Abstract
A major concern associated with the use of vaccines based on live-attenuated viruses is the possible and well documented reversion to pathogenic phenotypes. In the case of HIV, genomic deletions or mutations introduced to attenuate viral pathogenicity can be repaired by selection of compensating mutations. These events lead to increased virus replication rates and, eventually, disease progression. Because replication competence and degree of protection appear to be directly correlated, further attenuation of a vaccine virus may compromise the ability to elicit a protective immune response. Here, we describe an approach toward a safe attenuated HIV vaccine. The system is not based on permanent reduction of infectivity by alteration of important viral genomic sequences, but on strict control of replication through the insertion of the tetracycline (Tet) system in the HIV genome. Furthermore, extensive in vitro evolution was applied to the prototype Tet-controlled HIV to select for variants with optimized rather than diminished replication capacity. The final product of evolution has properties uniquely suited for use as a vaccine strain. The evolved virus is highly infectious, as opposed to a canonically attenuated virus. It replicates efficiently in T cell lines and in activated and unstimulated peripheral blood mononuclear cells. Most importantly, replication is strictly dependent on the nontoxic Tetanalogue doxycycline and can be turned on and off. These results suggest that this in vitro evolved, doxycycline-dependent HIV might represent a useful tool toward the development of a safer, live-attenuated HIV vaccine.
The majority of new HIV infections occurs in developing countries in which the recent advancements in antiretroviral therapy and prevention are the least affordable or applicable. This makes the development of an effective anti-HIV vaccine the best hope for containment of the spread of AIDS (1). Ongoing efforts are directed to the definition of the immunologic correlates of protection against HIV-1 infection, which, at the moment, are only partially understood (2–4). Most recent experimental evidence suggests that protective immunity eventually will result from a combination of an effective humoral (5, 6) and cellular immune response (7–9). In this perspective, a live-attenuated virus appears to be the optimal candidate for eliciting a protective response from both arms of the immune system (10, 11). Most attempts to develop such a vaccine have focused on the experimental model system of the pathogenic simian immunodeficiency virus (SIV) infection of macaques (8, 9, 12–15), and these attenuated vaccines appear to be promising candidates for future applications to HIV infection. Nonetheless, the possibility that a live-attenuated virus reverts to a pathogenic form has been substantiated extensively (16–18) and may pose serious limitations to any future experimentation in humans.
We have reported recently a genetic strategy to impose a genetic control over HIV-1 replication (19). The elements that control HIV-1 transcription, namely, the Tat protein and its TAR-binding site in the long terminal repeat (LTR) promoter, were inactivated and functionally replaced by components of the tetracycline (Tet) system (20). Although previous attempts by others led to a nonreplicating virus (21), our HIV-rtTA prototype virus is replication-competent, and viral replication depends on the nontoxic Tet analogue doxycycline (dox). Conditional replication of this HIV-rtTA virus provides a unique safety feature for a live-attenuated, virus-based vaccine. However, its replication capacity appeared to be decidedly low compared with the wild-type virus and possibly too low to elicit a protective immune response. We set out to improve its replicative properties by in vitro virus evolution (22). Here, we show that variants with optimal replication capacity were readily isolated in prolonged tissue culture infections. We demonstrate that the determinant of such improved replication is a major rearrangement in the LTR promoter region that was observed in several independent virus cultures. We also present experiments with the improved HIV-rtTA vaccine candidate, including replication studies in resting and activated peripheral blood mononuclear cells (PBMCs), infections in which virus replication is repeatedly turned on and off, and successful dox-mediated reactivation of an integrated provirus after a silent phase of several months in the absence of dox.
Methods
Primers and PCR.
Primers 5′CE (5′-TTGCTACAAGGGACTTTCCGCTGG-3′), ExtN4Not (5′-TCCGTTCGAAATAACTCCGAATTC-3′), XhoU3 (5′-CCGCTCGAGTGGAAGGGCTAATTCACT-3′), NEF.SEQ1 (5′-ATTCGCCACATACCTAGAAG-3′), and AntiU3att (5′-GGAGTGAATTAGCCCTTCCA-3′) were used in standard PCR amplifications with commercial reagents (Perkin–Elmer). Primer sets 1–2 and PCO3-PCO4, used for competitive PCR, were described earlier (23).
Plasmids and Molecular Clones.
The 2Δ tetO element obtained by virus evolution and the 2, 4, and 6 tetO elements derived from the corresponding pBlue3′LTR vectors (unpublished results) were cloned into the 3′ LTR of the HIV-rtTA molecular clone to substitute for the original 8 tetO element. The molecular clones constructed therefore differ only in the number of tetO elements present in the 3′ promoter, which will be inherited in the 5′ LTR after reverse transcription. A detailed description of the cloning strategy can be found elsewhere (19).
Cells and Transfections.
