Abstract
The assT gene encodes an arylsulfate sulfotransferase, an enzyme that catalyzes sulfuryl transfer from phenolic sulfate to a phenolic acceptor. In Salmonella enterica serovar Typhi IMSS-1, the assT gene is located upstream of the dsbL and dsbI genes, which are involved in a disulfide bond formation required for its activation. The assT-dsbL-dsbI gene cluster forms an operon transcribed by a LeuO-dependent promoter, in rich medium A (MA). Interestingly, in the absence of cloned leuO and in a ΔleuO background, two transcription start sites were detected for assT and two for dsbL-dsbI in minimal medium. The H-NS nucleoid protein repressed the expression of the assT-dsbL-dsbI LeuO-dependent operon, as well as of the assT transcriptional units. Thus, the expression of the assT-dsbL-dsbI gene cluster depends on the global regulatory proteins LeuO and H-NS, as well as on specific growth conditions.
INTRODUCTION
The assT gene encodes an arylsulfate sulfotransferase that is present in several organisms, including mammals and a wide spectrum of microbial genomes (2, 25, 55). Mammalian sulfotransferases are located in the cytosol and in the Golgi apparatus membrane; their activities have been detected in liver, brain, kidney, and intestinal epithelial cells (53, 55). These sulfotransferases use 3′-phosphoadenosine-5′-phosphosulfate (PAPS) as sulfate donor and are involved in a wide variety of biological processes such as cell communication, growth, development, defense, and detoxification (6, 19, 43, 63, 65). Unlike mammals, bacterial sulfotransferases are periplasmic enzymes that require phenolic sulfate esters as donor substrates, and it has been suggested that the intestinal microflora uses this enzyme to detoxify phenolic compounds (31, 32, 34, 35).
In Salmonella enterica serovar Typhi, the assT gene is located upstream of dsbL and dsbI, a couple of genes encoding a reduction-oxidation (red-ox) pair of proteins involved in the disulfide bond formation required for AssT activation in the periplasm (25, 40, 44). The nomenclature was adopted from the orthologous genes previously described in uropathogenic Escherichia coli (UPEC) and Salmonella enterica serovar Typhimurium (25, 40). In contrast to the highly conserved assT, the genomic organization of the assT-dsbL-dsbI gene cluster is only conserved in Enterobacter, Yersinia, Citrobacter, and Escherichia strains, as well as within the Salmonella genus (5, 12, 25, 40, 41, 62).
The crystal structures for AssT and DsbL proteins have been solved. AssT forms a homodimer, whose Cys-418 and Cys-424 are involved in disulfide bond formation and its His-436 undergoes the transient sulfurylation necessary for the ping-pong reaction mechanism (44). DsbL is a periplasmic monomer, forming a disulfide bond between Cys-29 and Cys-32, which is oxidized by the transmembrane protein DsbI (25). DsbI contains four predicted transmembrane helixes with two cysteine pairs (Cys-55 and Cys-58; Cys-127 and Cys-153).
In vivo, assT, dsbL, and dsbI expression has been detected in macrophages infected with Salmonella; additionally, a transcriptome assay showed assT and dsbL expression in the urinary tract during UPEC infection (20, 60). Moreover, in S. Typhimurium assT and dsbL have been reported as virulence factors in Nramp1-resistant mice, and a mutation in assT results in attenuated virulence in Edwardsiella tarda in a fish model (38, 45). However, assT mutants in S. Typhimurium did not affect the pathogenesis in BALB/c mice or assT or dsbL-dsbI gene deletions during UPEC colonization in the mouse urinary tract infection model (40, 62).
The assT-dsbL-dsbI cluster has been suggested to form an operon in UPEC and in Enterobacter amnigenus (25, 37). However, in S. Typhimurium, dsbL expression can occur independently from assT (40). We show that in S. Typhi IMSS-1, the transcriptional regulation of the assT-dsbL-dsbI cluster depends on global regulatory proteins and specific growth conditions. A LeuO-dependent promoter is necessary for induction of the complete assT-dsbL-dsbI operon in rich medium, whereas transcriptional start sites were determined for assT and the dsbL-dsbI transcriptional units in the absence of cloned leuO and in a ΔleuO background, in N-minimal medium which is used for induction of Salmonella SPI-2 (16). In addition, we also report that the global regulatory protein H-NS represses transcription of the assT-dsbL-dsbI gene cluster, both in rich and in minimal medium.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains and plasmids used are listed in Table S1 in the supplemental material. S. Typhi IMSS-1 (52) and the E. coli strains were grown aerobically at 37°C in LB (tryptone, 10 g; yeast extract, 5 g; NaCl, 10 g/liter) for overnight cultures. S. Typhi was grown in liquid MA (nutrient broth, 7 g; yeast extract, 1 g; glycerol, 2 g; 3.75 g of K2HPO4 and 1.3 g of KH2PO4/liter) (33) or in N-minimal medium [5 mM KCl, 7.5 mM (NH4)2SO4, 0.5 mM K2SO4, 1 mM KH2PO4, 100 mM Tris-HCl (pH 7.5), 10 μM MgCl2, 0.5% glycerol, 0.1% Casamino acids] (16). Kanamycin (Km) at 15 μg/ml, tetracycline (Tc) at 12 μg/ml, and ampicillin (Ap) at 100 μg/ml were added when required.
DNA and RNA manipulations.
Genomic DNA isolation was performed according to published protocols (54). Plasmid DNA was purified with the High-Pure plasmid isolation kit (Boehringer, Mannheim, Germany). Primers for PCR amplifications were provided by the oligonucleotide synthesis facility at our institute (see Table S2 in the supplemental material). Restriction enzymes, ligase, kinases, nucleotides and polymerases were obtained from New England Biolabs (Ipswich, MA) or Gibco-BRL. Sequencing was performed with an automatic Perkin-Elmer/Applied Biosystems 377-18 system. The one-step mutagenesis procedure described by Datsenko and Wanner (15) for bacterial chromosomal genes was used to generate gene deletions and replacements for antibiotic resistance markers.
For reverse transcription-PCR (RT-PCR), total RNA was isolated with Trizol from 1.5 ml of bacterial cells grown in 100 ml of MA medium to an optical density at 595 nm (OD595) of 0.6. For the 5′RACE (5′ rapid amplification of cDNA ends) and primer extension experiments, bacterial cells were grown in 50 ml of N-minimal medium for 12 h. The RNA extraction kit (Qiagen) was used to obtain total RNA samples for 5′RACE experiments, as established in the protocol. For primer extension experiments, 50 ml of N-minimal medium cell culture were treated with RNAlater (Fermentas) and used for RNA purification with acid-phenol extraction. The RNA concentration was determined by measuring the absorbance at 260 nm, and the integrity of RNA was verified by 1.5% agarose gel electrophoresis.
