Abstract
The head bobber transgenic mouse line, produced by pronuclear integration, exhibits repetitive head tilting, circling behavior, and severe hearing loss. Transmitted as an autosomal recessive trait, the homozygote has vestibular and cochlea inner ear defects. The space between the semicircular canals is enclosed within the otic capsule creating a vacuous chamber with remnants of the semicircular canals, associated cristae, and vestibular organs. A poorly developed stria vascularis and endolymphatic duct is likely the cause for Reissner’s membrane to collapse post-natally onto the organ of Corti in the cochlea. Molecular analyses identified a single integration of ~3 tandemly repeated copies of the transgene, a short duplicated segment of chromosome X and a 648 kb deletion of chromosome 7(F3). The three known genes (Gpr26, Cpxm2, and Chst15) in the deleted region are conserved in mammals and expressed in the wild-type inner ear during vestibular and cochlea development but are absent in homozygous mutant ears. We propose that genes critical for inner ear patterning and differentiation are lost at the head bobber locus and are candidate genes for human deafness and vestibular disorders.
Electronic supplementary material
The online version of this article (doi:10.1007/s10162-012-0316-5) contains supplementary material, which is available to authorized users.
Keywords: ear development, cloning, transgenic mouse, stria vascularis, semicircular canals
Introduction
Deafness is the most common sensory disorder in humans—approximately one in 1,000 children are born with permanent hearing impairment, and single-gene defects account for over half of all cases (Smith et al. 2005). Balance disorders affect about 30% of people over the age of 65 years (Colledge et al. 2002), but little is known about the genetic causes of vestibular dysfunction. The functional importance of many molecules expressed in the developing and adult ear have been determined by gene targeting, by the identification of affected genes in classical mouse mutants, or from human hearing and balance disease susceptibility loci (Anagnostopoulos 2002; Steel 1995). Insertional mutagenesis in mice has led to the identification and isolation of genes involved in normal growth and development (Meisler 1992), including normal hearing and balance. For example, an insertional mutation generated a new allele of Ames Waltzer (av), a circling and hearing-impaired mouse, which led to the identification of a novel protocadherin (PCDH) gene (Alagramam et al. 2001a). The human ortholog PCDH15 was cloned, and mutations in PCDH15 were discovered in families segregating Usher syndrome type 1F, a disease causing deafness and vestibular dysfunction (Ahmed et al. 2001; Alagramam et al. 2001b). Other spontaneous alleles of Ames Waltzer have been identified, allowing further delineation of the functional domains of this essential protein (Zheng et al. 2006). Thus, identification of novel hearing and balance gene mutations in mice can provide information about the pathways that specify auditory and vestibular function in humans.
The vertebrate ear is a complex organ consisting of an auditory system that includes external, middle, and inner ear structures, and a balance system, which is composed of inner ear vestibular otolithic and semicircular canal structures. Peripheral sensory information is conveyed through mechanosensory transducers in the form of hair cells localized in: cristae of the three semicircular canals used for detecting angular acceleration; maculae of the saccule and utricle used for detecting linear and angular acceleration and gravity; and in the organ of Corti used for detecting sound.
The morphogenetic development of the mammalian inner ear is a complex process, whose molecular and cellular details are still being defined (Bok et al. 2007; Fekete and Wu 2002; Wang and Lufkin 2005). The inner ear develops initially from a single layer of otic epithelium, the otic placode. The placode invaginates to form an otic vesicle that elongates into dorsal and ventral portions, which differentiate into components of the semicircular canals and vestibule, and cochlea, respectively. The cochlea is further divided into three compartments: the central cochlear duct (scala media) is separated from the scala vestibuli (above) by the Reissner’s membrane and from the scala tympani (below) by the basilar membrane. The scala media is filled with endolymph with high [K+] and the vestibuli and tympani compartments are filled with low [K+] perilymph, creating a chemical gradient essential for sensory cell function. The unique ionic composition of endolymph is maintained by the stria vascularis of the scala media through active reabsorption of sodium and active secretion of potassium against ionic gradients (Wangemann 2006). Genetic or non-genetic perturbation of any of these components can lead to auditory and/or vestibular dysfunction. We report here the physiological, developmental, and molecular studies of the head bobber mutation that causes defects in inner ear patterning, hyperactivity, circling behavior, and deafness.
Methods
Generation of head bobber mice and mouse husbandry
The head bobber mouse line was produced by a random transgene integration of exogenous DNA. Insertional mutation arising from the random integration of defined DNA sequences can be used for identifying genes with developmental roles. The integrated exogenous DNA serves two purposes: first, the integration of the transgene can confer a loss or a gain of function (depending if the integrated DNA interrupts or activates some gene) leading to a mutant phenotype; second, it acts as a molecular “tag” marking the integration locus. The classic transgene design includes an enhancer and promoter, the mRNA coding sequence and a complete set of poly-adenylation signals.
The transgene vector used to generate the head bobber mice (Figure 1A) contained exon 1 and the entire intron 1 (which excludes the ATG translation start codon in exon 2) of the human β-actin promoter (~3.3 kb). In detail, the vector (designated as p88) contained: bases 1–4,379 from the human β-actin gene (166,516 to 170,895 of gi9211523, an EcoRI-AluI fragment); bases 4,380–4,400 from pSP64 poly-linker; bases 4,401–6,600 derived from pcDVI, containing the pBR322 AmpR gene and the bacterial origin plus the SV40 late region polyadenylation signal; bases 6,600–10,000 derived from the PvuII-EcoRI fragment of the pSV2neo vector (accession no. U02434), containing the bacterial neo gene linked to the SV40 ori plus early promoter. Transgenic mice were generated by injection of the linearized (8,413 bp PvuI-HindIII fragment) vector into inbred FVB embryos. A mouse line OVE7 was kept due to the overt circling behavior and head tilting and was given the name head bobber. Congenic strains were obtained after >10 backcrosses to the recipient strain, counting the first hybrid generated as F1 and the following backcrosses as N2–N10 (Flaherty 1981). After the tenth generation, two carriers of the mutation were intercrossed, offspring homozygous for the mutation were selected, and the congenic strain was maintained by brother–sister mating. Moreover, since congenic strains may accumulate genetic differences over time due to unnoticed mutations that became fixed, they were backcrossed about four generations every 15–20 generations to mice of the parental inbred background so that sibling mating could be resumed. Matings of hb/hb males to +/hb females were used for phenotyping and cloning analyses. Some phenotypic analyses were previously described (see http://www.informatics.jax.org/searchtool/Search.do?query=MGI:2447989).
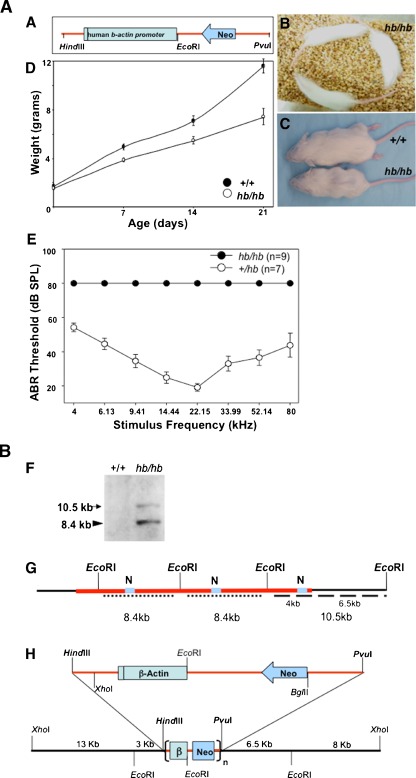
FIG. 1.
A, B Phenotype and molecular characterization of the genomic region interrupted by the transgene insertion in head bobber mice. A Diagram of the transgene fragment used to create head bobber line. B A composite of three images showing hb/hb mice display hyperactivity and circling behavior. C hb/hb mice are reduced size in adulthood and have a reduced growth compared with wild-type littermates (D). Black bar in human β-actin promoter represents exon 1. E Auditory brainstem response (ABR) thresholds were measured for hemizygotes (+/hb, n = 7) and homozygotes (hb/hb, n = 9) at P20. +/hb had normal hearing thresholds, but hb/hb mice had no responses at any frequencies tested. Data are presented as mean ± SEM in D and E. F Southern analysis of the hb locus with a neo gene probe (grey box, N in G) hybridized to two bands (10.5 and 8.4 kb) indicating a single insertion site (red line) of the transgene in the head bobber genome. The 11 kb band represents a single-copy transgene plus flanking genomic sequences (dashed line in G) and the 8.4 kb band represents the multicopy concatamer (dotted line in G). Appropriately, no bands were detected in the wild-type (wt) genome. G Restriction map of the transgene vector and genome allowed the creation of a map (H) of the genomic region surrounding the transgene integration site. β, human β-actin gene fragment; black line in human β-actin gene represents exon 1; n, multicopy concatamer. dB, decibels; SPL, sound pressure levels; kHz, kilohertz.