The SupT1 T cell line and the C33A cervical carcinoma cell line were maintained in RPMI 1640 medium and DMEM, respectively. The isolation, phytohemagglutinin (PHA) stimulation, and culturing of PBMCs were performed in RPMI medium as described previously (24). All media were supplemented with 10% FCS, penicillin (100 units/ml), and streptomycin (100 units/ml). C33A cells were transfected by a modification of the standard calcium-phosphate protocol (25). SupT1 cells and PBMCs were transfected by electroporation as described previously (24).
Viruses.
Virus stocks were produced by transfecting the appropriate molecular clones in C33A cells (25). Virus was quantitated by capsid (CA)-p24 antigen ELISA as described previously (26), and these values were used to normalize the amount of virus in subsequent infection assays. Infections were carried out as reported (19). Cells subsequently were washed to remove cell-free viral particles and kept in culture in the presence or absence of dox (1 μg/ml; Sigma). In the case of activated PBMCs, cells were infected on day 3 post-PHA stimulation and fresh, PHA-activated PBMCs were added after 1 week.
Infectivity Determination.
Infectivity of virus supernatants was expressed as 50% tissue culture infective dose/ml and determined in SupT1 cells with dox according to standard virological methods (27).
Virus Evolution.
SupT1 cells were transfected with the KYK-rtTA 8 tetO molecular clone (19) and split in 24 independent cultures. Infected cells were cultured in the presence of dox, and the virus was passaged cell-free at the peak of infection, as determined by the massive appearance of syncytia. Details of the evolution experiment will be provided elsewhere (unpublished results). Cells were collected after 3 months, and DNA was extracted (28). Proviral DNA sequences were PCR-amplified by using primer sets XhoU3/ExtN4Not and NEF.SEQ1/AntiU3att, amplifying the U3 region and the rtTA gene, respectively. PCR products were gel-purified and cloned into the TA cloning vector (Invitrogen). Multiple clones were sequenced with the 373 DNA sequencer and the Taq DyeDeoxy Terminator protocol (Applied Biosystems).
Virus Competition.
SupT1 cells were transfected by electroporation with a mixture of the HIV-rtTA 8-, 6-, 4-, 2-, and 2Δ tetO molecular clones (3 μg each). Viruses were passaged cell-free onto fresh SupT1 cells at the peak of infection. Cells were sampled at each virus passage for DNA extraction and PCR analysis of the promoter sequences by using primers 5′CE and ExtN4Not. After amplification, PCR products were resolved on a 8% nondenaturing polyacrylamide gel and stained with ethidium bromide.
DNA Quantification.
DNA was extracted from infected cells and quantitated by competitive PCR according to previously published methods (29). For competitive PCR experiments, fixed amounts of extracted DNA were mixed with increasing amounts of competitor DNA (23). Primer sets 1–2 and PCO3-PCO4 (23) were used to amplify and quantitate viral genomic DNA and cellular β-globin DNA, respectively, as an internal standard.
Results
Evolution of the dox-Dependent HIV.
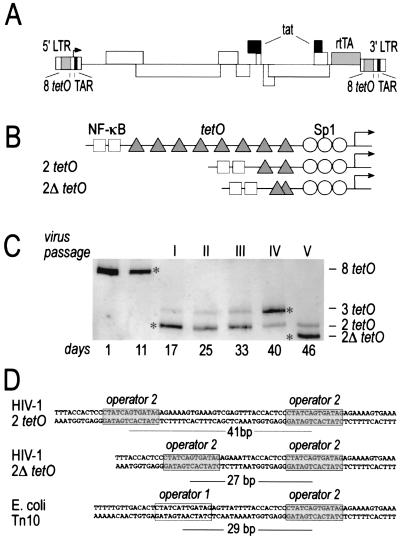
We have reported previously on the construction of an HIV variant of which the replication can be regulated by the nontoxic Tet derivative dox (19). This was achieved by inactivating the viral transcriptional Tat-TAR axis by multiple mutations and by introducing the rtTA gene in the viral genome together with 8 tetO-binding sites for this transactivator protein in the viral LTR promoter (Fig. 1A). Given its relatively low replication capacity that may hamper its efficacy as a vaccine candidate, we set out to improve this virus by means of in vitro evolution (22). The HIV-rtTA virus was passaged extensively in the presence of dox in 24 independent cultures, and we observed increased replication in all cultures within a few weeks. We monitored viral evolution by sequencing samples of viral DNA from each culture. The LTR promoter underwent a drastic rearrangement in all cultures in the form of a rapid shortening of the original 8 tetO elements to three or two binding sites. Furthermore, many of the viruses presented a novel configuration of the tetO elements because of a 14- to 15-bp deletion in the spacer region between the tetO elements (Fig. 1B). The striking convergence of similar mutations in the tetO-binding sites suggests that the altered LTR configuration is important for the establishment of rapid HIV-rtTA replication in vitro. A detailed example of the outcome of one such evolution experiment is shown in Fig. 1C, in which promoter shortening and appearance of the 2Δ tetO element are evident. The virus with 8 tetO motifs is present at the peak of infection of the transfected culture at day 11, but is lost after the first virus passage (day 17). The new, fast-migrating bands correspond to LTR promoters with 3 and 2 tetO elements. The virus with 3 tetO motifs gradually became predominant (day 40), but completely disappeared by day 46 when the virus with the 2Δ tetO promoter first appeared in the culture. Remarkably, promoter shortening was observed in all cultures, suggesting that this mutation provides a profound replication benefit for the virus. The deletion observed in the 2Δ tetO promoter takes place at a very specific position relative to the axis of the tetO operator. This 2Δ configuration is strikingly similar to that of the tetO elements in the natural Escherichia coli promoter, where two binding sites are separated by 29 bp, implying that the spacing between the two binding sites has a functional significance (Fig. 1D).
