free article] [PubMed] [Google Scholar]
- Johnson KR, Zheng QY, Weston MD, Ptacek LJ, Noben-Trauth K. The Mass1frings mutation underlies early onset hearing impairment in BUB/BnJ mice, a model for the auditory pathology of Usher syndrome IIC. Genomics. 2005;85:582–590. doi: 10.1016/j.ygeno.2005.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiernan AE, Steel KP. Mouse homologues for human deafness. Adv. Otorhinolaryngol. 2000;56:233–243. doi: 10.1159/000059107. [DOI] [PubMed] [Google Scholar]
- Maison SF, Liberman MC. Predicting vulnerability to acoustic injury with a noninvasive assay of olivocochlear reflex strength. J. Neurosci. 2000;20:4701–4707. doi: 10.1523/JNEUROSCI.20-12-04701.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashimo T, Erven AE, Spiden SL, Guenet JL, Steel KP. Two quantitative trait loci affecting progressive hearing loss in 101/H mice. Mamm. Genome. 2006;17:841–850. doi: 10.1007/s00335-004-2438-5. [DOI] [PubMed] [Google Scholar]
- Miller RA, Austad S, Burke D, Chrisp C, Dysko R, Galecki A, Monnier V. Exotic mice as models for aging research: polemic and prospectus. Neurobiol. Aging. 1999;20:217–231. doi: 10.1016/s0197-4580(99)00038-x. [DOI] [PubMed] [Google Scholar]
- Miller RA, Dolan D, Han M, Kohler W, Schacht J. Resistance of skin fibroblasts to peroxide and UV damage predicts hearing loss in aging mice. Aging Cell. 2011;10:362–363. doi: 10.1111/j.1474-9726.2010.00668.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RA, Harrison.D.E. Astle, C.M., Floyd RA, Flurkey K, Hensley KL, Javors MA, Leeuwenburgh C, Nelson JF, Ongini E, Nadon NL, Warner HR, Strong R. An aging interventions testing program: study design and interim report. Aging Cell. 2007;6:565–575. doi: 10.1111/j.1474-9726.2007.00311.x. [DOI] [PubMed] [Google Scholar]
- Miller RA, Jackson AU, Galecki AT, Burke DT. Genetic polymorphisms in mouse genes regulating age-sensitive and age-stable T cell subsets in mice. Genes and Immunity. 2003;4:30–39. doi: 10.1038/sj.gene.6363895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DI, Nelson RY, Concha-Barrientos M, Fingerhut M. The global burden of occupational noise-induced hearing loss. Am. J. Ind. Med. 2005;48:446–458. doi: 10.1002/ajim.20223. [DOI] [PubMed] [Google Scholar]
- Nichols TC, du Laney T, Zheng B, Bellinger DA, Nickols GA, Engleman W, Clemmons DR. Reduction in atherosclerotic lesion size in pigs by alphaVbeta3 inhibitors is associated with inhibition of insulin-like growth factor-I-mediated signaling. Circ. Res. 1999;85:1040–1045. doi: 10.1161/01.res.85.11.1040. [DOI] [PubMed] [Google Scholar]
- Noben-Trauth K, Johnson KR. Inheritance patterns of progressive hearing loss in laboratory strains of mice. Brain Res. 2009;1277:42–51. doi: 10.1016/j.brainres.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noben-Trauth K, Latoche JR, Neely HR, Bennett B. Phenotype and genetics of progressive sensorineural hearing loss (Snhl1) in the LXS set of recombinant inbred strains of mice. PLoS One. 2010;5:e11459. doi: 10.1371/journal.pone.0011459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noben-Trauth K, Zheng QY, Johnson KR. Association of cadherin 23 with polygenic inheritance and genetic modification of sensorineural hearing loss. Nat. Genet. 2003;35:21–23. doi: 10.1038/ng1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlemiller KK. Contributions of mouse models to understanding of age- and noise-related hearing loss. Brain Res. 2006;1091:89–102. doi: 10.1016/j.brainres.2006.03.017. [DOI] [PubMed] [Google Scholar]
- Ohlemiller KK. Age-related hearing loss: the status of Schuknecht's typology. Curr. Opin. Otolaryngol. Head Neck Surg. 2004;12:439–443. doi: 10.1097/01.moo.0000134450.99615.22. [DOI] [PubMed] [Google Scholar]
- Ohlemiller KK. Mechanisms and genes in human strial presbycusis from animal models. Brain Res. 2009;1277:70–83. doi: 10.1016/j.brainres.2009.02.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters LM, Anderson DW, Griffith AJ, Grundfast KM, San Agustin TB, Madeo AC, Friedman TB, Morell RJ. Mutation of a transcription factor, TFCP2L3, causes progressive autosomal dominant hearing loss, DFNA28. Hum. Mol. Genet. 2002;11:2877–2885. doi: 10.1093/hmg/11.23.2877. [DOI] [PubMed] [Google Scholar]