Growth curves
Weight data from a total of 23 animals resulting from the intercrossing of +/hb animals were collected at post-natal (P) days 0, 7, 14, and 21. Mice were genotyped and weight averages were plotted against time according to the genotype of the animals (Figure 1, D). Differences in weights were compared using t test statistics.
Histology and immunofluorescence analyses
The inner ears from animals (+/+, +/hb, and hb/hb) at P0, P3, P7, P16, and P30 were isolated, fixed in 4% paraformaldehyde (PFA), dehydrated, and paraffin-embedded. P7, P16, and P30 inner ears were decalcified in 100 mM EDTA for 3–7 days before dehydration. For histological analyses, serial sections (7 μm) were stained with hematoxylin and eosin. For immunofluorescence, citrate antigen retrieval (Vector Labs, CA) was performed prior to permeabilization (PBS containing 0.2% Triton X-100), blocked for 1 h in normal serum at room temperature, and incubated with primary antibodies: anti-myosin VIIa (1:6,000, Proteus Biosciences) (Hasson and Mooseker 1994), anti-fibronectin (1:200, A-21316 Molecular Probes, OR), and anti-cadherin (1:200, C1821 Sigma, MO) or the GSLI lectin (1:200, Vector Labs, CA) overnight at 4°C in a humidified chamber. Sections were washed (thrice in TBST) and incubated with appropriate secondary antibodies (1:1,000, Molecular Probes, OR) at 37°C for 2.5 h. After washing, sections were post-fixed with 4% PFA for 10–30 min, covered with Fluoromount with 4',6-diamidino-2-phenylindole (DAPI) and viewed and photographed using a Zeiss Axioplan 2 epi-fluorescence microscope.
Micro-computed tomography
The intact inner ears from P20 animals (+/+ and hb/hb) were isolated and fixed as above and imaged using a high-resolution Skyscan 1,172 micro-computed tomography imaging system (Skyscan, Kontich, Belgium). Each inner ear was pre-embedded in paraffin at a fixed orientation and scanned at 5 μm/pixel resolution with instrument settings of 40 kV voltage and 250 μA current. Four 1,280 × 1,024-pixel raw images were acquired every 0.2° over 180° and averaged to increase the signal-to-noise ratio. Coronal- and sagittal-oriented tomograms were reconstructed, resliced, and binarized from the raw images with NRecon CT Reconstruction and CT-Analyzer software packages (Skyscan, Kontich, Belgium) utilizing an adapted 3D cone beam reconstruction algorithm (Feldkamp et al. 1984). Volume rendering and analysis was performed using CT-Volume software package (Skyscan, Kontich, Belgium) with a voxel size of 10 μM.
Auditory brainstem responses
P20 mice of either sex were anesthetized using 45 mg/kg ketamine and 5.4 mg/kg xylazine, and their body temperature was maintained at 39°C. The auditory brainstem response (ABR) was recorded using needle electrodes positioned at the vertex of the skull and along the ventral surface of the tympanic bulla. A ground electrode was placed in the hind leg. The signals were amplified 10,000 times using a biological amplifier (HS4/DB4, Tucker-Davis Technologies, Alachua, FL), digitized at 200 kHz (RP-2, Tucker-Davis Technologies), and digitally band-pass filtered from 300 to 3,000 Hz. Sound stimuli were generated digitally, attenuated using programmable attenuators, and delivered via electrostatic speakers (RP-2, PA-5, ED-1, and EC-1, Tucker-Davis Technologies). The stimulus for eliciting the ABR was a 5 ms sine wave tone pip with cos2 envelope rise and fall times of 0.5 ms. The repetition time was 50 ms, and 250 trials were averaged. At each frequency, the peak ABR at stimulus intensities ranging from 10 to 80 dB sound pressure levels was measured in 10 dB steps and interpolated to find threshold four standard deviations above the noise floor. The speakers and microphone were calibrated across the frequency range of 1–80 kHz prior to each experiment using a probe-tip microphone (microphone type 8,192, NEXUS conditioning amplifier, Bruel and Kjar, Demark), after they were connected to a tube (an earbar) inserted into the animal’s ear canal.
Transgene copy number
Southern blots (Qiu et al. 1997) were prepared from genomic DNA extracted from whole E16 embryos (hb/hb or +/+) and restriction-digested with an enzyme (EcoRI) that cut the transgene once. Transgene copy number was estimated by comparing hybridization band intensities, measured by densitometry (Personal Densitometer SI, Molecular Dynamics), for the unique 5′ (neo) transgene region, which should represent a single-copy band continuous with chromosomal flanking DNA (10.5 kb, Figure 1, F), to the multiple copy concatenated region band (8.4 kb, Figure 1, F). Similar estimates of copy number were obtained with the 3′ (β-actin) region (data not shown). Restriction mapping and Southern blots were also used to characterize the alterations in the head bobber genome that occurred during insertion of the transgene.
Cloning, sequencing, and characterization of the transgene insertion site
To clone the transgene insertion site, genomic DNA (25–50 mg) taken from adult homozygous hb/hb circler mice brains were used. Polymerase chain reaction (PCR) was performed with transgene-specific primers (TSP1: tttatggtaataacgcgccggcc, TSP2: cccggctattctcgcaggatcagt, TSP3: aggatcagtcgacctgcagcccaa) that amplify the β-actin region and DNA Walking ACP primers (ACP1-4, DNA Walking Speedup Kit, Seegene), designed to capture genomic sequences flanking a transgene. Three sequential PCR reactions were performed as described in the manufacturer’s instructions. The third PCR reaction using TSP3 specifically amplified bands that were subcloned and sequenced (Figure 5, G). After identification of the hb locus on chromosome 7 (F3), a 1.66 Mb chromosome 7 genomic sequence (accession number: NT_039433.7, nucleotides 139,620,976 to 141,278,299) was obtained from Ensembl (www.ensembl.org) and studied to identify gene(s) in the region (Figure 5, B). For convenience, PCR primers for chromosome walking to extend and define the deleted portions were designed and named relative to the 1.66 Mb sequence (Electronic supplementary material Table I).
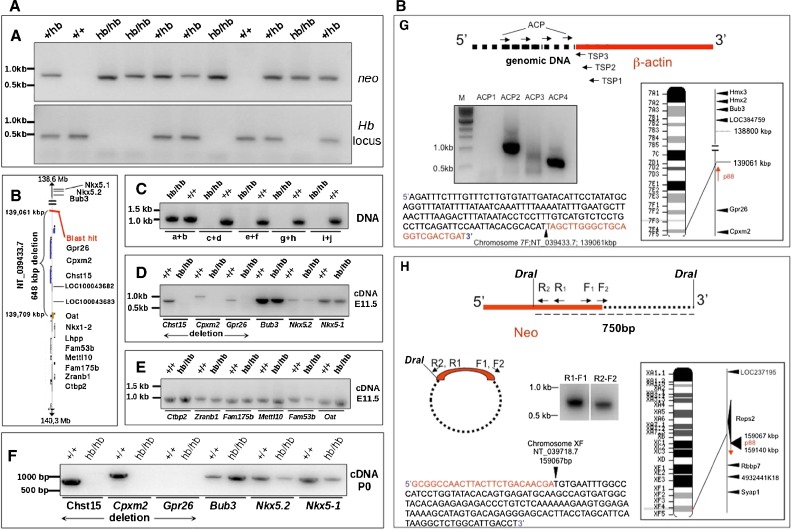
FIG. 5.