Figure 1.
In vitro evolution of the dox-dependent HIV-rtTA virus. (A) Schematic of the original HIV-rtTA virus genome that was used as the starting point of the evolution. The inactivated Tat/TAR elements (black) and the introduced rtTA/8 tetO elements of the Tet system (gray) are indicated. (B) Modified tetO configurations obtained by evolution of the original 8 tetO LTR in the SupT1 T cell line. (C) Representative example of an evolution experiment. The LTR promoter region was PCR-amplified at different times and resolved on gel. Asterisks indicate bands that were identified by sequencing. (D) Spacing of tetO elements in the E. coli Tn10 operon, the original HIV-rtTA virus, and the evolved 2Δ tetO elements.
Replication Capacity of the Novel HIV-rtTA 2Δ tetO.
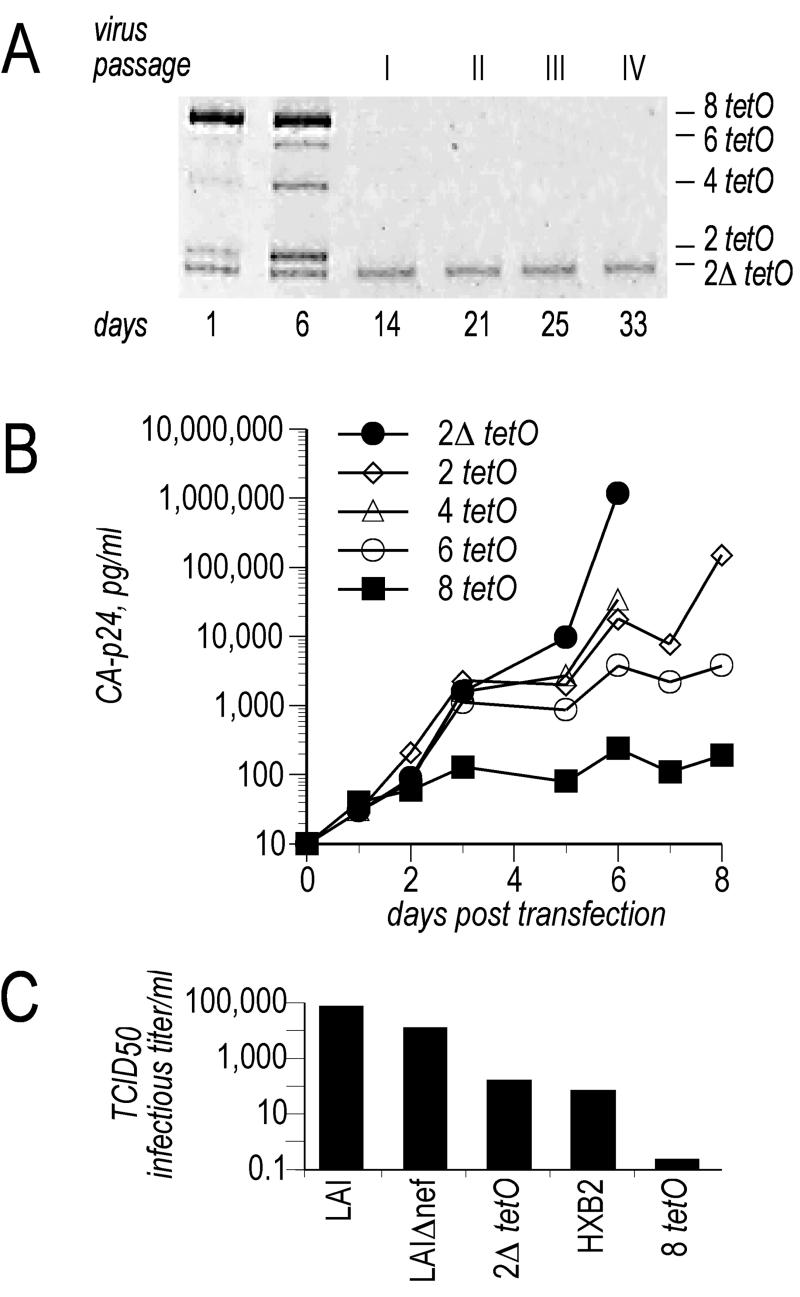
We performed a direct-competition experiment by using a panel of rtTA viruses with a variable number of tetO elements (Fig. 2A). An equimolar mixture of the infectious molecular clones with 8, 6, 4, 2, and 2Δ tetO-binding sites was introduced into SupT1 cells by transfection. The day 1 sample shows all promoter configurations, with a bias for the 8 tetO motifs because this is the actual 5′ LTR configuration that is present in all molecular clones but not inherited in the viral progeny after reverse transcription. Viruses were passaged at the peak of infection onto uninfected SupT1 cells, and the promoter composition of the virus population was analyzed by PCR. As evident in Fig. 2A, the virus containing the 2Δ tetO promoter became the only detectable species after just one passage. This result suggests that the novel dox-regulated element confers a remarkable advantage in terms of viral fitness, which subsequently was confirmed by comparing the replication curves of the individual virus variants of the panel (Fig. 2B). The original virus with the 8 tetO promoter showed the poorest replication capacity by far, whereas the novel HIV-rtTA 2Δ tetO was the most fit virus variant. It is also evident that the other product of virus evolution, namely, the virus with the 2 tetO configuration, replicates efficiently and better than the original 8 tetO virus. With the aim of quantifying the infectious capacity of the HIV-rtTA 2Δ tetO, we determined its 50% tissue culture infective dose/ml (TCID50; Fig. 2C). The 2Δ tetO promoter resulted in an almost 1,000-fold increase in TCID50 as compared with the original 8 tetO promoter, and its infectivity was similar to that of the laboratory isolate HIVHXB2.
