Abstract
Point mutations in the KIT receptor tyrosine kinase gene have recently been identified in mucosal, acral lentiginous, and chronically sun-damaged melanomas. We have identified the first human melanoma cell line with an endogenous L576P mutation, the most common KIT mutation in melanoma (∼30-40%). In vitro testing demonstrated that the cell viability of the L576P mutant cell line was not reduced by imatinib, nilotinib or sorafenib small molecule KIT inhibitors effective in non-melanoma cells with other KIT mutations. However, the viability of the mutant cells was reduced by dasatinib at concentrations as low as 10 nM (P =0.004). Molecular modeling studies found that the L576P mutation induces structural changes in KIT that reduce the affinity for imatinib (ΔΔGbind = -2.52 kcal/mol) but not for dasatinib (ΔΔGbind = +0.32 kcal/mol). Two metastatic melanoma patients with the L576P KIT mutation were treated with dasatinib, including one patient previously treated with imatinib. Both patients had marked reduction (>50%) and elimination of tumor FDG-avidity by PET imaging after dasatinib treatment. This data supports the selective inhibitory effect of dasatinib against cells harboring the most common KIT mutation in melanoma, and thus has therapeutic implications for acral lentiginous, chronic sun damaged, and mucosal melanomas.
Keywords: KIT, acral lentiginous, mucosal, chronic sun-damaged, melanoma, dasatinib
Introduction
Patients with metastatic melanoma have a median survival of 6-8 months (1). Unfortunately multiple clinical trials with chemotherapy, immunotherapy, and biochemotherapy have failed to significantly improve survival. Protein kinase inhibitors are beneficial in diseases with highly prevalent oncogenic events (e.g., CML, GIST), and within selected sub-populations (e.g., HER2-amplified breast cancer) (2). Over 50% of melanomas arising from areas without chronic sun damage harbor activating mutations of BRAF (3). However, BRAF mutations are extremely rare in other melanoma subtypes: acral lentiginous (AL), chronic sun-damaged (CSD) and mucosal (4). Interestingly, amplification of chromosomal region 4q12 was seen frequently and selectively in AL, CSD and mucosal melanomas. Interrogation of candidate genes in this region led to the discovery of frequent mutations and/or amplifications of the KIT tyrosine kinase receptor gene in these subtypes (5).
The identification of KIT mutations in melanoma has direct therapeutic implications. Activating KIT mutations are present in about 85-90% of gastrointestinal stromal tumors (GIST) (6). Treatment with the KIT inhibitor imatinib significantly improved survival in GIST patients, and it is now the standard of care for this disease (7). Three previous clinical trials using imatinib in unselected melanoma patients failed to demonstrate clinical benefit (8, 10). However, it is possible that KIT inhibitors will be beneficial to the subset of melanoma patients with KIT mutations. Recent case reports have described clinical responses following treatment with imatinib in two patients with metastatic mucosal melanoma with KIT K642E and KIT 577 PYDHKWE duplication mutations, respectively (11, 12). We reported a complete response in a metastatic mucosal melanoma patient with a KIT V560D mutation who was treated with a sorafenib-based regimen, which also inhibits KIT (13).
While these early responses are encouraging, there are characteristics of KIT mutations in melanoma that suggest imatinib resistance may be an issue in the treatment of these patients. In GIST, the majority (∼80%) of KIT mutations occur in the juxtamembrane regulatory domain encoded by exon 11. Most of these mutations have been characterized both in vitro and clinically as being imatinib-sensitive. In contrast, imatinib resistant mutations occur in exons 13 and 17, the kinase domains of the protein. These mutations are rare in GIST (exon 13, <1%; exon 17, 1%). The KIT mutations identified in melanomas occur in the same exons as those affected in GIST. However, there is a greater prevalence of mutations in exon 13 (20%) and exon 17 (10%) (5, 11-19). Additionally, in GIST >90% of the observed mutations are deletions or insertions, whereas >90% of the KIT mutations in melanoma are point mutations. The L576P KIT mutation, which is the most common KIT mutation reported to date in melanoma (∼30–40% of mutations), is located at the c-terminus of KIT exon 11, while KIT exon 11 deletions in GIST occur mostly at the n-terminus of KIT exon 11 (5). These differences in the type and localization of KIT mutations may impact drug efficacy.
Here we report the identification and characterization of the first human melanoma cell line (WM3211) with a L576P KIT mutation. The L576P mutation is the most common KIT mutation identified to date in melanoma, and represents approximately 30-40% of the reported point mutations. Despite the location of the L576P mutation in exon 11, which is generally associated with imatinib sensitivity in GIST, we observed that the WM3211 cells were markedly resistant to the growth inhibitory effects of imatinib. In contrast, the cell line was sensitive to dasatinib, a structurally distinct inhibitor of KIT. We also report here the results of molecular modeling studies to investigate the structural effects of the mutation on the interaction with KIT inhibitors, and describe the treatment of two metastatic mucosal melanoma patients with this mutation.