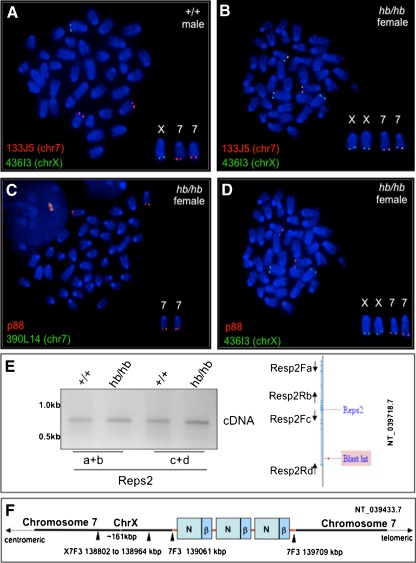
Head bobber genotyping, delineation of the deleted portion of chromosome 7, and identification of the transgene integration site. A PCR genotyping using Neo (top) and chromosome 7 (bottom) primer sets in +/+, hemizygous (+/hb), and head bobber (hb/hb) mice. B Haplotype diagram of the head bobber locus on chromosome 7 and gene loci interrogated by PCR analysis (C) for the extent of genomic deletion surrounding the hb locus (primers a–i are listed in Table I). Blast hit in B, initial sequence comparison location. D–F RT-PCR analyses for gene expression (primers are listed in Table II) in the head bobber region at E11.5 (D, E) and at P0 (F) from +/+ and head bobber (hb/hb) inner ears. G Using specific primers (TSP1, TSP2, TSP3, top panel) designed to the β-actin region and DNA Walking primers (top panel, ACP; designed to capture transgene flanking genomic sequences) specific bands were amplified (bottom panel) that were mapped to sequences on chromosome 7 (right panel). H Inverse PCR of DraI digested hb/hb genomic DNA (top panel) was circularized (middle panel), and nested primers (reverse R1, R2, and forward F1, F2) designed in the Neo gene were used to perform primary (R1–F1) and secondary (R2–F2) inverse PCR resulting in a approx. 750 bp band (bottom panel) that was mapped to chromosome X. Sequencing of cloned fragments allowed orientation of the transgene in the genome (arrows in chromosome maps).
Primers were used to amplify both +/+ and hb/hb DNA. The primers that identified the limits of the deletion were: 1160004F, 1160919 R and 1169679F, and 1170652 R. Verification of the 5′ region at the insertion site was determined by amplification of the correct size bands in both +/+ and hb/hb DNA using primers: 512800F: ggagagcagaaccctttgggaataagagtg, 514500R: cgctgctgctgcagccaggacaatt; 515820F: aaactgaatgctaagaaaccgaggaagc, 515245R: cagcaactgtatttaccaatcaaagcca. The sequence flanking the Neo-side (3′) or right arm of the transgene was studied using inverse PCR; briefly, 2.5 μg of genomic hb/hb DNA was digested with DraI (NEBiolabs, MA). The reaction was stopped by heating (65°C for 20 min) before the DNA fragments were self-circularized by dilution to 8.3 ng/μl and ligation (16°C for 16 h) using the T4 DNA ligase (NEBiolabs, MA). The volume of DNA was concentrated (to about 20 μl), and 2 μl was used to perform a primary PCR (50 μl reaction, 30 cycles with primers DraI F1: agaaaaataaacaaataggggttccgcg, DraI R1: cttattcccttttttgcggcattttgc). A secondary PCR was performed using 1 μl from the primary PCR; 10 mM dNTPs; 10 μmol each of forward and reverse primers (DraI F2: cccggcgtcaacacgggataatac, DraI R2: ttatgcagtgctgccataaccatga); 10× PCR buffer; 50 mM MgCl2; and 2.5 U Taq DNA polymerase mix (Stratagene, CA).
Routine PCR was performed using a DNA Engine (MJ Research, MA): Reactions were incubated at 94°C for 3 min, followed by 30 cycles of PCR amplification (denaturation, 94°C for 45 s; annealing, 55°C for 30 s; extension, 72°C for 3 min), 10 min extension at 72°C, and then maintained at 4°C. The amplification products were analyzed by agarose gel electrophoresis and visualized by ethidium bromide staining. Amplified DNA was sequenced with an ABI Analyzer, and sequences were compared against GenBank databases with Blastn. Both the primary and secondary PCR yielded a band that was subcloned (TOPO TA cloning, Invitrogen, CA), sequenced, and found to belong to the distal part of chromosome X. PCR for the chromosome X transgene hybrid was performed using a reverse primer from the transgene (agcatcttacggatggcatgaca) and a forward primer from the cloned region of chromosome X (tgccccagtagttaagagcactagctg).
FISH and aCGH analysis
Fluorescent in situ hybridization (FISH) was performed as described (Shaffer et al. 1997) using probes specific to the transgene (p88 vector) and BAC (bacterial artificial chromosome) probes for the genomic sequences (BAC clones RP23–390L14 and RP23–133J5 on chromosome 7 and RP23–436I3 on chromosome X) on metaphase spreads of spleen cells cultured from the head bobber line. BAC clones were identified from Ensembl (Ensembl.org) and purchased from BACPAC Resource Center (Oakland, CA). BAC DNA was isolated from the liquid bacterial cultures using BAC Prep Kit (GerardBiotech, OH).
Copy number variations were determined by performing a comparative genomic hybridization be-tween hb/hb and wild-type DNA using tail genomic DNA. The Baylor College of Medicine microarray core performed two biological replicate hybridizations on Agilent SurePrint G3 CGH 1 × 1M microarray high-resolution comparative genomic hybridization (aCGH) and copy number variation chips (Agilent, Santa Clara, CA) that have 963,261 features/probes that cover the current mouse genome NCBI build 37, UCSC mm9 (July 2007). Importantly, there are a significant number of probes covering the putative loss or gain in chromosomes regions in Hb mice, but there is a poor coverage of the Y-chromosome, which was excluded from final analysis. Data was analyzed using Nexus 6 copy number variation analysis software (BioDiscovery, El Segundo, CA). Frequency significance testing were performed on the aggregate replicates with default settings for areas of the genome with a statistically high frequency of aberration (Q-bound value ≤ 0.5 and G-score cut-off ≤1.0) corrected for multiple testing using false discovery rate correction (Benjamini and Hochberg 1995) and identified using the GISTIC (Genomic Identification of Significant Targets in Cancer) approach (Beroukhim et al. 2007).
Isolation of DNA and genotyping
Routinely, tail genomic DNAs were genotyped by PCR to amplify sequences specific to the transgene (800 bp neo-band, with primers NeoF: caagatggattgcacgcagg, NeoR: cccgctcagaagaactcgtc) to identify +/hb and hb/hb. Primers were also used to amplify a 450 bp (Hb locus band with primers HbF: ttctattaggaacagagcctgggc, HbR: ggcattctgggatgatgttgggtt) of the region in +/+ littermates or +/hb that was deleted in hb/hb mice (accession number of the region: NT-039433.7). Thus, +/+ animals lack a neo-band (no transgene) but have normal amplification of the Hb locus band; +/hb have both bands, and hb/hb mice have the neo-band only (Figure 5, A).
RNA isolation and RT-PCR analysis
Messenger RNA was purified from ears of wt and hb/hb P0 pups and from the region corresponding to the otic vesicle of wt and hb/hb E11.5 embryos using the RNAeasy mini kit (Qiagen, CA) and the RNase-free DNase treatment (DNA-free, Ambion, TX). One microgram of mRNA was used to generate cDNA according to the manufacturer’s instructions (RETROscript kit, Ambion, TX). PCR reactions were performed with 2 μl of the RT reaction and specific primers (Suppl. Table II). Expression of Gapdh, used as control for DNA contamination, and multiple genes surrounding the transgene integration site (Nkx5-1, Nkx5.2, Bub3, Gpr26, Cpxm2, Chst15, Oat, Fam53b, Mettl10, Fam175b, Zranb1, Ctbp2 and Resp2) were evaluated on 1% agarose gels.
Results
The head bobber transgenic mouse line (MGI: 2447989) was created by pronuclear microinjection of the transgene vector (Figure 1A, see Methods) into inbred FVB embryos resulting in three F0 founders. Normal litter sizes were obtained in established families and mice were genotyped by the presence of the neo resistance gene (see Methods). Hemizygous (+/hb) founder F0 and F1 animals were phenotypically indistinguishable from wild type littermates–being fertile, had similar growth rates and response to Preyer reflex (data not shown). However, +/hb intercrosses in one family (OVE7) produced some F2 offspring that displayed hyperactivity and circling (Figure 1B) and were reduced in size (Figure 1, C). To determine if the reduced size was due to reduced growth, +/+ mice (n = 9) and their +/hb littermates (n = 6) from two litters were weighed throughout the first 3 weeks of life (Figure 1, D). The weights were found to be statistically different at P7 (p < 0.001) and smaller mice never achieved wild type weight or size. The smaller animals displayed repetitive head tilting 7 to 9 days after birth and hyperactivity and circling behavior soon after. These animals were genotyped to be homozygous (hb/hb) for the head bobber allele (see Methods and Figure 5, A). Consistent with the possibility of inner ear defects, hb/hb mice were non-responsive for a Preyer reflex indicating impaired hearing. ABR thresholds analyzed at P20 showed hemizygous animals with normal thresholds whereas hb/hb mice had no ABR responses at all frequencies tested, which is consistent with a moderate to severe hearing loss (Figure 1, E), and any residual sensation, if present, would have minimal functional significance. The segregation of a phenotype in the F2 generation indicated that a mutation was transmitted in an autosomal recessive manner in the OVE7 family.