- Phillips SL, Henrich VC, Mace ST. Prevalence of noise-induced hearing loss in student musicians. Int. J. Audiol. 2010;49:309–316. doi: 10.3109/14992020903470809. [DOI] [PubMed] [Google Scholar]
- Sha SH, Kanicki A, Dootz G, Talaska AE, Halsey K, Dolan D, Altschuler R, Schacht J. Age-related auditory pathology in the CBA/J mouse. Hear. Res. 2008;243:87–94. doi: 10.1016/j.heares.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spongr VP, Flood DG, Frisina RD, Salvi RJ. Quantitative measures of hair cell loss in CBA and C57BL/6 mice throughout their life spans. J. Acoust. Soc. Am. 1997;101:3546–3553. doi: 10.1121/1.418315. [DOI] [PubMed] [Google Scholar]
- Strong R, Miller RA, Astle CM, Floyd RA, Flurkey K, Hensley KL, Javors MA, Leeuwenburgh C, Nelson JF, Ongini E, Nadon NL, Warner HR, Harrison DE. Nordihydroguaiaretic acid and aspirin increase lifespan of genetically heterogeneous male mice. Aging Cell. 2008;7:641–650. doi: 10.1111/j.1474-9726.2008.00414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadros SF, D'Souza M, Zhu X, Frisina RD. Apoptosis-related genes change their expression with age and hearing loss in the mouse cochlea. Apoptosis. 2008;13:1303–1321. doi: 10.1007/s10495-008-0266-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdar W, Solberg LC, Gauguier D, Burnett S, Klenerman P, Cookson WO, Taylor MS, Rawlins JN, Mott R, Flint J. Genome-wide genetic association of complex traits in heterogeneous stock mice. Nat. Genet. 2006;38:879–887. doi: 10.1038/ng1840. [DOI] [PubMed] [Google Scholar]
- Van Eyken E, Van Camp G, Van Laer L. The complexity of age-related hearing impairment: contributing environmental and genetic factors. Audiol. Neurootol. 2007;12:345–358. doi: 10.1159/000106478. [DOI] [PubMed] [Google Scholar]
- Van Laer L, Van Eyken E, Fransen E, Huyghe JR, Topsakal V, Hendrickx JJ, Hannula S, Maki-Torkko E, Jensen M, Demeester K, Baur M, Bonaconsa A, Mazzoli M, Espeso A, Verbruggen K, Huyghe J, Huygen P, Kunst S, Manninen M, Konings A, Diaz-Lacava AN, Steffens M, Wienker TF, Pyykko I, Cremers CW, Kremer H, Dhooge I, Stephens D, Orzan E, Pfister M, Bille M, Parving A, Sorri M, Van de Heyning PH, Van Camp G. The grainyhead like 2 gene (GRHL2), alias TFCP2L3, is associated with age-related hearing impairment. Hum. Mol. Genet. 2008;17:159–169. doi: 10.1093/hmg/ddm292. [DOI] [PubMed] [Google Scholar]
- Volkman SK, Galecki AT, Burke DT, Paczas MR, Moalli MR, Miller RA, Goldstein SA. Quantitative trait loci for femoral size and shape in a genetically heterogeneous mouse population. J. Bone Miner. Res. 2003;18:1497–1505. doi: 10.1359/jbmr.2003.18.8.1497. [DOI] [PubMed] [Google Scholar]
- White CH, Ohmen JD, Sheth S, Zebboudj AF, McHugh RK, Hoffman LF, Lusis AJ, Davis RC, Friedman RA. Genome-wide screening for genetic loci associated with noise-induced hearing loss. Mamm. Genome. 2009;20:207–213. doi: 10.1007/s00335-009-9178-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiltshire T, Pletcher MT, Batalov S, Barnes SW, Tarantino LM, Cooke MP, Wu H, Smylie K, Santrosyan A, Copeland NG, Jenkins NA, Kalush F, Mural RJ, Glynne RJ, Kay SA, Adams MD, Fletcher CF. Genome-wide single-nucleotide polymorphism analysis defines haplotype patterns in mouse. Proc. Natl. Acad. Sci. USA. 2003;100:3380–3385. doi: 10.1073/pnas.0130101100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf N, Galecki A, Lipman R, Chen S, Smith-Wheelock M, Burke D, Miller R. Quantitative trait locus mapping for age-related cataract severity and synechia prevalence using four-way cross mice. Invest. Ophthalmol. Vis. Sci. 2004;45:1922–1929. doi: 10.1167/iovs.03-0435. [DOI] [PubMed] [Google Scholar]
- Zheng QY, Ding D, Yu H, Salvi RJ, Johnson KR. A locus on distal chromosome 10 (ahl4) affecting age-related hearing loss in A/J mice. Neurobiol. Aging. 2009;30:1693–1705. doi: 10.1016/j.neurobiolaging.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng QY, Johnson KR. Hearing loss associated with the modifier of deaf waddler (mdfw) locus corresponds with age-related hearing loss in 12 inbred strains of mice. Hear. Res. 2001;154:45–53. doi: 10.1016/s0378-5955(01)00215-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng QY, Johnson KR, Erway LC. Assessment of hearing in 80 inbred strains of mice by ABR threshold analyses. Hear. Res. 1999;130:94–107. doi: 10.1016/s0378-5955(99)00003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]