Results
Identification and Characterization of L576P KIT Mutant Cell Line
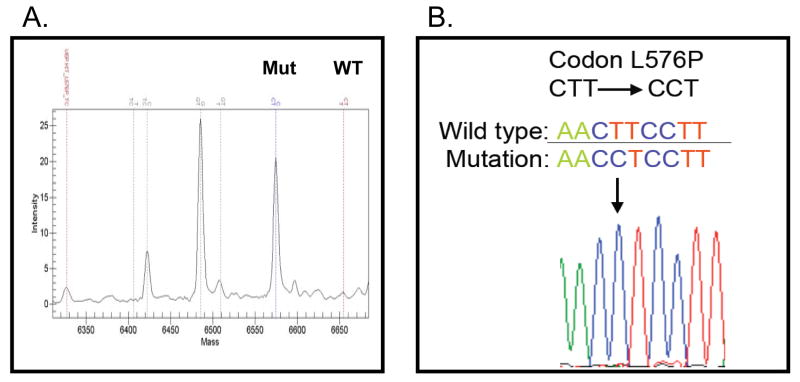
Mass-spectroscopy based genotyping was used to screen a panel of 65 human melanoma cell lines for point mutations in KIT previously reported in melanoma and other diseases (Supplemental Table 1). We identified one cell line, WM3211, that harbors a point mutation at nucleic acid residue 1727 in exon 11 of KIT, which results in the L576P amino acid substitution (Figure 1A). Sanger DNA sequencing of KIT exons 11, 13, and 17 confirmed this finding, and failed to identify any other mutations (Figure 1B). Additional analysis of the WM3211 cell line by mass-spectroscopy based genotyping failed to identify activating mutations in either BRAF or NRAS, which is consistent with the pattern of these mutations in melanoma (Supplemental Table 1).
Figure 1. L576P KIT Mutation in a Melanoma Cell Line.
(A) Mass spectroscopy-based detection of the L576P KIT mutant allele(s) in the WM3211 cell line. A peak is correlated with the L576P mutant KIT (Mut), whereas there is no peak for the wild-type KIT (WT). (B) Sanger sequencing of exon 11 of KIT in the WM3211 cells. Arrow, T-to-C base pair change.
Dasatinib, But Not Imatinib, Nilotinib, or Sorafenib Inhibits Cell Viability in WM3211 Cells
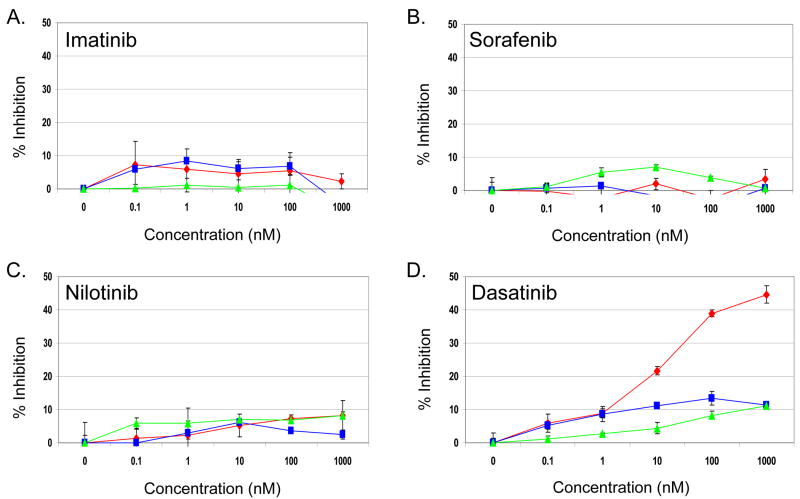
The WM3211 cell line was tested for sensitivity to a panel of FDA-approved, clinically utilized small molecule KIT inhibitors. The effect on cell viability in WM3211 cells were compared to effects on two human melanoma cell lines that have no mutations in KIT, A375 (V600E mutant BRAF, wild-type NRAS) and Mewo (wild-type BRAF and NRAS). Surprisingly, imatinib failed to inhibit the viability of WM3211 cells at concentrations up to 1 μM (<10% decrease compared to untreated cells) with similar results in the A375 and Mewo cell lines (Figure 2A). WM3211 also showed no statistically significant (P >0.05) difference in sensitivity to sorafenib and nilotinib versus the KIT wild-type melanoma cell lines (Figure 2B and C). However, dasatinib, a structurally distinct small molecule inhibitor of KIT, reduced WM3211 cell viability ∼25% (P =0.004 versus vehicle) at 10 nM and ∼50% (P = 0.00001) at 1 μM concentration (Figure 2D). In contrast, minimal inhibition of viability of the A375 and Mewo cell lines (≤15%, P = 0.9) was observed up to 1 μM concentration of dasatinib. Treatment of the T1 GIST cell line containing a heterozygous V560_Y579del in exon 11 confirmed that each of the inhibitors used for these experiments was active against a known sensitive KIT mutation (data not shown).