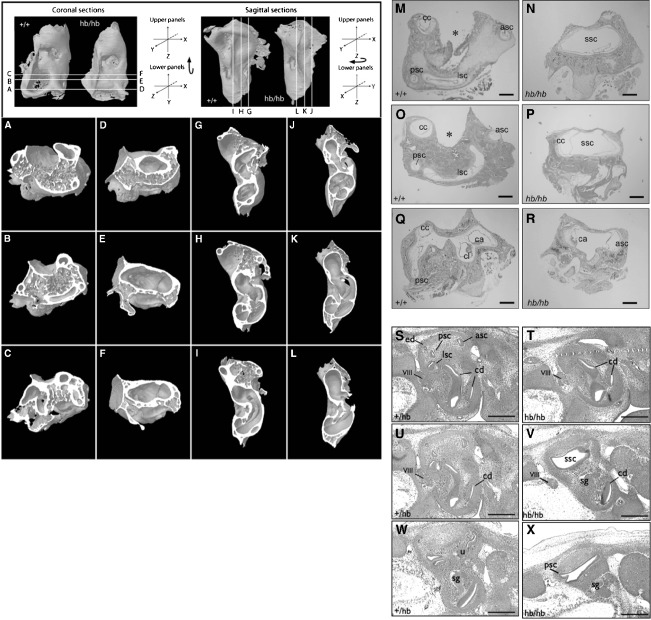
Whole mount micro-CT reconstructions and histological examination of inner ears from head bobber animals showed marked vestibular and cochlea defects. Whole-mount and sections at the vestibular level from wild type (Figure 2, A–C, G–I, and M, O, Q), which were indistinguishable to hemizygous inner ears (data not shown), showed the presence of semicircular canals and crista (Figure 2, Q). Ears of circling hb/hb mice had gross malformations of the semicircular canal region with absence of posterior and lateral canals and crista, and presented as a single vacuous chamber (SSC in Figure 2, D–F, J–L, and N, P), eliminating the arcs space bound by the semicircular canals where the cerebellar flocculus and paraflocculus normally reside (asterisks in WT Figure 2, A, B, G, H, and M, O). There was one reduced canal that, based on its position, is presumed to be the anterior semicircular canal with an associated crista (Figure 2, R) that was blind ending. The endolymphatic sac and vestibular ganglion were not identified at this stage but were present at E11 (data not shown) suggesting it degenerated. The saccule was malformed with a reduced saccular macula and the utricle and its associated macula and ganglia were not detected (data not shown and see Hardisty et al. 1997; Hughes et al. 1998; Somma et al. 2005). Indeed, the vestibular structures were maldeveloped at E13 (and not shown, as early as E11; Figure 2, S–X). Analysis of serial transverse and sagittal sections of hb/hb ears (n = 10) revealed little variation in the dysgenesis of the semicircular canals suggesting the defect occurs with complete penetrance.
FIG. 2.
Head bobber defects in semicircular canal and cochlear development. Micro-computed tomography coronal (A–F) and sagittal sections (I–J) of inner ears from P20 +/+ (A–C; G–I) and hb/hb (D–F; J–L), transverse sections from P7 +/+ (M–Q) and hb/hb (N–R) and E13 +/+ (S, U, W) and hb/hb (T, V, X) mice showing inner ear defects in hb/hb mice. hb/hb mice show severe malformation of the semicircular canal region (ssc): the anterior (asc), the posterior (psc), and lateral (lsc) canals, and crista are absent presenting a single vacuous chamber (D–F, J–L, and N,P). Panel R shows a reduced canal (presumed the anterior) with an associated crista. The absence of the SSCs is evident in hb/hb at E13 (compare V to U). cc, crus commune; ca, anterior canal crista; cd, cochlear duct; cl, lateral canal crista; ed, endolymphatic duct; sg, spiral ganglion; u, utricle; VIII, eighth cranial nerve. Scale bar = 100 μm (M–X).
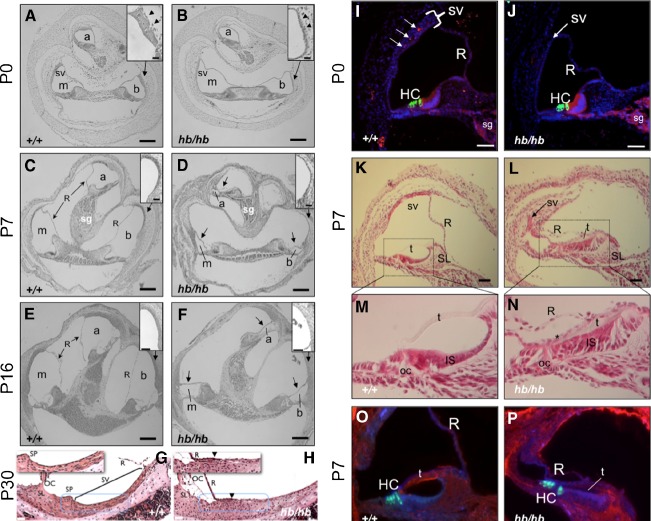
At the cochlear level, transverse sections of hb/hb ears at birth revealed an appropriately coiled cochlear duct (Figure 3B) as in the +/+ (Figure 3A) but with a poorly developed stria vascularis (compare arrowheads in Figure 3A to B and the area demarcated by dotted lines in Figure 3, G to H). It should be noted that occasionally a small portion of the stria is visible in the hb/hb mutant mice but this occurs in less than 1% of mutants tested. Further development resulted in a collapse of Reissner’s membrane by P3 (shown at P7, arrows in Figure 3, D and F) causing a reduction of the lumen of the scala media. The stria in the cochlear lateral wall is critical in endolymph formation and maintenance, is composed of marginal, intermediate and basal cells and is rich in capillary vessels (Spector and Carr 1979; Takeuchi et al. 2000; Wangemann 2006). Analysis of the vasculature revealed multiple capillaries of different diameters in the +/+ stria (Figure 3, I, arrows, also see 3G) that were scarce to absent in the hb/hb lateral wall (Figure 3, J and H). Importantly, the dense multiple-layered cell structure in the +/+ stria at P0 (bracket in Figure 3, I and dotted area in 3G) was diminished in the hb/hb lateral wall (Figure 3, J and dotted area in 3H). Only a closely packed marginal-like epithelial cell layer is evident in the hb/hb lateral wall (arrow in Figure 3, J, dotted area in 3H), which likely compromised strial function and collapse of Reissner’s membrane.
FIG. 3.
Defects in the stria and altered differentiation in the cochlea of head bobber ears. Transverse sections of inner ears are shown from P0 (A–B; I–J), P7 (C–D; K–P), P16 (E–F), and P30 (G–H), +/+ (A–O) and hb/hb (B–P). hb/hb have defects in the scala media. Sections through the cochlea show a poorly developed stria vascularis (sv, compare arrowheads in A and B insets) in hb/hb at P0. The scala media is further compromised by a collapse of Reissner’s membrane (R), shown at P7 (arrows in D) and P16 (arrows in F). The stria vascularis has completely lost the multilayer structure and appearing flat with reduced capillaries (compare insets in C to D and panels G to H). Moreover, blood vessels are readily visualized in wild-type (I) but severely reduced in the stria (sv) of hb/hb ears (J) as detected by anti-cadherin immunostaining (red) at P0. The stria in hb/hb (arrow in J) is significantly thinned compared with the DAPI-stained nuclei in the wild-type (bracketed in I). In the organ of Corti (oc), hb/hb hair cells (HC) and spiral ganglion (SG) express appropriate differentiation markers; compare myosin VII (green) and cadherin (red) in J to I. At P7, the hb/hb stria (arrow in L) is a remnant compared with the wild-type (K) stria at the apical turn and Reissner’s (R), and tectorial (t) membranes are collapsed onto the poorly differentiated inner sulcus (IS) (compare K to L, M to N and O to P). There also is a reduced expression of fibronectin (blue) in the hb/hb tectorial membrane (compare t in P to O). a, apical scala; b, basal scala; m, media scala; sg, spiral ganglion. GSLI lectin (red); SL, spiral limbus, SP, spiral prominence. Scale bar = 100 μm (A–F); 20 μm (insets in A, B; C, D; E, F, and in panels G–H); or 50 μm (I–P).