Construction of transcriptional reporter fusions.
Oligonucleotides (see Table S2 in the supplemental material) were designed to amplify by PCR different regions of the assT, dsbL, and dsbI genes. PCR fragments were double digested with BamHI-KpnI or BamHI-MluI and ligated into pKK232-9 (Km), which contains the promoterless cat gene. Transcriptional fusions in pKK232-8 (Ap) were generated from pKK232-9 (Km) digested with PstI to eliminate Km gene resistance. Fusions were sequenced in order to verify the correct DNA sequence of the PCR fragments. Each plasmid was then electroporated into different Salmonella strains to evaluate its transcriptional activity.
Deletions and substitutions.
In order to characterize the assT regulatory region, deletions and substitutions by overlap extension were performed according to the method of Sambrook, et al. (54). Two complementary oligonucleotides containing changes in the nucleotide sequence were used (see Table S2 in the supplemental material). The mutated PCR products were digested with the specific restriction enzymes, cloned into pKK232-9, and sequenced to verify the correct changes.
CAT assay.
S. Typhi strains were grown in 100 ml of MA medium or 50 ml of N-minimal medium for 12 h; 50 μM IPTG (isopropyl-β-d-thiogalactopyranoside) was added when required. Then, 1.5-ml portions of bacterial cultures were collected by centrifugation and washed with 0.8 ml of TDTT buffer (50 mM Tris-HCl [pH 7.8], 30 μM d,l-dithiothreitol [DTT]). Bacterial cells collected from MA medium and N-minimal medium were resuspended in 0.6 and 0.3 ml of TDTT, respectively, and sonicated on ice for 10-s intervals with 10-s rest periods until the extract was clear. The homogenate was centrifuged, and the supernatant was used for total protein and CAT activity measurement. For CAT assays, 5 μl of each extract were added in duplicate to a 96-well enzyme-linked immunosorbent assay (ELISA) plate, followed by the addition of 0.2 ml of a reaction mixture that contained 1 mM DTNB, 0.1 mM acetyl-CoA and 0.1 mM chloramphenicol in 0.1 M Tris-HCl (pH 7.8). The absorbance at 412 nm was measured every 5 s for 5 min using a scanning autoreader and the microplate workstation Ceres 900. In these studies, 1 μmol of chloramphenicol acetylated/min corresponds to 1 chloramphenicol acetyltransferase (CAT) unit. The protein concentration was determined using BCA protein assay reagent (Pierce). Protein values and the mean rate of product formation by cat were used to determine the CAT specific activity as μmol/min/mg protein. The results presented in the figures are the means of three independent experiments.
RT-PCR.
Five μg from total RNA was treated with DNase I (Fermentas), and 1.5 μg of RNA (DNA-free) was used for cDNA synthesis with random primers, as established in the Revert-Aid Minus M-MuLV RT protocol (Fermentas). PCR was performed from 1.5 μl of cDNA, and the reaction products were analyzed in a 1% agarose gel. As a positive control, a PCR was amplified for rsmC, the gene encoding for the 16S rRNA.
Primer extension analysis.
A total of 40 μg of total RNA isolated for bacteria grown in N-minimal medium was denatured at 95°C for 3 min and then slowly cooled to 45°C to anneal with the [γ-32P]ATP-labeled primers. DNA extensions were performed with reverse transcriptase at 42°C for 90 min. The extended products were purified by ethanol precipitation and analyzed by electrophoresis in 8% polyacrylamide–8 M urea gels alongside sequencing ladders. Sequencing ladders were generated from plasmids that contain the entire regulatory region of assT or dsbL.
5′RACE.
Total RNA was treated with DNase I and a RiboMinus transcriptome isolation kit (Invitrogen, Carlsbad, CA) to eliminate DNA and rRNA, respectively. The 5′-monophosphate RNA transcripts were removed by exonuclease treatment with Terminator 5′-phosphate-dependent exonuclease (Epicentre, Madison, WI). The enriched 5′-triphosphate end mRNAs were treated with tobacco acid pyrophosphatase (TAP; Epicentre) to generate 5′-monophosphate ends before ligation to a 5′ RNA adapter (5′-GUUCAGAGUUCUACAGUCCGACGAUC; Illumina, Inc., San Diego, CA) with T4 RNA ligase 1 (ssRNA ligase; New England Biolabs) as described previously (23, 56). A modified 5′RACE methodology (46) was used for the construction of cDNA libraries. A random primer, 5′-CAAGCAGAAGACGGCATACGANNNNNN (adapter 3′), was used for the generation of the cDNA. We performed PCR in order to enrich the yield of the cDNA, using small RNA sequencing primer and small RNA PCR primer 2, which are complementary to the random primer and the 5′ RNA adapter, respectively.
In addition, a second methodology was used to obtain the 5′ ends. Total RNA was used as a template to generate a cDNA library using an oligonucleotide attached to a hexanucleotide random sequence tag at its 3′ end (5′-GCCTTGCCAGCCCGCTCAGNNNNNN). Using terminal deoxynucleotidyltransferase (Fermentas), an adenine polynucleotide tag was incorporated into the 3′ end of the cDNA. A PCR was performed to enrich the cDNA library by using the oligonucleotides B1 (5′-GCCTTGCCAGCCCGCTCAG) and poly(T), which are complementary to the random primer and the 5′ RNA adapter, respectively.
Finally, a PCR was carried out to amplify the upstream region of the intended genes from both methodologies. The PCR products were separated by PAGE in 8% polyacrylamide gels, and DNA bands were isolated and sequenced.
H-NS purification.
Purification of H-NS-Myc-6×His proteins was performed with Ni-nitrilotriacetic acid resin (QIAExpress; Qiagen) according to the manufacturer's instructions and the modifications established by De la Cruz (18). Briefly, E. coli BL21(DE3) harboring the pMDHNS plasmid was grown in 100 ml of LB supplemented with Ap until an OD595 of 0.4 was reached. H-NS induction was performed with 0.1% l-arabinose (Sigma-Aldrich), and the cultures continued to grow for 4 h. The cells were collected and washed with 10 mM Tris-EDTA (pH 8). Cells were resuspended in 8 M urea (pH 8) and lysed by sonication. The cell debris was separated by centrifugation, and the supernatant was stored at −20°C. H-NS was collected with a Ni-nitrilotriacetic acid agarose column (Qiagen), washed with urea buffer at pH 8.0 and 6.5, and eluted with 8 M urea (pH 4.5). Fractions containing purified H-NS-Myc-6×His were resolved by SDS-PAGE. The selected fractions were dialyzed and stored in the H-NS buffer (50 mM Tris-HCl [pH 8], 10 mM MgCl2, 20% glycerol, 500 mM NaCl, 0.1% Triton X-100). Protein concentrations were determined by the Bradford procedure.