Figure 2. Effect of KIT inhibitors on WM3211 cell line viability.
WM3211 (◆), MEWO (■) and A375 (▲) human melanoma cells were treated with increasing doses of the KIT inhibitors (A) imatinib, (B) sorafenib, (C) nilotinib, or (D) dasatinib, x-axis, Drug concentration (nM); y-axis, Percent reduction in cell viability. Each data point represents the average of 3 replicates, error bars represent the standard deviation.
Modeling the Effects of the L576P KIT Mutation
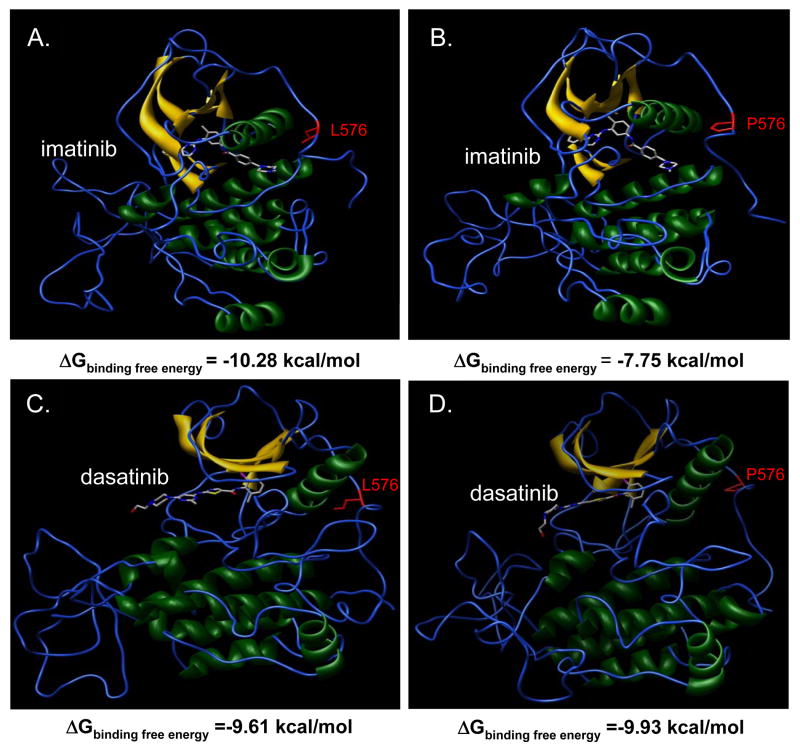
In order to investigate whether the differential sensitivity of imatinib and dasatinib observed in the WM3211 cell line may be due to altered KIT-binding affinity, molecular dynamic studies were performed to determine the effects of the L576P mutation on the structure of KIT and its interactions with the inhibitors. The free energy of binding of both inhibitors to wild-type KIT was relatively similar (ΔGbind, -10.28 kcal/mol for imatinib; ΔGbind, -9.61 kcal/mol for dasatinib) (Figures 3A, C). Modeling of the L576P mutant KIT protein demonstrated a structure that was similar to the active conformation of the molecule, which in previous studies has been associated with a diminished binding affinity for imatinib (20). Consistent with this data, imatinib had a less favorable free energy of binding for the L576P mutant form of KIT (ΔGbind, -7.75 kcal/mol) as compared to the wild-type protein, whereas the free energy of binding of dasatinib was essentially unchanged (ΔGbind, -9.93 kcal/mol) (Figures 3C, D).
Figure 3. Effects of L576P KIT mutation on protein structure and drug binding.
The interaction of imatinib with (A) wild-type versus (B) L576P KIT reveals a ΔΔGbind = -2.52 kcal/mol, reflecting a decrease in affinity induced by the mutation. The interaction of dasatinib with (C) wild-type versus (D) L576P KIT reveals a ΔΔGbind = +0.32 kcal/mol. The free binding energy for L576P KIT is more favorable for dasatinib (-9.93 kcal/mole) than for imatinib (-7.75 kcal/mole), indicating a greater affinity for dasatinib.