Analysis of the organ of Corti revealed the presence of differentiating inner and outer hair cells at P0 (Figure 3, I and J) and even after collapse of Reissner’s membrane (Figure 3, O and P). However, there was a delay in development of the inner sulcus (IS) in the apical turn of hb/hb cochlear ducts (Figure 3, L and N) compared with the +/+ (Figure 3, K and M). The tectorial membrane (t in Figure 3, P), which now resided between the collapsed Reissner’s, an unidentified epithelial layer (asterisk in Figure 3, N) and the inner sulcus cells, stained with reduced intensity for fibronectin (compare Figure 3, P and O). Taken together, the morphological and histological findings in the hb/hb inner ear are consistent with functional problems of hearing and balance.
Since the head bobber line has been maintained through >10 generations of reciprocal backcrosses (hb/hb x FVB) and the changes in inner ear development are only seen in homozygotes and the hemizygous animals are phenotypically normal, we assumed that the hb/hb phenotype was the result of recessive insertional inactivation of a locus essential for development of the inner ear. We proceeded to characterize the transgene integration site and flanking genomic regions by Southern analysis. Using a restriction enzyme (EcoRI) that cut once within the transgene in combination with a probe to the neo gene (N in Figure 1, G) we detected two bands indicating a single integration site in the hb/hb genome (Figure 1, F). The 8.4 kb band (Figure 1, F) was produced by a multicopy head-to-tail concatamer of the transgene (Figure 1, G) while the 10.5 kb band (Figure 1, F) contained one copy of the 5′ transgene fragment plus flanking genomic sequences (Figure 1, G). From densitometric comparison of the band intensities we estimated that there were ~3 copies of the transgene integrated in the genome. We could also conclude that there was an EcoRI site about 6.5 kb upstream from the transgene (Figure 1, G). Using multiple restriction digests and probes that hybridized either with the 5′ or the 3′ region of the transgene (data not shown), we next constructed a restriction map of the region flanking the transgene insertion site (Figure 1, H).
To identify the head bobber integration site and the potential causative gene(s), we cloned the genomic sequences flanking the transgene insertion site. We used PCR-based chromosome walking using pools of primers (Figure 5, G) and specific β-actin primers (Figure 5, G) to amplify and clone flanking sequences from hb/hb genomic DNA. One sequence corresponded to the multiple copy transgene insert (Figure 5, G, 1 kb band with ACP2) and the other (Figure 5, G, 500 bp band with ACP4) contained the transgene and flanking sequence (junction at nucleotide 139,061,196) from the distal part of chromosome 7(F3) region (Figure 5, G), which was encouraging since preliminary FISH (Fluorescence In Situ Hybridization) analysis had revealed a single integration site on chromosome 7 (data not shown). These data also revealed that the transgene inserted in a head-to-tail fashion with the β-actin at the 5′ proximal and neo in the 3′ distal region relative to the centromere. No annotated genes are located within the 100,000 bp that flank the transgene sequence (at the β-actin side) (Figs. 4, F and 5, B).
FIG. 4.
The head bobber locus on chromosome 7 contains a duplicated fragment of chromosome X. A Chromosome-7- (133J5, red) and chromosome-X (436I3, green)-specific BAC clones are seen to hybridize to their respective chromosomal loci in wild-type (wt) male spleen cells, whereas in female hb/hb cells, the chromosome 7 BAC clone (436J3, red) also co-localizes with the chromosome X (133J5, green) clone (B). Both chromosome 7 (green in C) and chromosome X (green in D) BAC clones co-localize with the p88 vector on the chromosome 7 from hb/hb spleen cells. E Splice variants of the Reps2 gene are appropriately expressed in hb/hb ears as measured by RT-PCR (see “Methods” for primers used). Blast hit, sequence comparison location. F Diagram of the head bobber locus (red line) showing nucleotide positions of the transgene integration site and junctions on chromosome 7 having three copies of the transgene (green boxes, neo; blue boxes, β-actin cassette). chr7, chromosome 7; chrX, chromosome X.
Since the chromosome walking primers were unsuccessful to amplify the 3′ Neo flanking sequences, we next employed inverse PCR with primers specific to the neo gene (Figure 5, H) and a DraI digest (Figure 5, H) to successfully amplify a 750 bp fragment. This sequence mapped the 3′ flanking sequences to the distal part of chromosome X (F4) with the junction at nucleotide 159,067,369 (NT_039718.7) of intron 1 of the RESP2 gene (Figure 5, H). Our results suggested that the sequence flanking the site of integration at the 5′ region was in chromosome 7 (F3) while the 3′ region was flanked by sequence from the X chromosome (F4) in the head bobber genome. This was unexpected since Southern analyses (Figure 1, F) and metaphase FISH analyses suggested a single integration site on chromosome 7 (Figure 4).
To confirm that we had cloned the site of integration and to clarify how sequences from chromosome X flanked the transgene on chromosome 7 we performed FISH analysis on metaphase spreads of spleen cells from wild type or hb/hb mice using the transgene vector (p88) and bacterial artificial chromosomes (BACs) for chromosome 7 (F3) (RP23–390L14 or RP23–133J5) and chromosome X (F4) (RP23–436I3) as probes. We first confirmed that the BAC clones for chromosome 7 (F3) (RP23–133J5) and chromosome X (F4) (RP23–436I3) uniquely mapped to the appropriate chromosome sub-regions in wild type (wt) chromosomes from a male mouse (Figure 4A). The same BAC clones were then hybridized to chromosomes from an hb/hb female (Figure 4B). The chromosome X BAC clone (RP23–436I3) hybridized not only to chromosome X but also to chromosome 7 and co-localized with the chromosome 7 BAC clone (RP23–133J5). These results indicated that the hb/hb line had a normal chromosome X with a duplication and insertion of a fragment of the chromosome XF4 to chromosome 7F3. We also confirmed that both chromosome 7 BAC clones (RP23–390L14 and RP23–133J5 from chr7F3) colocalized with the transgene vector (p88) (RP23–390L14 shown in Figure 4C) and that the transgene vector (p88) also colocalized with the chromosome X BAC clone (RP23–436I3, Figure 4D) on female hb/hb chromosomes at chromosome 7(F3).
By comparing the signal intensities labeled by the chromosome X BAC clone (RP23–436I3) on the chromosome X and chromosome 7 in hb/hb cells, we estimated that the translocated chromosome X fragment was smaller than the full-length chromosome X BAC clone. This was confirmed by direct PCR chromosome walking of genomic DNA flanking the insertion site and of the Reps2 gene, which yielded a hybrid junction fragment (transgene and intron 1 of REPS2 sequences, Figure 4E) using DNA from hemizygote or hb/hb mice and as expected no band was detected from wild type mice (data not shown). We also determined that sequences on chromosome X proximal and distal (up to 50 kb) to the insertion site were present in hb/hb cells by PCR, suggesting that a duplication existed on chromosomal 7 (data not shown and see aCGH results). This was supported by confirmation that the Reps2 gene was appropriately transcribed in hb/hb ear tissues. Reverse transcription polymerase chain reaction (RT-PCR) analysis was conducted on P0 ear tissues, and both Reps2 splice variants were expressed in hb/hb just as in wild-type ears (Figure 4E) indicating that Resp2 is not responsible for the head bobber phenotype. These data indicate the hb/hb mice have a normal chromosome X in addition to the abnormal chimeric chromosome 7 (Figure 4F).