Electrophoretic mobility shift assay (EMSA).
The DNA probes used for nonradioactive EMSA were obtained by PCR using the primers described in Table S2 in the supplemental material. A total of 40 ng of each probe was mixed with the protein buffer (10X: 400 mM HEPES, 80 mM MgCl2, 500 mM KCl, 10 mM DTT, 0.5% NP-40, 1 mg of bovine serum albumin/ml) and increasing amounts of H-NS (0 to 600 nM). The DNA fragments and H-NS mixtures were incubated for 20 min at room temperature and analyzed by electrophoresis in 6% native polyacrylamide gels in 0.5× Tris-borate-EDTA buffer. The DNA bands were visualized by ethidium bromide staining. The structural ler gene from enteropathogenic Escherichia coli (EPEC) was used as a negative control.
RESULTS
The LysR family transcriptional regulator LeuO controls assT-dsbL-dsbI expression in S. Typhi IMSS-1.
Previous reports suggested that the assT, dsbL, and dsbI genes are transcribed as an operon in UPEC and Enterobacter amnigenus (25, 37); however, no transcriptional mechanism of expression has been defined for either organism. More recently, in S. Typhimurium it has been shown that dsbL expression is independent of assT transcription (40). Previous studies performed in our laboratory with the S. Typhi IMSS-1 wild-type strain showed that the assT transcriptional expression depends on the transcriptional regulator LeuO and, by primer extension experiments, a LeuO-dependent promoter (PLeuO), located 164 bp upstream of the putative assT translation start site, was reported (26).
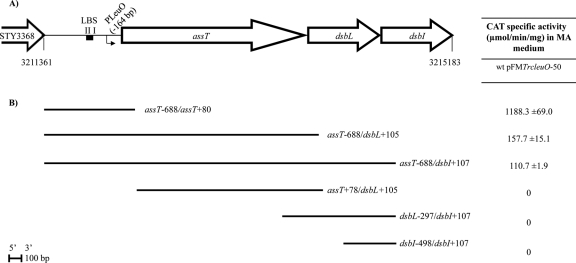
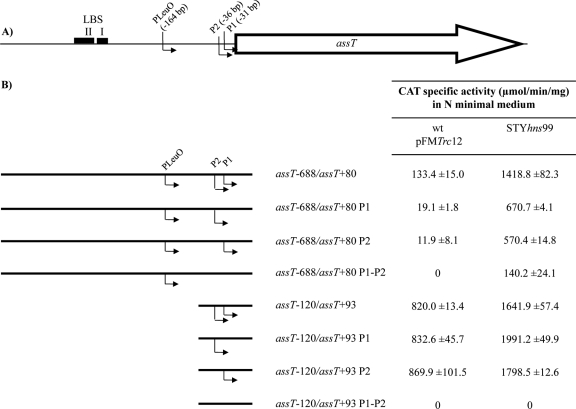
To determine whether assT, dsbL, and dsbI form a LeuO-dependent operon, several cat transcriptional fusions were designed (Fig. 1), and their expression was evaluated in MA medium, in S. Typhi IMSS-1 harboring the pFMTrc12 control vector or the pFMTrcleuO-50 plasmid (pFMTrc12 derivative plasmid encoding leuO under an IPTG-inducible ptrc promoter; see Table S1 in the supplemental material) (18, 21). As previously reported (26), assT was not expressed in the wild-type strain with the control vector (data not shown), although it was indeed induced in a fusion that contains 688 bp upstream and 80 bp downstream from the assT translation start site (pKK232-9 assT−688/assT+80), upon LeuO overexpression. In this respect, the coordinates in all fusions are relative to the translational start sites for each gene. The transcriptional fusions assT−688/dsbL+105 and assT−688/dsbI+107, containing the assT LeuO-dependent promoter, as well as the upstream regions of dsbL and dsbI, respectively, also showed null transcriptional activity in the wild-type strain with the plasmid vector (data not shown). In contrast, transcriptional expression was detected in the presence of pFMTrcleuO-50 induced with 50 μM IPTG (Fig. 1), albeit at lower levels compared to assT−688/assT+80. Elimination of the assT regulatory region, including the LeuO-dependent promoter (assT+78/dsbL+105, dsbL−297/dsbI+107, and dsbI−498/dsbI+107 cat fusions), abolished the transcriptional activation generated by LeuO, indicating that the regulatory region of assT is essential for assT-dsbL-dsbI expression mediated by LeuO, and suggesting a LeuO-dependent operon in S. Typhi IMSS-1.
Fig 1.
(A) assT-dsbL-dsbI gene cluster organization in S. Typhi. The LeuO-dependent promoter (PLeuO), as well as the LeuO binding sites I and II (LBS), is shown. The location of the assT-dsbL-dsbI cluster is annotated according to the S. enterica serovar Typhi CT18 genome (51). (B) DNA fragments fused to the cat reporter gene in the 3′ end. The coordinates in each fragment are according to the upstream or downstream distance in nucleotides from the start codon of the corresponding open reading frame. The right column indicates the CAT specific activity obtained from each construction in S. Typhi IMSS-1 wild type with pFMTrcleuO-50, induced with 50 mM IPTG, in MA medium at 12 h.
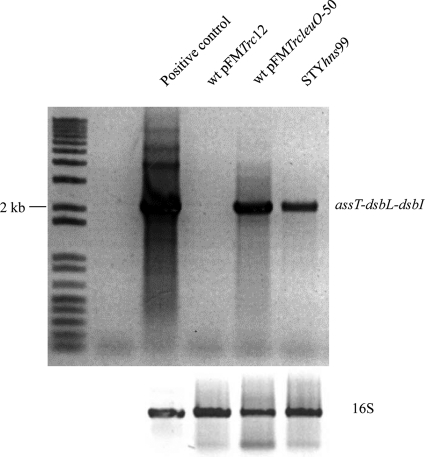
Further support of the assT, dsbL, and dsbI operon organization comes from the RT-PCR amplification of a 2,025-bp fragment that contained most of the assT structural gene, the dsbL, and part of the dsbI genes (dsbL−1232 to dsbI+107) detected upon LeuO overexpression (Fig. 2). As expected, no transcription of the operon was observed from S. Typhi wild type with the pFMTrc12 empty vector. These data indicate that assT, dsbL, and dsbI are functionally organized as a LeuO-dependent operon in the rich MA medium.