Treatment of 2 Patients with L576P KIT Mutant Melanoma
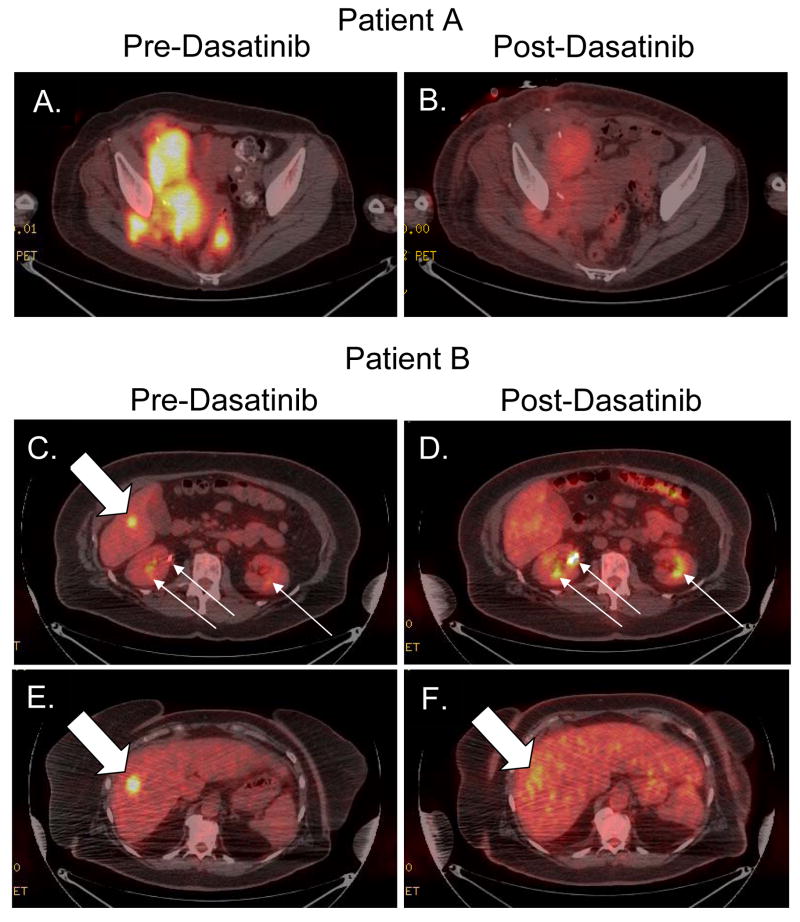
Patient A is a 55 year old woman who had resection of a labial mucosal melanoma. She received adjuvant interferon therapy for one year, but developed a recurrence in the inguinal lymph node bed, and underwent lymphadenectomy. Molecular analysis of a lymph node metastasis identified the presence of a L576P KIT mutation. The patient was then treated with adjuvant imatinib 400 mg daily, with a dose reduction to 300 mg daily after three months for periorbital edema. After eight additional months of adjuvant imatinib therapy a PET/CT showed a new FDG-avid right internal iliac lymph node representing a recurrence of melanoma while on imatinib. The patient was subsequently treated with several standard and experimental regimens for metastatic melanoma, including CRO11-vcMMAE (Anti-GPNMB antibody conjugated to auristatin E), high-dose bolus IL-2, sorafenib and temsirolimus, and combination chemotherapy with CVD (cisplatin, vinblastine, and dacarbazine). A repeat biopsy after progression on the sorafenib and temsirolimus regimen confirmed continued presence of the L576P KIT mutation, and failed to identify any secondary mutations in KIT. Based on the clinical activity of dasatinib in imatinib-resistant or –intolerant patients in other cancers, the decision was made to treat the patient off clinical protocol with dasatinib. After undergoing a pre-treatment PET scan, the patient was started on dasatinib 70 mg twice daily. After one month, the patient reported a marked improvement in her symptoms, which primarily consisted of inguinal/pelvic pain. Repeat PET/CT imaging demonstrated an overall 53.5% reduction in average SUVmax (Figure 4A, B). Unfortunately, despite this early response and clinical benefit, the patient developed progressive disease at the same sites in the pelvis after four months of therapy. No secondary KIT mutations were identified in material obtained from a biopsy of the deep pelvic tumor.
Figure 4. Treatment of L576P KIT mucosal melanoma patients with dasatinib.
FDG-PET/CT images of Patient A (A-B) and Patient B (C-F), metastatic mucosal melanoma patients, with L576P KIT mutant tumors. Images were obtained before (A, C, E) and after (B, D, F) treatment with 70 mg twice daily of dasatinib for one month. Solid block white arrows indicate FDG-avid tumors, Narrow white arrows indicate physiologic FDG excretion in renal collecting system.