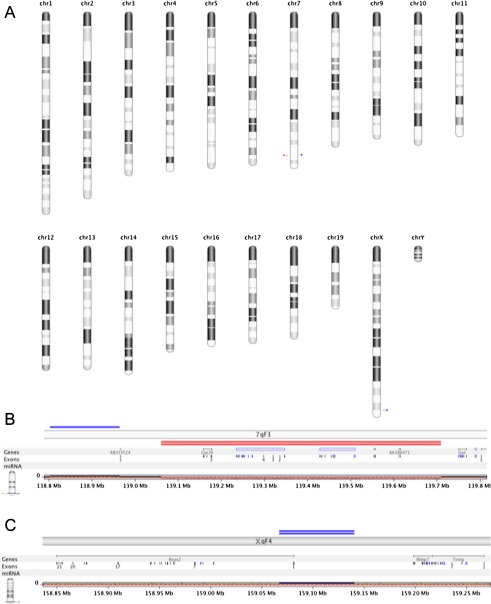
Cloning and identification of the insertion site and junctions allowed us to design a simple PCR strategy to genotype the head bobber litters (Figure 5, A and see “Methods”). We next characterized the integration site on chromosome 7 to determine which gene(s), if any, were affected. We performed a systematic chromosome walk in the 5′ and 3′ directions flanking the insertion site to determine the limits of the deletion (see “Methods”) and defined it to be about 648 kbp spanning from 139,061 to 139,709 kbp of chromosome 7 (Figure 5, B, C). These genomic changes were confirmed and refined by performing aCGH between hb/hb and wild-type genomic DNA on Agilent aCGH microarrays (see “Methods” and Figure 6). Statistical frequency testing identified copy number variations with homozygous loss of chromosome 7 region (ch7/qF3 139,060,510–139,709,028, GISTIC Q-bound of 0.0 and G-score of 9.477) spanning the deletion. Two other significant copy number variations were identified: a gain of ch7/qF3 138,802,774–138,964,543 and a gain of chX/qF4 159,067,363–159,140,649. There were no other regions with statistically significant variation between the genotypes. There are three known genes (Gpr26, Cpxm2, and Chst15) and four predicted open reading frames (AK080057, AK086971, and GM10584 AK0421584) within the deleted region of chromosome 7. These genes and multiple genes proximal and distal to the deletion site were analyzed for expression in ear tissues at E11.5 and P0 by semi-quantitative RT-PCR analysis. The predicted genes were not detected in either +/+ or hb/hb ear tissues at any stage (data not shown). RT-PCR analysis at E11.5 detected Gpr26, Cpxm2, and Chst15 transcripts in the developing +/+ ear, but no transcripts were found in hb/hb ears (Figure 5, D). Genes proximal (Nkx5-1, Nkx5.2, Bub3) and distal (Oat, Fam53b, Mettl10, Fam175b, Zranb1, Ctb2) to the site of integration and deletion were expressed in both +/+ and hb/hb ears at E11.5 (Figure 5, D, E). At P0, Cpxm2 and Chst15 were also expressed in the +/+ ear but were not detected in hb/hb, whereas Gpr26 was absent in both +/+ and hb/hb ear tissues (Figure 5, F). At P0, Nkx5.2 and Nkx5-1 expressions were detected but were consistently lower in hb/hb ears compared with Bub3, likely reflecting some variable development of the inner ear tissue.
FIG. 6.
Comparative genomic analysis (aCGH) of hb/hb and +/+ genomes. A All chromosome view of significant genomic variations in the hb/hb genome showing loss (red arrowhead) and gain (blue arrowhead) in chromosome 7 and a gain (blue arrowhead) on the X chromosome. B View of chromosome 7qF3 region showing copy number variations. Single blue line depicts gain (ch7/qF3 138,802,774–138,964,543) and double red lines depict homozygous loss of genomic region encompassing chr7qF2: 139,060,510–139,709,028. C View of chromosome XqF4 showing a gain of chX/qF4 159,067,363–159,140,649.
Discussion
Head bobber is a unique mouse model of deafness and vestibular disorders. Homozygous transgenic mice in Family OVE7 exhibit a distinctive behavioral phenotype that includes persistent movements of the head starting at P7, hyperactive circling about 2 weeks after birth, and deafness. We named the mutation head bobber. Since the mutant phenotype segregates with homozygosity for the transgene, we have characterized the transgenic insertion site at the molecular level. The transgene, along with a small segment of mouse chromosome X, was found to have integrated toward the telomeric end of mouse chromosome 7. It is also possible that a regulatory element was deleted affecting expression of upstream or downstream targets. Likewise, the transgene promoter may have an effect in cis that is more pronounced in the homozygous mutant mice. Integration was accompanied by deletion of nearly 650 kb of genomic DNA, including three known genes—Gpr26, Cpxm2, and Chst15. These genes are therefore candidate genes for deafness and normal inner ear development.
Many transgenic mouse lines have been described in which the random integration of foreign DNA has disrupted endogenous genes that are essential for normal embryonic development (Jaenisch et al. 1983; Mark et al. 1985). While some microinjected fragments produce simple insertions (Palmiter and Brinster 1986), deletions around the insertion site are quite common (Meisler 1992). Rearrangements at the site of insertion, including inversions (Ratty et al. 1992), translocations (Francke et al. 1992), and duplications (Morgan et al. 1998) can occur. Multi-copy integrations are typically arranged in tandem, usually head-to-tail (Gordon and Ruddle 1985). The co-integration of chromosome X sequences on chromosome 7 was unexpected but suggests the transgene vector initially integrated in chromosome X but was subsequently translocated on chromosome 7 bringing along a duplicated fragment of chromosome X.
The head bobber locus, and genes therein, are conserved in many species (human, rat, chicken, Zebrafish, among others). This region of mouse chromosome 7 is syntenic to the distal part of human chromosome 10 (10q26.13). There are no known human hearing loss loci in this region. Gpr26 is an orphan member of the G protein-coupled receptors (GPCR) 1 family of integral membrane proteins with homology with transducin (Lee et al. 2000, 2001). Gpr26 has been identified to be epigenetically silenced in human gliomas, but a function has not been ascribed (Boulay et al. 2009). The GPCRs have diverse biological roles that transduce signals from a range of stimuli, including neurotransmitters (Bresnick et al. 2003; Lee et al. 2000; Lee et al. 2001). Recently, a GPCR, sphingocine-1-phosphate receptor S1P2, was demonstrated to be essential for proper functioning of the auditory and vestibular systems. Moreover, vascular defects in the stria vascularis were implicated to be a primary mechanism leading to deafness in the S1P2 receptor-null mice (Kono et al. 2007; MacLennan et al. 2006). Cpmx2 is a member of the metallocarboxypeptidase A family of digestive enzymes that are synthesized as inactive molecules with inhibitory propeptides that are cleaved to activate the enzyme. However, Cpxm2 lacks several of the predicted active site residues required for enzyme activity and may function instead as a phospholipid-binding protein (Xin et al. 1998a, b, 1997). The Chst15 gene has highest sequence identity to human and rat type II transmembrane N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase (GalNAc4S6ST) found in the Golgi system required for biogenesis of glycoproteins (Ohtake et al. 2001). It catalyzes the transfer of sulfate from 3′-phosphoadenosine 5′-phosphosulfate to position 6 of the non-reducing GalNAc(4SO(4)) residues contained in proteins such as chondroitin sulfate A and dermatan sulfate (Ohtake et al. 2001, 2003). The extent of contribution of each of these genes to the head bobber phenotype will require further investigation. The malformations of the semicircular canal structures and absence of vestibular ganglion are consistent with the hyperactivity and circling behavior, but a possible involvement of the CNS cannot be ruled out since Gpr26 has extensive expression in the brain (Lee et al. 2000).
Similar to head bobber mice, single- and double-deletion mutations in the Hmx3 (Nkx5-1) and Hmx2 (Nkx5.2) genes result in mice that have hyperactivity, head tilting, and circling behaviors but do not have defects in the stria or collapse of Reissner’s membrane (Hadrys et al. 1998; Wang et al. 2001, 2004, 1998). In the ear, the defects affect cell fate and vestibular morphogenesis, including development of the vestibular ganglion (Hadrys et al. 1998; Wang et al. 2001, 2004, 1998). Since the anterior canal is first to develop and the lateral last (Martin and Swanson 1993; Sher 1971), development of the remnant anterior canal in hb/hb suggests that semicircular canal development was initiated but was not maintained. Consistent with this hypothesis, the hmx genes were initially expressed at normal levels in hb/hb ears at the onset of semicircular canal development but are not maintained at normal levels, which may be a result of the lack of developing vestibular tissues or that Hmx3 and Hmx2 each contribute to the vestibular defects in head bobber mice. Nonetheless, because these transcripts are never absent in hb/hb ears and Hmx2 and Hmx3 mutants do not have similar cochlear defects, the head bobber gene or genes functionally act upstream.
The deafness in head bobber mice is categorized as severe (71–90-db loss) cochlear dysfunction and can be explained by the cochlear defects, in particular, collapse of the scala media. The formation of the endolymph is a multistep process involving ion pumps, transporters, and channels in the stria vascularis (Hibino and Kurachi 2006; Steel 1995; Takeuchi et al. 2000; Wangemann 2006). Similar alterations of the scala media that resulted in deafness were reported in mice lacking the Kir4.1 potassium channel subunit of the strial intermediate cells (Rozengurt et al. 2003) and the NKCC1 co-transporter of the strial marginal cells (Flagella et al. 1999). Thus, deafness in head bobber is likely due to the absence of a functional stria vascularis, which is indispensable for the generation and maintenance of the endocochlear potential, and its dysfunction results in hearing deficiencies in rodent models (Ohlemiller 2009) and in humans (Boulay et al. 2009; Ishiyama et al. 2007).