Fig 2.
RT-PCR of the assT-dsbL-dsbI gene cluster from S. Typhi IMSS-1 wild type with either the pFMTrc12 empty plasmid vector or the pFMTrcleuO-50 induced with 50 mM IPTG and from IMSS-1 STYhns99 in MA medium. As a gel loading control, a fragment of the gene encoding the 16S rRNA was amplified. As a full-length positive control, a PCR from genomic DNA was amplified.
In order to determine whether the assT PLeuO promoter (26) generates the assT-dsbL-dsbI polycistronic mRNA, A/T-to-G/C substitutions in the −10 box (TACTAT to CGCCGC) were generated in the transcriptional fusions assT−688/assT+80, assT−688/dsbL+105, and assT−688/dsbI+107. The transcription assay showed null expression when the TATA box was mutated in all of these fusions in the presence of overexpressed LeuO (data not shown), showing that PLeuO indeed transcribes assT, dsbL, and dsbI as an operon in MA medium.
Effect of deletions and point mutations in the LeuO binding sites of the assT-dsbL-dsbI operon.
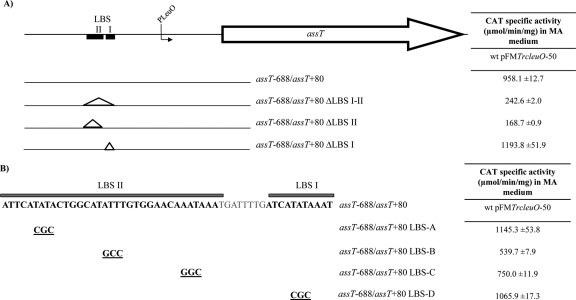
Transcription experiments indicated that assT-dsbL-dsbI expression is dependent on LeuO induction in rich medium. Previously, we determined by DNase protection footprinting experiments the presence of two LeuO binding sites (LBS I and LBS II), located at bp −276 to −286 and bp −295 to −328 upstream of the assT translation start site (26). In order to identify the nucleotides involved in the assT LeuO-dependent activation, deletions and substitution in the LBS were generated (Fig. 3), and their transcriptional expression evaluated in wild-type S. Typhi IMSS-1 harboring the pFMTrc12 or the pFMTrcleuO-50 plasmid.
Fig 3.
(A and B) Deletions (A) and substitutions (B) in the LeuO binding sites (LBS I and II) generated in the assT−688/assT+80 fusion. The right columns indicate CAT specific activity from each fusion in S. Typhi IMSS-1 containing the pFMTrcleuO-50 in MA medium induced with 50 mM IPTG at 12 h.
In the absence of cloned leuO, null transcriptional activity was observed in all constructions analyzed (data not shown). Interestingly, compared to assT−688/assT+80, CAT activity levels in the presence of LeuO decreased more than 75% when the LBS II or both the LBS I-II were eliminated (Fig. 3). In contrast, a null effect was observed with the elimination of LBS I, showing that the LBS II nucleotide sequence is crucial for assT transcriptional activation mediated by LeuO.
Because LeuO still activates assT when both LBS I and II have been deleted (assT−688/assT+80ΔLBS I-II construction), although at <25% of the wild-type levels, we analyzed, by footprinting of a DNA fragment lacking both LBS I and II sites, whether there are other LeuO-binding sites in this region. However, no DNase protection was observed (data not shown). As a control, a wild-type DNA fragment from assT was used for footprinting assays and, as reported previously (26), the two LeuO binding sites were detected. This result suggests that there are no other high-affinity LeuO binding sites in this region.
In order to determine the nucleotides in LBS II involved in assT activation by LeuO, substitutions were generated on the assT−688/assT+80 gene fusion (Fig. 3B). Three regions with high A+T content were replaced by G+C, considering that LeuO binds preferentially to sites with high A+T (9, 10, 11, 26). The substitutions assT−688/assT+80 LBS-B and assT−688/assT+80 LBS-C, decreased LeuO-mediated assT activation by 53 and 35%, respectively (Fig. 3), whereas no changes in transcriptional activity were detected from assT−688/assT+80 LBS-D, containing a 3-bp substitution in LBS I. These results indicate, in agreement with data obtained from LBS deletions, that the nucleotides of LBS II are more relevant for LeuO transcriptional activation.
assT transcriptional activation in N-minimal medium and repression by H-NS.
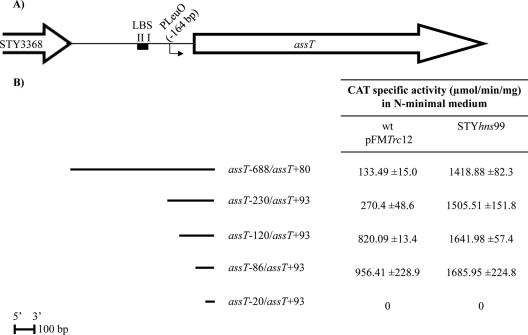
In vivo, assT, dsbL, and dsbI expression has been detected in macrophages infected with Salmonella (20). In order to determine assT activation in this environmental condition, the transcriptional activity of cat gene fusions, containing different lengths of the 5′ assT upstream region, were analyzed in N-minimal medium, a laboratory growth condition widely used for induction of SPI-2 pathogenicity island expression (16).
Surprisingly, assT expression was observed in wild-type pFMTrc12 from the assT−688/assT+80 transcriptional fusion (Fig. 4). Interestingly, the elimination of nucleotides −230 to −120 (assT−120/assT+93), showed the highest transcriptional levels of assT gene expression in S. Typhi IMSS-1 wild type, containing the empty vector pFMTrc12, and similar CAT activity levels were obtained for fusion assT−86/assT+93, whereas no activity could be detected from the assT−20/assT+93 cat fusion. The data indicate that assT is induced in N-minimal medium in the absence of LeuO, since the same activity was observed in all of the constructions in a ΔleuO isogenic background (data not shown). Moreover, the results indicate the presence of a promoter closer to the ATG (between bp −86 and −20 regarding the assT translation start site), as well as a negative regulatory elements located between bp −230 and −120 upstream of the assT translational start site.
Fig 4.
(A) assT gene organization in S. Typhi. The LeuO-dependent promoter (PLeuO), as well as the LeuO binding sites I and II (LBS), is shown. (B) DNA fragments fused to the cat reporter gene in the 3′ end. The right columns indicate the CAT specific activity obtained from each construction in S. Typhi IMSS-1 wild type with pFMTrc12 and in IMSS-1 STYhns99 in N-minimal medium at 12 h.