Figure 2.
Replication capacity conferred by the 2Δ tetO promoter element. (A) Virus-competition experiment. Infectious molecular clones differing only in the number of tetO elements (8, 6, 4, 2, 2Δ) were cotransfected (3 μg each) in SupT1 cells in the presence of dox. The composition of the virus population was determined at the indicated days posttransfection by PCR amplification as described in Fig. 1C. (B) Replication curves of this panel of viruses. SupT1 cells were transfected with 5 μg of the indicated infectious molecular clone, and virus production was monitored by measuring CA-p24 protein in the supernatant. (C) Infectivity of HIV-rtTA 8 tetO 2Δ tetO virus was compared with that of the original HIV-rtTA 8 tetO, the HIV isolates HXB2 and LAI, and the LAI Δnef mutant.
Strict Replication Control by dox.
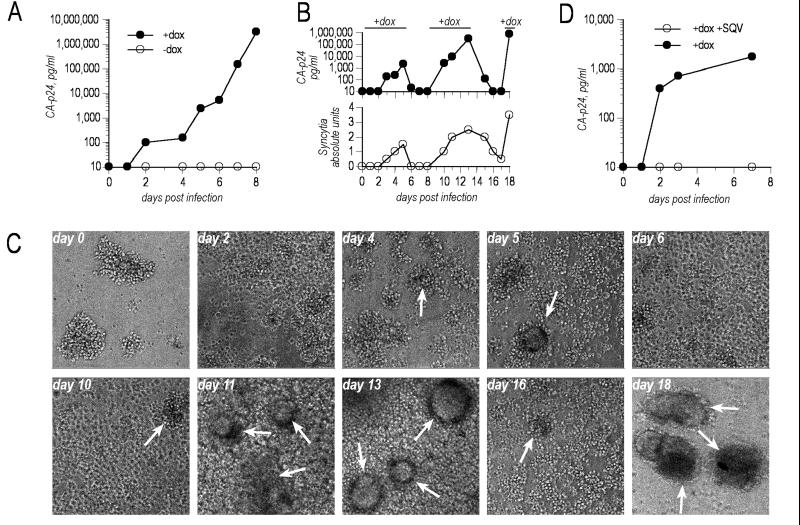
An important issue was to determine to which extent the evolved virus would be regulated by dox despite its increased fitness. This is critical for any subsequent application as an attenuated vaccine candidate. SupT1 cells were infected with HIV-rtTA 2Δ tetO, and dox dependency was tested in different experimental settings (Fig. 3). When dox was omitted from the culture, cells became readily infected as verified by PCR amplification of proviral DNA (results not shown), but no virus production could be detected (Fig. 3A). Dox also was added repeatedly and removed from the infected culture, and each withdrawal led to a rapid and complete loss of virus replication (Fig. 3B Upper). Later additions of dox resulted in a prompt reactivation of the virus. Interestingly, the appearance of syncytia in the infected cell cultures also regressed as dox was removed from the culture, strengthening the conclusion that viral gene expression effectively is switched off in the absence of dox (Fig. 3B Lower and C). Finally, we also studied the sensitivity of HIV-rtTA toward currently used antiretroviral agents such as Saquinavir (SQV; Fig. 3D) and AZT (not shown). Similar to wild-type HIV, replication of the dox-activated HIV-rtTA 2Δ tetO is completely repressed by these HIV inhibitors.
Figure 3.