In conclusion, we have described a new mouse locus that is crucial for normal development of the semicircular canals, the vestibule, and the stria vascularis. We have used PCR-based strategies to define the critical region in the genome and have identified at least three candidate genes that are expressed at the appropriate stage of inner ear development and thus are candidate genes responsible for the deafness and vestibular dysfunction observed in the head bobber mice.
Electronic supplementary material
(DOC 64 kb)
Acknowledgments
We would like to thank all members of the Pereira lab and Dr. Venkatesh Govindarajan for critique of the manuscript. We would also like to thank Karen Steel for communicating unpublished data. This work was supported by Huffington Center on Aging funds and a pilot grant from the NOHR Foundation. We thank the Baylor College of Medicine Genome and RNA profiling core for microarray and data analysis.
References
- Ahmed ZM, Riazuddin S, Bernstein SL, Ahmed Z, Khan S, Griffith AJ, Morell RJ, Friedman TB, Wilcox ER. Mutations of the protocadherin gene PCDH15 cause Usher syndrome type 1F. Am J Hum Genet. 2001;69(1):25–34. doi: 10.1086/321277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alagramam KN, Murcia CL, Kwon HY, Pawlowski KS, Wright CG, Woychik RP. The mouse Ames Waltzer hearing-loss mutant is caused by mutation of Pcdh15, a novel protocadherin gene. Nat Genet. 2001;27(1):99–102. doi: 10.1038/83837. [DOI] [PubMed] [Google Scholar]
- Alagramam KN, Yuan H, Kuehn MH, Murcia CL, Wayne S, Srisailpathy CR, Lowry RB, Knaus R, Laer L, Bernier FP, Schwartz S, Lee C, Morton CC, Mullins RF, Ramesh A, Camp G, Hageman GS, Woychik RP, Smith RJ, Hagemen GS. Mutations in the novel protocadherin PCDH15 cause Usher syndrome type 1F. Hum Mol Genet. 2001;10(16):1709–1718. doi: 10.1093/hmg/10.16.1709. [DOI] [PubMed] [Google Scholar]
- Anagnostopoulos AV. A compendium of mouse knockouts with inner ear defects. Trends Genet. 2002;18(10):499. doi: 10.1016/S0168-9525(02)02753-1. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. “Controlling the false discovery rate: a practical and powerful approach to multiple testing”. J R Stat Soc Ser Methodol. 1995;57(1):289–300. [Google Scholar]
- Beroukhim R, et al. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc Natl Acad Sci U S A. 2007;104(50):20007–20012. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bok J, Chang W, Wu DK. Patterning and morphogenesis of the vertebrate inner ear. Int J Dev Biol. 2007;51(6–7):521–533. doi: 10.1387/ijdb.072381jb. [DOI] [PubMed] [Google Scholar]
- Boulay JL, Ionescu MC, Sivasankaran B, Labuhn M, Dolder-Schlienger B, Taylor E, Morin P, Jr, Hemmings BA, Lino MM, Jones G, Maier D, Merlo A. The 10q25.3-26.1 G protein-coupled receptor gene GPR26 is epigenetically silenced in human gliomas. Int J Oncol. 2009;35(5):1123–1131. doi: 10.3892/ijo_00000428. [DOI] [PubMed] [Google Scholar]
- Bresnick JN, Skynner HA, Chapman KL, Jack AD, Zamiara E, Negulescu P, Beaumont K, Patel S, McAllister G. Identification of signal transduction pathways used by orphan g protein-coupled receptors. Assay Drug Dev Technol. 2003;1(2):239–249. doi: 10.1089/15406580360545053. [DOI] [PubMed] [Google Scholar]
- Colledge N, Lewis S, Mead G, Sellar R, Wardlaw J, Wilson J. Magnetic resonance brain imaging in people with dizziness: a comparison with non-dizzy people. J Neurol Neurosurg Psychiatry. 2002;72(5):587–589. doi: 10.1136/jnnp.72.5.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekete DM, Wu DK. Revisiting cell fate specification in the inner ear. Curr Opin Neurobiol. 2002;12(1):35–42. doi: 10.1016/S0959-4388(02)00287-8. [DOI] [PubMed] [Google Scholar]
- Feldkamp LA, Davis LC, Kress JW (1984) “Practical cone-beam algorithm,” J. Opf. Soc. Amer. A, vol. 1, no. 6, pp 612–619
- Flagella M, Clarke LL, Miller ML, Erway LC, Giannella RA, Andringa A, Gawenis LR, Kramer J, Duffy JJ, Doetschman T, Lorenz JN, Yamoah EN, Cardell EL, Shull GE. Mice lacking the basolateral Na-K-2Cl cotransporter have impaired epithelial chloride secretion and are profoundly deaf. J Biol Chem. 1999;274(38):26946–26955. doi: 10.1074/jbc.274.38.26946. [DOI] [PubMed] [Google Scholar]
- Flaherty L. Congenic strains. In: Foster HL, Small JD, Fox JG, editors. The mouse in biomedical research. New York: Academic; 1981. pp. 215–222. [Google Scholar]
- Francke U, Hsieh CL, Kelly D, Lai E, Popko B. Induced reciprocal translocation in transgenic mice near sites of transgene integration. Mamm Genome. 1992;3(4):209–216. doi: 10.1007/BF00355721. [DOI] [PubMed] [Google Scholar]
- Gordon JW, Ruddle FH. DNA-mediated genetic transformation of mouse embryos and bone marrow—a review. Gene. 1985;33(2):121–136. doi: 10.1016/0378-1119(85)90087-3. [DOI] [PubMed] [Google Scholar]
- Hadrys T, Braun T, Rinkwitz-Brandt S, Arnold HH, Bober E. Nkx5-1 controls semicircular canal formation in the mouse inner ear. Development. 1998;125(1):33–39. doi: 10.1242/dev.125.1.33. [DOI] [PubMed] [Google Scholar]
- Hardisty RE, Hughes DC, Steel KP.The characterization of the head bobber mouse mutant Br J Audiol 199731273–132. 10.3109/030053640000000129183385 [DOI] [Google Scholar]
- Hasson T, Mooseker MS. Porcine myosin-VI: characterization of a new mammalian unconventional myosin. J Cell Biol. 1994;127(2):425–440. doi: 10.1083/jcb.127.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibino H, Kurachi Y. Molecular and physiological bases of the K+ circulation in the mammalian inner ear. Physiology (Bethesda) 2006;21:336–345. doi: 10.1152/physiol.00023.2006. [DOI] [PubMed] [Google Scholar]
- Hughes D, Hardisty R, Steel KP. Characterisation of the mouse mutant head bobber (hb) Hered Deaf News. 1998;15:35. [Google Scholar]
- Ishiyama G, Tokita J, Lopez I, Tang Y, Ishiyama A. Unbiased stereological estimation of the spiral ligament and stria vascularis volumes in aging and Meniere’s disease using archival human temporal bones. J Assoc Res Otolaryngol. 2007;8(1):8–17. doi: 10.1007/s10162-006-0057-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R, Harbers K, Schnieke A, Lohler J, Chumakov I, Jahner D, Grotkopp D, Hoffmann E. Germline integration of moloney murine leukemia virus at the Mov13 locus leads to recessive lethal mutation and early embryonic death. Cell. 1983;32(1):209–216. doi: 10.1016/0092-8674(83)90511-1. [DOI] [PubMed] [Google Scholar]
- Kono M, Belyantseva IA, Skoura A, Frolenkov GI, Starost MF, Dreier JL, Lidington D, Bolz SS, Friedman TB, Hla T, Proia RL. Deafness and stria vascularis defects in S1P2 receptor-null mice. J Biol Chem. 2007;282(14):10690–10696. doi: 10.1074/jbc.M700370200. [DOI] [PubMed] [Google Scholar]
- Lee DK, Lynch KR, Nguyen T, Im DS, Cheng R, Saldivia VR, Liu Y, Liu IS, Heng HH, Seeman P, George SR, O’Dowd BF, Marchese A. Cloning and characterization of additional members of the G protein-coupled receptor family. Biochim Biophys Acta. 2000;1490(3):311–323. doi: 10.1016/s0167-4781(99)00241-9. [DOI] [PubMed] [Google Scholar]
- Lee DK, Nguyen T, Lynch KR, Cheng R, Vanti WB, Arkhitko O, Lewis T, Evans JF, George SR, O’Dowd BF. Discovery and mapping of ten novel G protein-coupled receptor genes. Gene. 2001;275(1):83–91. doi: 10.1016/S0378-1119(01)00651-5. [DOI] [PubMed] [Google Scholar]