Since previous reports have shown that H-NS negatively regulates assT transcription in S. Typhi IMSS-1 (26, 42, 49), the assT transcriptional fusions were analyzed in an hns background, in N-minimal medium (Fig. 4). Interestingly, derepression of assT was observed and similar transcriptional levels were obtained for all fusions (with the exception of the assT−20/assT+93 cat fusion in which no activity could be detected), indicating that H-NS is an assT major negative regulator in this culture condition. The results also showed that H-NS represses expression from the transcription start site located proximal to the ATG (Fig. 4).
Determination of the assT transcriptional start site in N-minimal medium.
Transcriptional fusions suggested that an assT promoter, located between nucleotides −20 and −86 regard to the translation start site, directed assT expression in N-minimal medium. Primer extension assays and 5′RACE were used to identify such promoter.
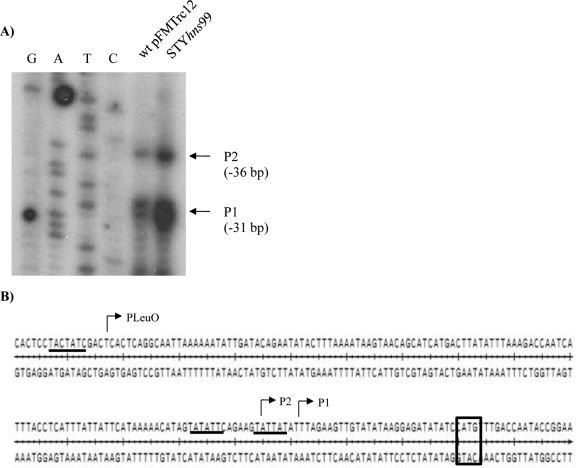
For primer extension assays, RNA was isolated from the S. Typhi IMSS-1 wild type harboring the pFMTrc12 empty vector or S. Typhi STYhns99, both containing the assT−120/assT+93 fusion. These strains were selected due to the high transcriptional levels detected in the CAT expression assays in N-minimal medium (Fig. 4). Primer extension analyses showed two 5′ ends located at bp −31 (P1) and −36 (P2) from the assT translation start site (Fig. 5), with a higher level of transcription in an hns mutant, according with the data obtained from transcriptional assays (Fig. 4).
Fig 5.
assT promoter region in N-minimal medium. (A) Primer extension assay performed in N-minimal medium at 12 h from S. Typhi IMSS-1 wild type with pFMTrc12 and from IMSS-1 STYhns99, with both containing the assT−120/assT+93 gene fusion. (B) Upstream regulatory sequence of assT. The location of the PLeuO (−164) promoter observed in MA medium and the two transcription start sites, P1 (−31) and P2 (−36), observed in N-minimal medium are shown, as well the corresponding −10 TATA boxes, which are underlined. The initiation codon for assT is indicated in a rectangle.
The 5′RACE experiments were performed from total RNA extracted from cells grown in N-minimal medium, using wild-type pFMTrc12, and using two strategies: polynucleotide addition by terminal transferase and ligation of an oligonucleotide adapter. Interestingly, only a 5′ end located at bp −30 upstream of the assT ATG was identified, which differs by 1 bp from P1, identified by primer extension (see Fig. S1 in the supplemental material). Moreover, the same 5′ end was identified in STYhns99 (not shown).
To validate the transcription start sites identified by primer extension, substitutions in the −10 box (A/T to G/C) from each promoter, P1 and P2, or a double substitution P1-P2, were generated in fusions assT−688/assT+80 and assT−120/assT+93, and transcriptional activity assays were performed in N-minimal medium (Fig. 6). The substitutions in the −10 region of P1 or P2 in the assT−688/assT+80 fusions generated lower transcriptional levels in either wild-type pFMTrc12 or STYhns99 mutant, whereas no CAT activity was detected from a double promoter substitution in a wild-type background, indicating that P1 and P2 are the assT promoters expressed in N-minimal medium. In contrast, low levels of assT expression were detected in the absence of P1 and P2, in a STYhns99 strain, suggesting that the upstream PLeuO promoter activity is also repressed by the H-NS global regulator in N-minimal medium.
Fig 6.
(A) assT gene organization in S. Typhi. The three assT promoters (P1, P2, and PLeuO), as well as the LeuO binding sites I and II (LBS), are shown. (B) DNA fragments fused to the cat reporter gene in the 3′ end with substitution in the −10 box of the P1 and/or P2 promoters in the regulatory region of assT, in the constructions containing the region assT−688/assT+80 or assT−120/assT+93. For P1, the nucleotides TATTAT were changed for CGCCGC, whereas for P2, the nucleotides TATATT were changed for CGCGCC. The right columns indicate the CAT specific activity obtained from each construction in S. Typhi IMSS-1 wild type with pFMTrc12 and in IMSS-1 STYhns99 in N-minimal medium at 12 h.
Transcriptional assays from assT−120/assT+93 derivatives showed no changes in expression in independent promoter substitutions in wild-type pFMTrc12, and STYhns99 (Fig. 6); in contrast, double promoter substitutions generated no transcriptional activity in N-minimal medium. These results validate the data obtained by primer extension, showing two assT transcription start sites, located at bp −31 and −36 upstream the assT translation start site, preferentially expressed in N-minimal medium and negatively regulated by H-NS.
Regulation of dsbL and dsbI expression in N-minimal medium.
As previously mentioned, S. Typhimurium expressed assT, as well as dsbL and dsbI, during macrophage infection (20); however, the mechanisms of the transcriptional regulation of dsbL and dsbI have not been elucidated. As shown above, transcriptional assays and RT-PCR showed LeuO-dependent activation of the assT-dsbL-dsbI operon in rich MA medium, whose expression was enhanced under a LeuO-dependent promoter (Fig. 1 and 2). Moreover, RT-PCR performed in an hns background in MA medium generates a 2-kb PCR product, which corresponds in length to the polycistronic fragment (Fig. 2), showing that the assT-dsbL-dsbI operon is regulated negatively by the H-NS global regulator. However, because assT expression could be detected in the absence of LeuO from P1 and P2 in N-minimal medium, indicating a different regulatory mechanism, it was thus of interest to analyze the transcriptional organization of the assT-dsbL-dsbI gene cluster in N-minimal medium, for which the appropriate cat transcriptional fusions were designed (Fig. 1 and Table 1).
Table 1.