Strict dox control of the evolved HIV-rtTA 2Δ tetO. (A) Absence of replication without dox. After infection of SupT1 cells with HIV-rtTA 2Δ tetO (5 ng CA-p24), the culture was split in two and maintained in the absence or presence of 1 μg/ml dox. Virus replication was monitored throughout the experiment by measuring CA-p24 production. (B) Repeated virus activation. After infection of SupT1 cells, dox was added and removed repeatedly (+dox: days 0, 8, and 17; −dox: day 5 and 13) from the culture medium. The experiment was stopped at day 18 because of massive syncytia formation. Virus replication is measured by means of CA-p24 production and by scoring the extent of syncytia formation. (C) Microphotographs of infected SupT1 corresponding to the experiment in B. (D) Sensitivity of HIV-rtTA 2Δ tetO to antiretroviral agents. Infected cells were grown in the presence of dox and either with or without Saquinavir (SQV, 200 nM).
Long-Term Reactivation.
An important safety concern is whether the virus remains dox-dependent after extensive culturing and whether the integrated, silent provirus remains susceptible to dox activation at considerable time intervals from the infection event. To address these issues, cells were infected with HIV-rtTA 2Δ tetO, as well as with HIV-rtTA 8 tetO for comparison, and maintained in culture without dox. No signs of viral replication could be detected in either culture for up to 6 months after infection (ongoing experiment, results not shown). At days 3, 15, 36, and 90 postinfection, dox was added to an aliquot of the culture and virus production in the medium was quantitated. HIV-rtTA 2Δ tetO could be readily reactivated by dox at all times tested, although a gradual delay was apparent (Fig. 4A). The original HIV-rtTA 8 tetO could be rescued from latency only at day 3, but not at later times. Importantly, both cultures that were maintained without dox contained equivalent amounts of viral DNA throughout the experiment (Fig. 4B), indicating that these distinct behaviors reflect a real difference in viral gene expression rather than a gradual loss of provirus-containing cells over time.
Figure 4.
Proviral persistence and reactivation in prolonged cultures. (A) SupT1 cells infected with HIV-rtTA 2Δ tetO (Left) or HIV-rtTA 8 tetO (right) were kept in culture in the absence of dox for up to 6 months (experiment still in progress). Samples of the cultures were treated with dox on days 3, 15, 36, and 90, and virus production was measured by CA-p24 ELISA. (B) The average number of HIV proviral DNA molecules in the cells was monitored by competitive PCR. Values were normalized for the number of cellular diploid genomes as measured by competitive PCR on the cellular single-copy gene β-globin.
Replication in Primary T Cells.
Wild-type HIV can productively infect both activated and resting CD4+ T lymphocytes, the main natural target of HIV infection, and the nef gene is thought to play an important role in the infection of primary cells. Because our dox-dependent HIV was derived from a primary isolate, HIVLAI, in which the nef gene was replaced by the rtTA-coding region, we set out to test whether HIV-rtTA can infect primary T cells. For comparison, we included a LAI mutant in which we had introduced the same nef deletion present in the dox-dependent variant (LAI Δnef) and the T cell line-adapted nef − HIVHXB2 isolate. We compared the replication kinetics of this panel of viruses in SupT1 T cells (Fig. 5A), PHA-activated PBMCs (Fig. 5B), and unstimulated PBMCs (Fig. 5C). A limiting amount of virus was used for the infection to maximize replication differences, resulting in poor replication of the original HIV-rtTA 8 tetO virus. In contrast, the evolved 2Δ tetO virus replicated efficiently, with a fitness similar to that of HIVHXB2. The lack of nef function is not the only factor responsible for the difference in replication between HIVLAI and the novel dox-dependent virus, because the LAI Δnef variant replicated more efficiently than HIV-rtTA 2Δ tetO (see also Fig. 2C). In all cases, the virus with the 2Δ tetO configuration showed a strong replicative advantage over the original virus with 8 tetO elements. Because resting CD4+ T cells are estimated to be the vast majority of circulating CD4+ cells (30), these results suggest that the optimized HIV-rtTA is able to establish a productive infection in vivo in circulating T cells. Replication of this virus in primary cells was also completely dox-dependent (results not shown).
Figure 5.
Replication kinetics in primary cells. Equivalent amounts (25 ng of CA-p24) of the indicated viruses were used to infect the SupT1 T cell line (A), PBMCs after phytohemagglutinin activation (B), and unstimulated PBMCs (C). Virus replication was monitored by measuring CA-p24 production in the supernatant.
Discussion
Several molecular approaches can be used to attenuate the retroviral replication capacity to limit the pathogenic potential of a live-attenuated candidate vaccine. Nonessential segments have been deleted from the viral genome, either individually or in combination (12, 31, 32). Alternatively, the function of essential viral proteins or regulatory RNA/DNA signals can be attenuated or altered by mutation (33, 34). However, evidence of a clear attenuation threshold in SIV-vaccination studies suggests the existence of a direct relationship between replication competence and degree of protection conferred. If the efficacy of vaccination depends on the replication competence of the virus, the success of further attenuation strategies might be limited (9, 17, 35). Furthermore, reversion to a pathogenic phenotype poses serious concerns about the application of such attenuated virus vaccines to human disease (16–18). In the case of HIV, an additional complication of the design of an immunizing, nonpathogenic virus is that the exact mechanisms responsible for pathogenicity are only partially understood.