- MacLennan AJ, Benner SJ, Andringa A, Chaves AH, Rosing JL, Vesey R, Karpman AM, Cronier SA, Lee N, Erway LC, Miller ML. The S1P2 sphingosine 1-phosphate receptor is essential for auditory and vestibular function. Hear Res. 2006;220(1–2):38–48. doi: 10.1016/j.heares.2006.06.016. [DOI] [PubMed] [Google Scholar]
- Mark WH, Signorelli K, Lacy E. An insertional mutation in a transgenic mouse line results in developmental arrest at day 5 of gestation. Cold Spring Harb Symp Quant Biol. 1985;50:453–463. doi: 10.1101/SQB.1985.050.01.057. [DOI] [PubMed] [Google Scholar]
- Martin P, Swanson GJ. Descriptive and experimental analysis of the epithelial remodellings that control semicircular canal formation in the developing mouse inner ear. Dev Biol. 1993;159(2):549–558. doi: 10.1006/dbio.1993.1263. [DOI] [PubMed] [Google Scholar]
- Meisler MH. Insertional mutation of ‘classical’ and novel genes in transgenic mice. Trends Genet. 1992;8(10):341–344. doi: 10.1016/0168-9525(92)90278-C. [DOI] [PubMed] [Google Scholar]
- Morgan D, Turnpenny L, Goodship J, Dai W, Majumder K, Matthews L, Gardner A, Schuster G, Vien L, Harrison W, Elder FF, Penman-Splitt M, Overbeek P, Strachan T. Inversin, a novel gene in the vertebrate left-right axis pathway, is partially deleted in the inv mouse. Nat Genet. 1998;20(2):149–156. doi: 10.1038/2450. [DOI] [PubMed] [Google Scholar]
- Mouse Genome Informatics (MGI) 2447989: http://www.informatics.jax.org/javawi2/servlet/WIFetch?page=alleleDetail&key=22145http://www.informatics.jax.org/searchtool/Search.do?query=MGI:2447989
- Ohlemiller KK. Mechanisms and genes in human strial presbycusis from animal models. Brain Res. 2009;1277:70–83. doi: 10.1016/j.brainres.2009.02.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtake S, Ito Y, Fukuta M, Habuchi O. Human N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase cDNA is related to human B cell recombination activating gene-associated gene. J Biol Chem. 2001;276(47):43894–43900. doi: 10.1074/jbc.M104922200. [DOI] [PubMed] [Google Scholar]
- Ohtake S, Kimata K, Habuchi O. A unique nonreducing terminal modification of chondroitin sulfate by N-acetylgalactosamine 4-sulfate 6-o-sulfotransferase. J Biol Chem. 2003;278(40):38443–38452. doi: 10.1074/jbc.M306132200. [DOI] [PubMed] [Google Scholar]
- Palmiter RD, Brinster RL. Germ-line transformation of mice. Annu Rev Genet. 1986;20:465–499. doi: 10.1146/annurev.ge.20.120186.002341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y, Pereira FA, DeMayo FJ, Lydon JP, Tsai SY, Tsai MJ. Null mutation of mCOUP-TFI results in defects in morphogenesis of the glossopharyngeal ganglion, axonal projection, and arborization. Genes Dev. 1997;11(15):1925–1937. doi: 10.1101/gad.11.15.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratty AK, Matsuda Y, Elliott RW, Chapman VM, Gross KW. Genetic mapping of two DNA markers, D16Ros1 and D16Ros2, flanking the mutation site in the chakragati mouse, a transgenic insertional mutant. Mamm Genome. 1992;3(1):5–10. doi: 10.1007/BF00355834. [DOI] [PubMed] [Google Scholar]
- Rozengurt N, Lopez I, Chiu CS, Kofuji P, Lester HA, Neusch C. Time course of inner ear degeneration and deafness in mice lacking the Kir4.1 potassium channel subunit. Hear Res. 2003;177(1–2):71–80. doi: 10.1016/S0378-5955(02)00799-2. [DOI] [PubMed] [Google Scholar]
- Shaffer LG, Kennedy GM, Spikes AS, Lupski JR. Diagnosis of CMT1A duplications and HNPP deletions by interphase FISH: implications for testing in the cytogenetics laboratory. Am J Med Genet. 1997;69(3):325–331. doi: 10.1002/(SICI)1096-8628(19970331)69:3<325::AID-AJMG20>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Sher AE. The embryonic and postnatal development of the inner ear of the mouse. Acta Otolaryngol Suppl. 1971;285:1–77. [PubMed] [Google Scholar]
- Smith RJ, Bale JF, Jr, White KR. Sensorineural hearing loss in children. Lancet. 2005;365(9462):879–890. doi: 10.1016/S0140-6736(05)71047-3. [DOI] [PubMed] [Google Scholar]
- Somma G, Alger H, Stankiewicz P, Yatsenko SA, Overbeek, PA, Pereira FA (2005) Association for Research in Otolaryngology mid-winter meeting, New Orleans, Louisiana, Feb.19–24
- Spector GJ, Carr C. The ultrastructural cytochemistry of peroxisomes in the guinea pig cochlea: a metabolic hypothesis for the stria vascularis. Laryngoscope. 1979;89(6 Pt 2 Suppl 16):1–38. doi: 10.1288/00005537-197906001-00001. [DOI] [PubMed] [Google Scholar]
- Steel KP. Inherited hearing defects in mice. Annu Rev Genet. 1995;29:675–701. doi: 10.1146/annurev.ge.29.120195.003331. [DOI] [PubMed] [Google Scholar]
- Takeuchi S, Ando M, Kakigi A. Mechanism generating endocochlear potential: role played by intermediate cells in stria vascularis. Biophys J. 2000;79(5):2572–2582. doi: 10.1016/S0006-3495(00)76497-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Lufkin T. Hmx homeobox gene function in inner ear and nervous system cell-type specification and development. Exp Cell Res. 2005;306(2):373–379. doi: 10.1016/j.yexcr.2005.03.016. [DOI] [PubMed] [Google Scholar]
- Wang W, Water T, Lufkin T. Inner ear and maternal reproductive defects in mice lacking the Hmx3 homeobox gene. Development. 1998;125(4):621–634. doi: 10.1242/dev.125.4.621. [DOI] [PubMed] [Google Scholar]
- Wang W, Chan EK, Baron S, Water T, Lufkin T. Hmx2 homeobox gene control of murine vestibular morphogenesis. Development. 2001;128(24):5017–5029. doi: 10.1242/dev.128.24.5017. [DOI] [PubMed] [Google Scholar]
- Wang W, Grimmer JF, Water TR, Lufkin T. Hmx2 and Hmx3 homeobox genes direct development of the murine inner ear and hypothalamus and can be functionally replaced by Drosophila Hmx. Dev Cell. 2004;7(3):439–453. doi: 10.1016/j.devcel.2004.06.016. [DOI] [PubMed] [Google Scholar]
- Wangemann P. Supporting sensory transduction: cochlear fluid homeostasis and the endocochlear potential. J Physiol. 2006;576(Pt 1):11–21. doi: 10.1113/jphysiol.2006.112888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin X, Varlamov O, Day R, Dong W, Bridgett MM, Leiter EH, Fricker LD. Cloning and sequence analysis of cDNA encoding rat carboxypeptidase D. DNA Cell Biol. 1997;16(7):897–909. doi: 10.1089/dna.1997.16.897. [DOI] [PubMed] [Google Scholar]
- Xin X, Day R, Dong W, Lei Y, Fricker LD. Cloning, sequence analysis, and distribution of rat metallocarboxypeptidase Z. DNA Cell Biol. 1998;17(4):311–319. doi: 10.1089/dna.1998.17.311. [DOI] [PubMed] [Google Scholar]
- Xin X, Day R, Dong W, Lei Y, Fricker LD. Identification of mouse CPX-2, a novel member of the metallocarboxypeptidase gene family: cDNA cloning, mRNA distribution, and protein expression and characterization. DNA Cell Biol. 1998;17(10):897–909. doi: 10.1089/dna.1998.17.897. [DOI] [PubMed] [Google Scholar]
- Zheng QY, Yu H, Washington JL, 3rd, Kisley LB, Kikkawa YS, Pawlowski KS, Wright CG, Alagramam KN. A new spontaneous mutation in the mouse protocadherin 15 gene. Hear Res. 2006;219(1–2):110–120. doi: 10.1016/j.heares.2006.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC 64 kb)