CAT specific activity in N-minimal medium at 12 h from S. Typhi IMSS-1 wild type with the pFMTrc12 empty vector or STYhns99
| PCR fused to cata | Mean CAT sp act (μmol/min/mg) ± SD in N-minimal medium |
|
|---|---|---|
| Wild-type pFMTrc12 | STYhns99 | |
| assT+78/dsbL+105 | 392.8 ± 48.2 | 343.0 ± 44.4 |
| dsbL−297/dsbI+107 | 30.3 ± 11.6 | 320.0 ± 57.4 |
| dsbI−498/dsbI+107 | 0 | 0 |
The PCR fragments fused to the cat reporter gene, containing the upstream region of dsbL and/or dsbI, are indicated.
The transcriptional assays performed in N-minimal medium showed dsbL expression in the wild-type strain containing pFMTrc12 from the assT+78/dsbL+105 fusion, which does not contain the assT promoters or sequences upstream of the assT translation start site (the construction begins at 1,738 bp upstream of the dsbL translation start site). The results suggest an alternative promoter for dsbL, independent of assT expression that is activated in N-minimal medium. It is noteworthy that expression of dsbL (assT+78/dsbL+105) was not influenced by the hns mutation (Table 1).
Transcriptional activity could be detected in N-minimal medium in the wild-type pFMTrc12 from cat fusion dsbL−297/dsbI+107, containing the 5′ regulatory region and the whole structural dsbL gene and a portion of the structural dsbI gene (Table 1). Higher transcriptional expression was observed in the STYhns99 strain. Interestingly, no transcriptional activity was detected with the elimination of the dsbL regulatory region, in dsbI−498/dsbI+107 in the two backgrounds analyzed. These data indicate that dsbL and dsbI are transcribed as an operon in N-minimal medium.
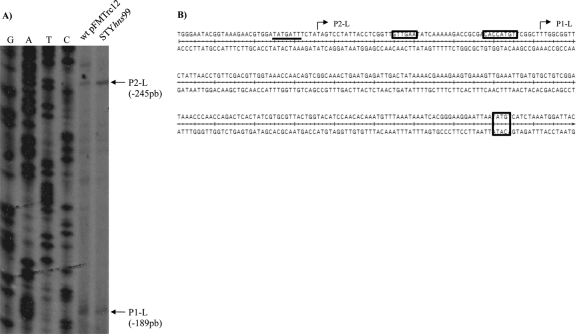
To determine the dsbL transcriptional start sites expressed in N-minimal medium, primer extension assays were performed from S. Typhi wild-type strain harboring the pFMTrc12 plasmid or in S. Typhi STYhns99, both containing the assT+78/dsbL+105 fusion (Fig. 7). Two putative transcriptional initiation sites were determined: P1-L and P2-L, located at bp −189 and −245 upstream of the dsbL translation start codon, respectively. Neither promoter exhibited activation in the hns mutant, in accord with the expression values from Table 1. Interestingly, P1-L contains a recognition sequence for σ32, which is involved in heat shock stress (13); however, the promoter was activated in wild-type pFMTrc12 and in STYhns99 backgrounds in the absence of a heat shock induction. On the other hand, σ32 induction has been detected inside the macrophage during Salmonella Typhimurium infection (20), a condition that should be explored in future studies.
Fig 7.
dsbL promoter region in N-minimal medium. (A) Primer extension assay results for dsbL, performed in N-minimal medium at 12 h from S. Typhi IMSS-1 wild type with pFMTrc12 and from IMSS-1 STYhns99, both containing the assT+78/dsbL+105 gene fusion. (B) Upstream regulatory sequence of dsbL. The locations of the two transcription start sites (P1-L and P2-L) are shown. The putative −10 and −35 σ32 promoter sequences for P1-L are shown in rectangles, whereas the −10 TATA box for the P2-L σ70 promoter is underlined. The initiation codon for the dsbL gene is indicated in a rectangle.
H-NS binding sites in the assT-dsbL-dsbI gene cluster.
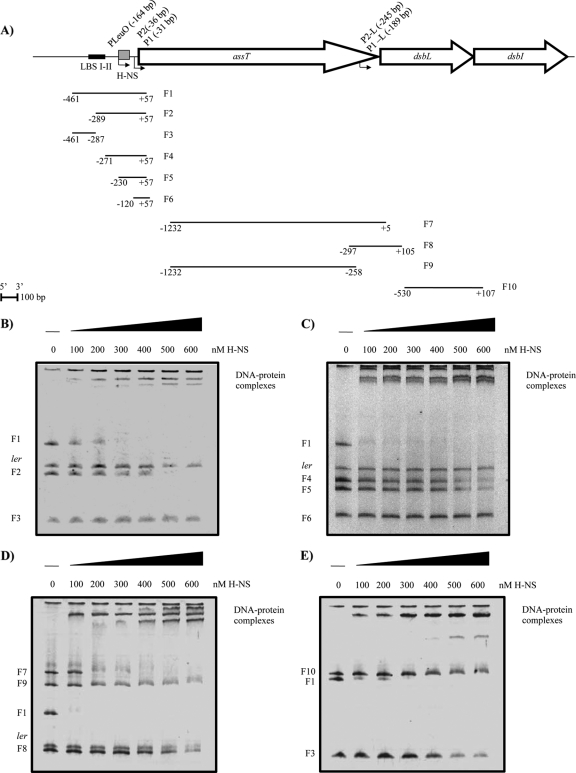
In order to define whether H-NS regulates directly assT and dsbL, EMSAs were performed with the 5′ upstream region of each gene and the purified H-NS protein (Fig. 8). A sequence from the structural ler gene from EPEC was used as a negative control (22). H-NS bound to the complete 5′ region of assT (F1) at a concentration of ≥200 nM H-NS (Fig. 8B). To narrow down the H-NS binding site, F1 was divided into two DNA fragments, F2 and F3. An H-NS-F2 complex was observed at 500 nM protein, while no interaction with H-NS was observed for F3.
Fig 8.
EMSAs of H-NS with various DNA fragments from the assT-dsbL-dsbI cluster. (A) DNA fragments utilized. The coordinates of the fragments according to the ATG of each gene are shown. EMSAs were performed with increasing amounts of H-NS (0 to 600 nM) for assT (B and C), dsbL (D), and dsbI (E). The structural ler gene from EPEC was used as a negative control for some experiments.