Patient B is a 61 year old woman with anal melanoma who received neoadjuvant radiation therapy (5 Gy) in five fractions followed by tumor resection. A PET/CT performed just prior to radiation therapy did not show any evidence of metastatic disease. A follow-up PET/CT four months after surgery revealed two FDG-avid lesions in the right lobe of the liver. DNA sequencing of the original biopsy demonstrated the presence of a L576P KIT mutation. After one month of dasatinib therapy the lesion in the inferior aspect of the right liver lobe (SUVmax = 8.5) could not be identified (Figure 5C, D) and the lesion in the superior aspect of the right liver lobe was no longer well-circumscribed and showed a 54% reduction in SUVmax (SUVmax 11.1 → 5.1) (Figure 5E, F). The patient tolerated dasatinib therapy well, with the only complication being the development of asymptomatic pleural effusions. The patient was maintained on dasatinib for three months then treatment was stopped for the development of progressive disease at the same sites within the liver.
Methods
Cell Culture and Reagents
The WM3211 human melanoma cell line was provided by Dr. Meenhard Herlyn. A375 and MEWO human melanoma cell lines were obtained from ATCC. All cells were maintained in RPMI media with 5% FCS. The imatinib, dasatinib, nilotinib, and sorafenib were purchased from Eurasias Chemicals (Fort Oglethorpe, GA).
Mutation Detection
A mass spectroscopy–based approach evaluating single nucleotide polymorphisms (SNPs) (21) was used to detect Kit mutations (A829P, D816H, D816V, K642E, L576P, N556D, R634W, V559A, V559D, V560D, V825A, Y553N). PCR and extension primers for KIT were designed using Sequenom, Inc. Assay Design (Sequenom, San Diego, CA). PCR-amplified DNA was cleaned using EXO-SAP (Sequenom), and primer was extended by IPLEX chemistry, desalted using Clean Resin (Sequenom), and spotted onto Spectrochip matrix chips using a nanodispenser (Samsung). Chips were run in duplicate on a Sequenom MassArray MALDI-TOF MassArray system. Sequenom Typer Software and visual inspection were used to interpret mass spectra. Reactions where >15% of the resultant mass ran in the mutant site in both reactions were scored as positive. Traditional Sanger sequencing of exons 11, 13, and 17 of KIT was performed as previously described (6). Genomic DNA samples were isolated from paraffin-embedded or frozen tissue, polymerase chain reaction was performed, and mutations were identified by a 3730 9 1 DNA Analyzer (Applied Biosystems, Foster City, CA) at the MDACC Nucleic Acid Core Facility.
In Vitro Inhibitor Testing
Cells were plated in 96 well plates and treated the following day with increasing concentrations of imatinib, nilotinib, sorafenib, dasatinib, (0 – 1 μM doses), or equamolar DMSO in triplicate. After 48 hours, cell viability was determined by the Cell Titer-Blue Cell Viability Assay (Promega, Madison, WI) per the manufacturer's instructions. The relative growth of each cell line for each treatment was determined relative to mock treated control. DMSO vehicle at equamolar concentrations had no significant effect on cell viability in all lines. Calculations and graphs were made using Microsoft Excel. Statistical comparison was made using Student's t-test.
18FDG PET Imaging
Patients underwent 18FDG PET imaging as described previously (22). Images were interpreted using volumetric and multiple orthogonal projection analysis then quantified using vendor specific software. Maximal standardized uptake value (SUVmax) was measured from a region(s) of interest (ROI) representative of tumor on pretreatment images and corresponding ROIs on posttreatment images. Images were reviewed and measured by an experienced radiologist (D.A.P.). PET response was defined as >25% decrease in SUVmax (23).
Molecular Dynamics Simulations
Molecular dynamics (MD) simulations were carried out using the AMBER 9 (sander and pmemd module) suite of programs and the parm99 all-atom force field working in parallel on 64 processors of the IBM/BCX calculation cluster of the CINECA calculation centre of Bologna, as described previously (24, 25). Briefly, starting geometries for both wild-type and mutant simulations used the crystallographic coordinates of the active KIT structure in complex with imatinib. The crystallographic structure of the complex was then modified by substituting imatinib with dasatinib. The binding free energy (ΔGbind) for each inhibitor is calculated using the molecular mechanics/Poisson-Boltzmann surface area method as the sum of the electrostatic, van der Waals, polar solvation, non-polar solvation, and entopic contributions. (See Supplemental Methods).
Discussion
More effective treatments are desperately needed for patients with metastatic melanoma. The recent discovery of KIT mutations in acral lentiginous and mucosal melanoma tumors and those arising in chronically sun-damaged skin suggests that small molecule KIT inhibitors may have efficacy in these tumor types. The assumption of general imatinib-sensitivity in these tumors is largely based on the clinical experience in GIST, which is also characterized by KIT mutations. The data presented here suggests that the management of KIT-mutant melanoma may need to be further refined.