We have used a strategy to impose a strict control on HIV-1 replication. Moreover, we have applied the method of in vitro evolution (22) to obtain an improved version of the dox-dependent HIV (19) with optimized replication properties. The original virus, which encodes the E. coli-derived rtTA transactivator protein and, therefore, depends on the presence of dox for activation of transcription, was passaged for several months in the presence of dox to select for improved variants. Because of the low fidelity of the viral reverse transcriptase, mutant viruses readily arose in the culture and viruses with mutations that conferred a replicative advantage became the predominant genotype over time. Major rearrangements in the LTR promoter were apparent in all independent evolution experiments. The original 8 tetO-binding sites for rtTA, placed between the NF-κB and Sp1 sites of the viral LTR, were reduced to only three or two sites, and in many instances a 14- to 15-bp deletion appeared between the 2 tetO elements. This novel 2Δ tetO promoter configuration is solely responsible for the observed 3-log increase in viral fitness, as measured by recloning the modified LTR in the original virus. It is interesting to note that the spacing of the tetO elements with the deletion closely resembles that of the E. coli Tn10 operon from which these elements were originally derived (36). The altered spacing between the operators is likely to affect the binding of the rtTA transactivator and, thus, may explain the observed increase in viral fitness. Further molecular studies are underway to determine the characteristics of this novel dox-dependent promoter in terms of rtTA-binding affinity, cooperativity, and transcriptional activation.
A virus preparation of the improved HIV-rtTA 2Δ tetO virus had an infectious titer 1,000-fold higher than that of the original HIV-rtTA 8 tetO. Moreover, infectivity of the HIV-rtTA 2Δ tetO virus was similar to that of the laboratory isolate nef− HIVHXB2, although lower than that of HIVLAI. The evident replication advantage of the LAI isolate is not solely a result of the nef gene, which is lacking in the dox-dependent virus. A LAI isolate with the same nef deletion replicated better than the HIV-rtTA 2Δ tetO virus. It should be noted that we deleted the nef gene primarily to accommodate the rtTA ORF and not only to attenuate viral pathogenicity. In fact, recent studies in SIV-infected monkeys (37–39) and results from the Sidney blood bank cohort (40, 41) show that nef inactivation is not sufficient to ensure stable attenuation. Most importantly, replication of our HIV-rtTA variant is still completely dependent on dox despite the increased infectivity. Without dox, no sign of virus replication could be detected for up to at least 6 months after the initial infection. However, addition of dox to the cultures resulted in a rapid reactivation of the silent, integrated provirus. Control of viral replication is so effective that the virus can be turned on and off by repeated addition/withdrawal of dox. The activation or repression of HIV gene expression was measured both at the level of virus production in the supernatant and, consequently, by the appearance/disappearance of syncytia in the cultures. Regression of syncytia is likely to result from a combination of cell death and outgrowth of noninfected cells in the absence of viral replication.
The ability to repeatedly activate the virus, including at time points distant from the initial infection event, is particularly important for vaccine applications. In fact, a key issue with any attenuated vaccine is the likelihood that solid immunity, if achieved, will be observed only as long as the attenuated virus is retained and able to preserve the immunological memory (4). The issue of immunological memory is still controversial in the literature, but if continued antigenic stimulation is required, the dox-dependent virus will allow one to preserve a protective state of immunity by boost dox reactivation of the dormant virus. Such boost reactivations also would serve the function of building up a more profound immune response. Additional advantages of the dox-dependent virus are that, because of its widespread use, much is known on the pharmacokinetics of dox in humans, and dox is inexpensive, nontoxic, and easy to administer (42). The concentration of dox required for activation of the virus is far lower than the blood concentration commonly reached in clinical practice. Dox can be easily administered orally and reaches peak concentrations in the blood within hours. The relatively short half-life of the compound (15–22 h) allows a rapid shut-off of the doxycycline-dependent system in vivo, as demonstrated in animals with transgenes expressed by a dox-regulated promoter (43).
Based on the current experimental observation in other attenuated vaccine systems, it can be envisaged that such a dox-dependent, but otherwise fully replication-competent, HIV may be able to confer a protective immunity. Studies in monkeys (11, 44) suggest that it should be possible to induce sufficient protective immunity against HIV to down-modulate a second infection, although these studies fail to indicate whether such partial immunity will be protective against disease. The potential of the HIV-rtTA 2Δ virus as a vaccine candidate now should be evaluated in an animal model. Homologous SIV constructs are under construction to test the ability to confer protective protection in vivo and to analyze in more detail the safety of such a putative vaccine virus.
Acknowledgments
We thank A. T. Das and W. A. Paxton for critical reading of this manuscript. We are grateful to H. Bujard (University of Heidelberg) for providing several reagents of the Tet system and for helpful comments and discussion on the Tet HIV project. Research within the Berkhout laboratory is sponsored by the Dutch AIDS Fund (AIDS Fonds, Amsterdam), the Dutch cancer society (Amsterdam), the Technology Foundation (Utrecht, The Netherlands), and the National Institutes of Health (Bethesda, MD). G.M. and K.V. are supported by Human Frontier Science Program and European Molecular Biology Organization fellowships.