Additional DNA fragments were designed with the elimination of 5′ upstream sequences contained in F1. Thus, F4, F5, and F6 were generated: F4 contains the region between assT−271 and assT+57, F5 contains the region between assT−230 and assT+57, and F6 contains the region between assT−120 and assT+57 (Fig. 8C). H-NS bound to F4 and F5 at the same concentration (500 nM); interestingly, the elimination of bp −120 to −230 lowered the DNA-protein interaction (F6), a finding in agreement with the derepression observed in the transcriptional assays (Fig. 4). Hence, a full shift was only obtained with the full 5′ regulatory fragment (F1), indicating that the whole region is necessary for the strongest H-NS interaction.
A DNA fragment of the dsbL 5′ region was amplified (F7), containing 1,232 bp upstream and 5 bp downstream of the dsbL ATG (or +584 of assT on the left end). H-NS bound to F7, resulting in a full interaction at 400 nM (Fig. 8D). Interestingly, this fragment is the most similar to the one in fusion assT+78/dsbL+105 whose expression was not influenced by the hns mutation in the transcriptional assays and the primer extension (Table 1 and Fig. 7). Nevertheless, when F7 was divided into two smaller fragments (F8 and F9), containing the 5′ and 3′ regions of F7, respectively, no H-NS affinity for both F8 (−297 to +105) and F9 (−1232 to −258) was detected, suggesting nonspecific binding of H-NS to F7 in accordance with the null effect of the hns mutation on dsbL expression (Table 1 and Fig. 7) and suggesting that H-NS may act indirectly on the dsbL−297/dsbI+107 fusion (Table 1), which contains the whole dsbL gene and the initial portion of dsbI.
The H-NS interaction also was evaluated for dsbI (dsbI−530/dsbI+107) and no DNA-protein interaction could be observed, which is in agreement with previous results where neither promoters nor regulatory regions were detected close to the translation start site of dsbI (Fig. 8E). It is clear that more studies will be necessary to further understand the regulatory mechanisms for this operon.
DISCUSSION
The assT-dsbL-dsbI cluster is conserved in a small number of Enterobacteriaceae (5, 25, 41, 62). The assT gene encodes an arylsulfate sulfotransferase, and dsbL-dsbI encodes an oxidoreductase system whose function is to generate disulfide bonds in the AssT protein in the periplasm.
The Salmonella and pathogenic E. coli genomes encode a major oxidoreductase system composed of DsbA and DsbB, paralogues of DsbL and DsbI, respectively (3, 4, 14). DsbA is a periplasmic monomer that generates disulfide bonds in periplasmic proteins and contains, as well as DsbL, a CXXC motif characteristic of the thioredoxin family of proteins (24, 25). DsbB is an inner membrane protein with four transmembrane helices and two periplasmic loops, each one containing a cysteine pair, whose function is to keep DsbA in its active form by reoxidation (4, 28, 30). DsbA plays a central role in periplasmic protein folding, and diverse protein substrates for DsbA have been described, including proteins involved in transport of amino acids and peptides, RNA and protein degradation, a flagellar and type III secretion system apparatus, and some proteins such as toxins and chaperons (27, 29, 47, 50, 66). In addition to the similarity in protein structure between DsbA and DsbL, differences in the active site and the hydrogen bond network might limit potential substrate for the DsbL-DsbI oxidoreductase system (25). However, overexpression of the DsbL-DsbI system restored the absence of the major red-ox system in some DsbA targets, such as PhoA, FlgI, and PapD (40, 62).
Contrasting with the well-characterized structure and function of AssT, DsbL, and DsbI, the transcriptional regulation of the corresponding gene cluster has not been thoroughly studied. In S. enterica serovar Typhi IMSS-1, we showed that the LeuO-dependent promoter (PLeuO), located at bp −164 upstream of the assT translation start site (26), is activated only in the presence of LeuO, resulting in the transcription of assT, dsbL, and dsbI as an operon (Fig. 1 and 2). Moreover, negative regulation was observed by the global transcriptional factor H-NS (Fig. 2). Interestingly, LeuO and H-NS coregulation have been observed for several genes (8, 26, 36, 39, 57, 61, 64).
LeuO has been described recently as a global transcriptional regulator (26, 58, 59); however, a consensus DNA-binding sequence has not been established for this protein. The analysis of the LBS showed a deficient LeuO activation when LBS II, which is the farthest from PLeuO, is eliminated from the assT regulatory region (Fig. 3). Moreover, substitutions in the LBS II affect assT activation mediated by LeuO. Similar results were obtained for ompS1, where changes in the LBS located farthest from the transcription start site affect LeuO derepression and binding (18).
Multiple promoters have been identified for several genes; more than 10% of the genes contain two promoters, whereas in some cases more than four promoters per gene have been found (46). In this respect, aside from PLeuO, the regulatory region of assT contains two additional promoters (P1 and P2), located 36 and 31 bp upstream the assT translation start site, whose activation occurs in the absence of cloned leuO and in a ΔleuO background and whose repression is mediated by H-NS upon growth in N-minimal medium (Fig. 5 and 6). The binding site for H-NS was ascribed to the −230 to −120 assT region, slightly upstream of P1 and P2 and encompassing PLeuO. This site was based on the activity results in Fig. 4, where removal of such region results in lesser fold induction in the hns background. However, the facts that subdivisions of the whole assT 5′ regulatory region into fragments F2, F4, and F5 results in lower binding and F3, encompassing the furthest 5′ upstream region, does not bind H-NS (Fig. 8) suggest that the whole segment is needed for the tightest binding, and yet the initial nucleation site appears to be at −230 to −120.
Moreover, in the upstream region of dsbL two promoters were also identified (P1-L and P2-L) in N-minimal medium (Fig. 7), which were active in the absence of cloned leuO and in a ΔleuO background. Interestingly, no promoter was observed in the upstream region of dsbI, although transcriptional activity was detected in a dsbI gene fusion that contains both P1-L and P2-L, indicating that dsbL-dsbI form an operon that is expressed in this growth condition (Table 1). However, it cannot be excluded that the assT-dsbL-dsbI gene cluster is also transcribed as an operon in N-minimal medium. P1-L contains a putative σ32 promoter sequence, whereas P2-L is very likely a σ70 promoter (7, 13). In this respect, it is interesting that no heat shock stress was utilized for dsbL expression; however, growth in N-minimal medium could be an artificial stress signal for the dsbL activation under σ32, since rpoH activation has been detected when Salmonella is inside macrophages (20).
N-minimal medium, which contains low concentrations of magnesium and phosphate, has been utilized to activate virulence gene expression in Salmonella pathogenicity island 2 (SPI-2), which is preferentially expressed inside the macrophage when Salmonella replicates in the vacuole (1, 16, 17, 20). Despite the activation of assT, dsbL, and dsbI in N-minimal medium, a mutation in the gene encoding the major SPI-2 global regulator, i.e., PhoP (48), had no considerable effect on transcription of assT or dsbL-dsbI (data not shown). Hence, it will be of interest to elucidate the regulators that allow such activation.