We have identified a human melanoma cell line (WM3211) with the L576P KIT mutation, the most common KIT mutation in melanoma. This mutation occurs in exon 11, a region in which the vast majority of mutations in GIST occur, and to date have been associated with imatinib sensitivity. Surprisingly, we found that the WM3211 cell line was resistant to the growth inhibitory effects of imatinib and to structurally similar inhibitors. The resistance to imatinib is at least in part due to conformational changes induced by the L576P substitution that reduces the affinity of imatinib for its binding pocket within the receptor. In contrast, the viability of WM3211 cells was reduced by nanomolar concentrations of dasatinib, a small-molecule inhibitor used in other diseases in the setting of imatinib-resistance and/or -intolerance. Despite the similar binding energy for both the wild-type and L576P KIT, dasatinib had no significant effect on the viability of Mewo or A375 melanoma cell lines that express only the wild-type KIT protein. These findings are congruent with those of Buettner et al., who showed that dasatinib had no effect on the cell viability of multiple melanoma cell lines (26). These data support the functional dependence upon dasatinib-sensitive signaling in this KIT mutant melanoma.
The differential effects of dasatinib and imatinib on melanoma cells with the L576P KIT mutation are consistent with the report of Antonescu et al. who demonstrated that stable transfection of the L576P KIT protein into murine pro-B-cell Ba/F3 cells yielded clones that were more sensitive to dasatinib than imatinib (14). Ba/F3 cells exogenously expressing the V559D KIT mutation in this model showed no difference in sensitivity between dasatinib and imatinib, indicating the specificity of the effect of dasatinib on the L576P mutant. These results correspond with the recent observation that human melanoma cells harboring a KIT V559A mutation showed growth inhibition to imatinib (27).
Dasatinib is a multi-kinase inhibitor with targets in addition to KIT. It is possible that inhibition of these additional targets along with KIT may synergize to produce the growth suppression observed in the WM3211. This possibility is an ongoing area of research. However, we did not observe significant growth inhibition in the WM3211, or a differential effect as compared the wild-type KIT melanoma cell lines, following treatment with a small molecule inhibitor of Src family kinases (data not shown), which are known targets of dasatinib. The critical nature of KIT as a target is also supported by the fact that in the work by Antonescu, et al. dasatinib only inhibited the Ba/F3 cells that were transfected with the mutant KIT constructs (personal communication).
We did not see a complete elimination of cell viability of the WM3211 with dasatinib treatment. It is possible that dasatinib-induced cell death is cell-cycle specific and thus only kills a fraction of the melanoma cells while having a cytostatic effect on cells in other phases of the cell cycle. Alternatively, the WM3211 cell line may be composed of a heterogeneous collection of cells with differential sensitivity to dasatinib. Combinatorial approaches may be required to achieve optimal results in patients with KIT mutations. Consistent with this, both metastatic melanoma patients with L576P KIT mutations treated with dasatinib demonstrated initial marked reduction in PET positivity, but had reemergence of metabolic activity and tumor growth after ∼4 months of treatment. The identification of a melanoma cell line with the most common KIT mutation in this disease presents a powerful tool for future investigations.
Patient A had a melanoma recurrence in the setting of adjuvant imatinib therapy without evidence of a secondary mutation in KIT. Given the infrequency of KIT L576P mutations in GIST (<1% of cases), there are few reported cases of treatment with imatinib, and none with dasatinib, for GIST patients with this mutation. Of the cases reported, stable disease or partial response was achievable, but often at higher doses of imatinib (28, 30). As the experience with treating KIT-mutant melanoma is in its infancy, it will be critical to treat patients in the setting of clinical trials, with documented evaluation of the responsiveness of different mutations allowing for the optimization of patient management in the future.
In summary, we have identified the first melanoma cell line that has a naturally occurring L576P KIT mutation, the most common KIT mutation observed in melanoma. Our in vitro and clinical data demonstrate that melanoma cells with this mutation are sensitive to dasatinib, but they are relatively resistant to imatinib. Our results also suggest that resistance to therapy with KIT inhibitors is likely to be an important issue in melanoma. This data has implications for the development of therapeutic strategies for AL, CSD, and mucosal melanoma patients. The results also support previous data that dasatinib may have selective activity in specific subsets of melanoma patients, which should be investigated in the setting of ongoing clinical trials with this agent. Importantly, this work represents an example of the application of bench-to-bedside research to develop new therapeutic approaches. We believe that similar interrogations of the functional alterations that are identified in this disease will be the key to improving outcomes in patients.
Supplementary Material
Acknowledgments
We are indebted to Drs. Maurie Markman and Waun Ki Hong for providing funding and commentary in support of this project.