Abbreviations
- Tet
tetracycline
- dox
doxycycline
- PBMC
peripheral blood mononuclear cell
- SIV
simian immunodeficiency virus
- LTR
long terminal repeat
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Mills J, Desrosiers R, Rud E, Almond N. AIDS Res Hum Retroviruses. 2000;16:1453–1461. doi: 10.1089/088922200750005976. [DOI] [PubMed] [Google Scholar]
- 2.Letvin N L. Science. 1998;280:1875–1880. doi: 10.1126/science.280.5371.1875. [DOI] [PubMed] [Google Scholar]
- 3.McMichael A J, Hanke T. Nat Med. 1999;5:612–614. doi: 10.1038/9455. [DOI] [PubMed] [Google Scholar]
- 4.Paul W E. Cell. 1995;82:177–182. doi: 10.1016/0092-8674(95)90304-6. [DOI] [PubMed] [Google Scholar]
- 5.Mascola J R, Stiegler G, VanCott T C, Katinger H, Carpenter C B, Hanson C E, Beary H, Hayes D, Frankel S S, Birx D L, et al. Nat Med. 2000;6:207–210. doi: 10.1038/72318. [DOI] [PubMed] [Google Scholar]
- 6.Baba T W, Liska V, Hofmann-Lehmann R, Vlasak J, Xu W, Ayehunie S, Cavacini L A, Posner M R, Katinger H, Stiegler G, et al. Nat Med. 2000;6:200–206. doi: 10.1038/72309. [DOI] [PubMed] [Google Scholar]
- 7.Allen T M, O'Connor D H, Jing P, Dzuris J L, Mothe B R, Vogel T U, Dunphy E, Liebl M E, Emerson C, Wilson N, et al. Nature (London) 2000;407:386–390. doi: 10.1038/35030124. [DOI] [PubMed] [Google Scholar]
- 8.Wyand M S, Manson K, Montefiori D C, Lifson J D, Johnson R P, Desrosiers R C. J Virol. 1999;73:8356–8363. doi: 10.1128/jvi.73.10.8356-8363.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson R P, Lifson J D, Czajak S C, Cole K S, Manson K H, Glickman R, Yang J, Montefiori D C, Montelaro R, Wyand M S, et al. J Virol. 1999;73:4952–4961. doi: 10.1128/jvi.73.6.4952-4961.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Desrosiers R C. Nat Med. 1999;5:723–725. doi: 10.1038/10439. [DOI] [PubMed] [Google Scholar]
- 11.Nathanson N, Mathieson B J. J Infect Dis. 2000;182:579–589. doi: 10.1086/315707. [DOI] [PubMed] [Google Scholar]
- 12.Almond N, Kent K, Cranage M, Rud E, Clarke B, Stott E J. Lancet. 1995;345:1342–1344. doi: 10.1016/s0140-6736(95)92540-6. [DOI] [PubMed] [Google Scholar]
- 13.Daniel M D, Kirchhoff F, Czajak S C, Sehgal P K, Desrosiers R C. Science. 1992;258:1938–1941. doi: 10.1126/science.1470917. [DOI] [PubMed] [Google Scholar]
- 14.Lohman B L, McChesney M B, Miller C J, McGowan E, Joye S M, Van Rompay K K, Reay E, Antipa L, Pedersen N C, Marthas M L. J Virol. 1994;68:7021–7029. doi: 10.1128/jvi.68.11.7021-7029.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shibata R, Siemon C, Czajak S C, Desrosiers R C, Martin M A. J Virol. 1997;71:8141–8148. doi: 10.1128/jvi.71.11.8141-8148.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berkhout B, Verhoef K, van Wamel J L, Back N K. J Virol. 1999;73:1138–1145. doi: 10.1128/jvi.73.2.1138-1145.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gundlach B R, Lewis M G, Sopper S, Schnell T, Sodroski J, Stahl-Hennig C, Uberla K. J Virol. 2000;74:3537–3542. doi: 10.1128/jvi.74.8.3537-3542.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Whatmore A M, Cook N, Hall G A, Sharpe S, Rud E W, Cranage M P. J Virol. 1995;69:5117–5123. doi: 10.1128/jvi.69.8.5117-5123.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Verhoef K, Marzio G, Hillen W, Bujard H, Berkhout B. J Virol. 2001;75:979–987. doi: 10.1128/JVI.75.2.979-987.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baron U, Bujard H. Methods Enzymol. 2000;327:401–421. doi: 10.1016/s0076-6879(00)27292-3. [DOI] [PubMed] [Google Scholar]
- 21.Xiao Y, Kuwata T, Miura T, Hayami M, Shida H. Virology. 2000;269:268–275. doi: 10.1006/viro.2000.0213. [DOI] [PubMed] [Google Scholar]
- 22.Das A T, Land A, Braakman I, Klaver B, Berkhout B. Virology. 1999;263:55–69. doi: 10.1006/viro.1999.9898. [DOI] [PubMed] [Google Scholar]
- 23.Comar M, Marzio G, D'Agaro P, Giacca M. AIDS Res Hum Retroviruses. 1996;12:117–126. doi: 10.1089/aid.1996.12.117. [DOI] [PubMed] [Google Scholar]
- 24.Back N K, Nijhuis M, Keulen W, Boucher C A, Oude Essink B O, van Kuilenburg A B, van Gennip A H, Berkhout B. EMBO J. 1996;15:4040–4049. [PMC free article] [PubMed] [Google Scholar]
- 25.Das A T, Klaver B, Berkhout B. J Virol. 1999;73:81–91. doi: 10.1128/jvi.73.1.81-91.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jeeninga R E, Hoogenkamp M, Armand-Ugon M, de Baar M, Verhoef K, Berkhout B. J Virol. 2000;74:3740–3751. doi: 10.1128/jvi.74.8.3740-3751.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aldovini A, Walker B D. Techniques in HIV Research. New York: Stockton; 1990. [Google Scholar]
- 28.Das A T, Klaver B, Klasens B I, van Wamel J L, Berkhout B. J Virol. 1997;71:2346–2356. doi: 10.1128/jvi.71.3.2346-2356.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marzio G, Tyagi M, Gutierrez M I, Giacca M. Proc Natl Acad Sci USA. 1998;95:13519–13524. doi: 10.1073/pnas.95.23.13519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Finzi D, Silliciano R F. Cell. 1998;93:665–671. doi: 10.1016/s0092-8674(00)81427-0. [DOI] [PubMed] [Google Scholar]
- 31.Kestler H W, III, Ringler D J, Mori K, Panicali D L, Sehgal P K, Daniel M D, Desrosiers R C. Cell. 1991;65:651–662. doi: 10.1016/0092-8674(91)90097-i. [DOI] [PubMed] [Google Scholar]
- 32.Wyand M S, Manson K H, Garcia-Moll M, Montefiori D, Desrosiers R C. J Virol. 1996;70:3724–3733. doi: 10.1128/jvi.70.6.3724-3733.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berkhout B. Adv Pharmacol. 2000;48:29–73. doi: 10.1016/s1054-3589(00)48003-8. [DOI] [PubMed] [Google Scholar]
- 34.Reitter J N, Means R E, Desrosiers R C. Nat Med. 1998;4:679–684. doi: 10.1038/nm0698-679. [DOI] [PubMed] [Google Scholar]
- 35.Ten Haaft P, Verstrepen B, Uberla K, Rosenwirth B, Heeney J. J Virol. 1998;72:10281–10285. doi: 10.1128/jvi.72.12.10281-10285.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hillen W, Berens C. Annu Rev Microbiol. 1994;48:345–369. doi: 10.1146/annurev.mi.48.100194.002021. [DOI] [PubMed] [Google Scholar]
- 37.Baba T W, Jeong Y S, Pennick D, Bronson R, Greene M F, Ruprecht R M. Science. 1995;267:1820–1825. doi: 10.1126/science.7892606. [DOI] [PubMed] [Google Scholar]
- 38.Baba T W, Liska V, Khimani A H, Ray N B, Dailey P J, Penninck D, Bronson R, Greene M F, McClure H M, Martin L N, et al. Nat Med. 1999;5:194–203. doi: 10.1038/5557. [DOI] [PubMed] [Google Scholar]
- 39.Wyand M S, Manson K H, Lackner A A, Desrosiers R C. Nat Med. 1997;3:32–36. doi: 10.1038/nm0197-32. [DOI] [PubMed] [Google Scholar]
- 40.Greenough T C, Sullivan J L, Desrosiers R C. N Engl J Med. 1999;340:236–237. doi: 10.1056/NEJM199901213400314. [DOI] [PubMed] [Google Scholar]
- 41.Learmont J C, Geczy A F, Mills J, Ashton L J, Raynes-Greenow C H, Garsia R J, Dyer W B, McIntyre L, Oelrichs R B, Rhodes D I, et al. N Engl J Med. 1999;340:1715–1722. doi: 10.1056/NEJM199906033402203. [DOI] [PubMed] [Google Scholar]
- 42.Joshi N, Miller D Q. Arch Intern Med. 1997;157:1421–1428. [PubMed] [Google Scholar]
- 43.Harding T C, Geddes B J, Murphy D, Knight D, Uney J B. Nat Biotechnol. 1998;16:553–555. doi: 10.1038/nbt0698-553. [DOI] [PubMed] [Google Scholar]
- 44.Novembre F J, Saucier M, Anderson D C, Klumpp S A, O'Neil S P, Brown C R, II, Hart C E, Guenthner P C, Swenson R B, McClure H M. J Virol. 1997;71:4086–4091. doi: 10.1128/jvi.71.5.4086-4091.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]