Supported in part by MDACC Melanoma Spore Development Grant (MAD, KBK, AJL, JCT); Carol Cogdell Courtney Fellowship (SEW, MAD); Ruth L. Kirschstein National Research Service Award, NIH (SEW); ASCO Young Investigator Award (MAD); Institutional Physician-Scientist Award (JCT and AJL), NIH/NCI grant 1K23CA109060-04 (JCT); The DNA Sequencing Core Facility is supported by an NCI Cancer Center Support Grant CA-16672.
References
- 1.Tsao H, Atkins MB, Sober AJ. Management of cutaneous melanoma. N Engl J Med. 2004 Sep 2;351(10):998–1012. doi: 10.1056/NEJMra041245. [DOI] [PubMed] [Google Scholar]
- 2.Davies M, Hennessy B, Mills GB. Point mutations of protein kinases and individualised cancer therapy. Expert Opin Pharmacother. 2006 Nov;7(16):2243–61. doi: 10.1517/14656566.7.16.2243. [DOI] [PubMed] [Google Scholar]
- 3.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002 Jun 27;417(6892):949–54. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 4.Curtin JA, Fridlyand J, Kageshita T, et al. Distinct sets of genetic alterations in melanoma. N Engl J Med. 2005 Nov 17;353(20):2135–47. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- 5.Beadling C, Jacobson-Dunlop E, Hodi FS, et al. KIT gene mutations and copy number in melanoma subtypes. Clin Cancer Res. 2008 Nov 1;14(21):6821–8. doi: 10.1158/1078-0432.CCR-08-0575. [DOI] [PubMed] [Google Scholar]
- 6.McAuliffe JC, Lazar AJ, Yang D, et al. Association of intratumoral vascular endothelial growth factor expression and clinical outcome for patients with gastrointestinal stromal tumors treated with imatinib mesylate. Clin Cancer Res. 2007 Nov 15;13(22 Pt 1):6727–34. doi: 10.1158/1078-0432.CCR-07-0895. [DOI] [PubMed] [Google Scholar]
- 7.Blanke CD, Demetri GD, von Mehren M, et al. Long-term results from a randomized phase II trial of standard- versus higher-dose imatinib mesylate for patients with unresectable or metastatic gastrointestinal stromal tumors expressing KIT. J Clin Oncol. 2008 Feb 1;26(4):620–5. doi: 10.1200/JCO.2007.13.4403. [DOI] [PubMed] [Google Scholar]
- 8.Ugurel S, Hildenbrand R, Zimpfer A, et al. Lack of clinical efficacy of imatinib in metastatic melanoma. Br J Cancer. 2005 Apr 25;92(8):1398–405. doi: 10.1038/sj.bjc.6602529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim KB, Eton O, Davis DW, et al. Phase II trial of imatinib mesylate in patients with metastatic melanoma. Br J Cancer. 2008 Sep 2;99(5):734–40. doi: 10.1038/sj.bjc.6604482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wyman K, Atkins MB, Prieto V, et al. Multicenter Phase II trial of high-dose imatinib mesylate in metastatic melanoma: significant toxicity with no clinical efficacy. Cancer. 2006 May 1;106(9):2005–11. doi: 10.1002/cncr.21834. [DOI] [PubMed] [Google Scholar]
- 11.Hodi FS, Friedlander P, Corless CL, et al. Major response to imatinib mesylate in KIT-mutated melanoma. J Clin Oncol. 2008 Apr 20;26(12):2046–51. doi: 10.1200/JCO.2007.14.0707. [DOI] [PubMed] [Google Scholar]
- 12.Lutzky J, Bauer J, Bastian BC. Dose-dependent, complete response to imatinib of a metastatic mucosal melanoma with a K642E KIT mutation. Pigment Cell Melanoma Res. 2008 Aug;21(4):492–3. doi: 10.1111/j.1755-148X.2008.00475.x. [DOI] [PubMed] [Google Scholar]
- 13.Quintas-Cardama A, Lazar AJ, Woodman SE, Kim K, Ross M, Hwu P. Complete response of stage IV anal mucosal melanoma expressing KIT Val560Asp to the multikinase inhibitor sorafenib. Nat Clin Pract Oncol. 2008 Oct 21; doi: 10.1038/ncponc1251. [DOI] [PubMed] [Google Scholar]
- 14.Antonescu CR, Busam KJ, Francone TD, et al. L576P KIT mutation in anal melanomas correlates with KIT protein expression and is sensitive to specific kinase inhibition. Int J Cancer. 2007 Jul 15;121(2):257–64. doi: 10.1002/ijc.22681. [DOI] [PubMed] [Google Scholar]
- 15.Curtin JA, Busam K, Pinkel D, Bastian BC. Somatic activation of KIT in distinct subtypes of melanoma. J Clin Oncol. 2006 Sep 10;24(26):4340–6. doi: 10.1200/JCO.2006.06.2984. [DOI] [PubMed] [Google Scholar]
- 16.Rivera RS, Nagatsuka H, Gunduz M, et al. C-kit protein expression correlated with activating mutations in KIT gene in oral mucosal melanoma. Virchows Arch. 2008 Jan;452(1):27–32. doi: 10.1007/s00428-007-0524-2. [DOI] [PubMed] [Google Scholar]
- 17.Went PT, Dirnhofer S, Bundi M, et al. Prevalence of KIT expression in human tumors. J Clin Oncol. 2004 Nov 15;22(22):4514–22. doi: 10.1200/JCO.2004.10.125. [DOI] [PubMed] [Google Scholar]
- 18.Willmore-Payne C, Holden JA, Hirschowitz S, Layfield LJ. BRAF and c-kit gene copy number in mutation-positive malignant melanoma. Hum Pathol. 2006 May;37(5):520–7. doi: 10.1016/j.humpath.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 19.Willmore-Payne C, Holden JA, Tripp S, Layfield LJ. Human malignant melanoma: detection of BRAF- and c-kit-activating mutations by high-resolution amplicon melting analysis. Hum Pathol. 2005 May;36(5):486–93. doi: 10.1016/j.humpath.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 20.Mol CD, Fabbro D, Hosfield DJ. Structural insights into the conformational selectivity of STI-571 and related kinase inhibitors. Curr Opin Drug Discov Devel. 2004 Sep;7(5):639–48. [PubMed] [Google Scholar]
- 21.Davies MA, Stemke-Hale K, Tellez C, et al. A novel AKT3 mutation in melanoma tumours and cell lines. Br J Cancer. 2008 Oct 21;99(8):1265–8. doi: 10.1038/sj.bjc.6604637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McAuliffe JC, Hunt KK, Lazar AJ, et al. A Randomized, Phase II Study of Preoperative plus Postoperative Imatinib in GIST: Evidence of Rapid Radiographic Response and Temporal Induction of Tumor Cell Apoptosis. Ann Surg Oncol. 2008 Oct 25; doi: 10.1245/s10434-008-0177-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Young H, Baum R, Cremerius U, et al. Measurement of clinical and subclinical tumour response using [18F]-fluorodeoxyglucose and positron emission tomography: review and 1999 EORTC recommendations. European Organization for Research and Treatment of Cancer (EORTC) PET Study Group. Eur J Cancer. 1999 Dec;35(13):1773–82. doi: 10.1016/s0959-8049(99)00229-4. [DOI] [PubMed] [Google Scholar]
- 24.Tamborini E, Gabanti E, Lagonigro MS, et al. KIT/Val654 Ala receptor detected in one imatinib-resistant GIST patient. Cancer Res. 2005 Feb 1;65(3):1115. author reply. [PubMed] [Google Scholar]
- 25.Tamborini E, Pricl S, Negri T, et al. Functional analyses and molecular modeling of two c-Kit mutations responsible for imatinib secondary resistance in GIST patients. Oncogene. 2006 Oct 5;25(45):6140–6. doi: 10.1038/sj.onc.1209639. [DOI] [PubMed] [Google Scholar]
- 26.Buettner R, Mesa T, Vultur A, Lee F, Jove R. Inhibition of Src family kinases with dasatinib blocks migration and invasion of human melanoma cells. Mol Cancer Res. 2008 Nov;6(11):1766–74. doi: 10.1158/1541-7786.MCR-08-0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang X, Zhou J, Yuen NK, et al. Imatinib targeting of KIT-mutant oncoprotein in melanoma. Clin Cancer Res. 2008 Dec 1;14(23):7726–32. doi: 10.1158/1078-0432.CCR-08-1144. [DOI] [PubMed] [Google Scholar]
- 28.Antonescu CR, Besmer P, Guo T, et al. Acquired resistance to imatinib in gastrointestinal stromal tumor occurs through secondary gene mutation. Clin Cancer Res. 2005 Jun 1;11(11):4182–90. doi: 10.1158/1078-0432.CCR-04-2245. [DOI] [PubMed] [Google Scholar]
- 29.Tabone S, Theou N, Wozniak A, et al. KIT overexpression and amplification in gastrointestinal stromal tumors (GISTs) Biochim Biophys Acta. 2005 Jun 30;1741(1-2):165–72. doi: 10.1016/j.bbadis.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 30.Heinrich MC, Corless CL, Blanke CD, et al. Molecular correlates of imatinib resistance in gastrointestinal stromal tumors. J Clin Oncol. 2006 Oct 10;24(29):4764–74. doi: 10.1200/JCO.2006.06.2265. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.