In conclusion, the transcriptional regulation of the assT-dsbL-dsbI gene cluster in Salmonella enterica serovar Typhi is dependent on growth culture conditions, where the global regulators LeuO and H-NS are involved in positive and negative regulation, respectively. The complex regulation observed suggests that these genes need to be expressed with high precision in space and time during infection.
Supplementary Material
ACKNOWLEDGMENTS
We thank J. L. Puente, D. Georgellis, J. Caballero, M. Olvera, A. Mendoza-Vargas, R. Oropeza, C. Guadarrama, F. J. Santana, A. Medrano-López, and A. Huerta-Saquero for stimulating discussions and E. Villa, E. López, S. Becerra, J. Yáñez, and P. Gaytán for technical help.
This research was supported by grants to E.C. from CONACyT, Mexico (grant 82383), DGAPA/UNAM (IN216310), I.H.-L. from CONACyT, Mexico (grants 89337 and 127298), DGAPA/UNAM (IN214808), and E.M. from CONACyT, Mexico (grant 83686), and by the National Institutes of Health (GM071962-03).
Footnotes
Published ahead of print 17 February 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Abshire KZ, Neidhardt FC. 1993. Analysis of proteins synthesized by Salmonella Typhimurium during growth within a host macrophage. J. Bacteriol. 175:3734–3743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Baek MC, Kwon AR, Chung YJ, Kim BK, Choi EC. 1998. Distribution of bacteria with the arylsulfate sulfotransferase activity. Arch. Pharm. Res. 21:475–477 [DOI] [PubMed] [Google Scholar]
- 3. Bardwell JC, McGovern K, Beckwith J. 1991. Identification of a protein required for disulfide bond formation in vivo. Cell 67:581–589 [DOI] [PubMed] [Google Scholar]
- 4. Bardwell JC, et al. 1993. A pathway for disulfide bond formation in vivo. Proc. Natl. Acad. Sci. U. S. A. 90:1038–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bonacorsi SP, et al. 2000. Identification of regions of the Escherichia coli chromosome specific for neonatal meningitis-associated strains. Infect. Immun. 68:2096–2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bullock SL, Fletcher JM, Beddington RS, Wilson VA. 1998. Renal agenesis in mice homozygous for a gene trap mutation in the gene encoding heparan sulfate 2-sulfotransferase. Genes Dev. 12:1894–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Burgess RR, Travers AA, Dunn JJ, Bautz EK. 1969. Factor stimulating transcription by RNA polymerase. Nature 221:43–46 [DOI] [PubMed] [Google Scholar]
- 8. Chen CC, Fang M, Majumder A, Wu HY. 2001. A 72-base pair AT-rich DNA sequence element functions as a bacterial gene silencer. J. Biol. Chem. 276:9478–9485 [DOI] [PubMed] [Google Scholar]
- 9. Chen CC, et al. 2003. LeuO-mediated transcriptional derepression. J. Biol. Chem. 278:38094–38103 [DOI] [PubMed] [Google Scholar]
- 10. Chen CC, Chou MY, Huang CH, Majumder A, Wu HY. 2005. A cis-spreading nucleoprotein filament is responsible for the gene silencing activity found in the promoter relay mechanism. J. Biol. Chem. 280:5101–5112 [DOI] [PubMed] [Google Scholar]
- 11. Chen CC, Wu HY. 2005. LeuO protein delimits the transcriptionally active end repressive domains on the bacterial chromosome. J. Biol. Chem. 280:15111–15121 [DOI] [PubMed] [Google Scholar]
- 12. Ciria R, Abreu-Goodger C, Morett E, Merino E. 2004. GeConT: gene context analysis. Bioinformatics 20:2307–2308 [DOI] [PubMed] [Google Scholar]
- 13. Cowing DW, et al. 1985. Consensus sequence for Escherichia coli heat shock gene promoters. Proc. Natl. Acad. Sci. U. S. A. 82:2679–2683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dailey FE, Berg HC. 1993. Mutants in disulfide bond formation that disrupt flagellar assembly in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 90:1043–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Deiwick J, Nikolaus T, Erdogan S, Hensel M. 1999. Environmental regulation of Salmonella pathogenicity island 2 gene expression. Mol. Microbiol. 31:1759–1773 [DOI] [PubMed] [Google Scholar]
- 17. Deiwick J, Hensel M. 1999. Regulation of virulence genes by environmental signals in Salmonella Typhimurium. Electrophoresis 20:813–817 [DOI] [PubMed] [Google Scholar]
- 18. De la Cruz MA, et al. 2007. LeuO antagonizes H-NS and StpA-dependent repression in Salmonella enterica ompS1. Mol. Microbiol. 66:727–743 [DOI] [PubMed] [Google Scholar]
- 19. Elkin M, et al. 2001. Heparanase as mediator of angiogenesis: mode of action. FASEB J. 15:1661–1663 [DOI] [PubMed] [Google Scholar]
- 20. Eriksson S, Lucchini S, Thompson A, Rhen M, Hinton JC. 2003. Unraveling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol. Microbiol. 47:103–118 [DOI] [PubMed] [Google Scholar]
- 21. Fernández-Mora M, Puente JL, Calva E. 2004. OmpR and LeuO positively regulate the Salmonella enterica serovar Typhi ompS2 porin gene. J. Bacteriol. 186:2909–2920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Flores-Valdez MA, Puente JL, Calva E. 2003. Negative osmoregulation of the Salmonella ompS1 porin gene independently of OmpR in an hns background. J. Bacteriol. 185:6497–6506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gama-Castro S, et al. 2011. RegulonDB version 7.0: transcriptional regulation of Escherichia coli K-12 integrated within genetic sensory response units (Gensor units). Nucleic Acids Res. 39(Database Issue):D98–D105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Grauschopf U, et al. 1995. Why is DsbA such an oxidizing disulfide catalyst? Cell 83:947–955 [DOI] [PubMed] [Google Scholar]
- 25. Grimshaw JP, et al. 2008. DsbL and DsbI form a specific dithiol oxidase system for periplasmic arylsulfate sulfotransferase in uropathogenic Escherichia coli. J. Mol. Biol. 380:667–680 [DOI] [PubMed] [Google Scholar]
- 26. Hernández-Lucas I, et al. 2008. The LysR-type transcriptional regulator LeuO controls expression of several genes in Salmonella enterica serovar Typhi. J. Bacteriol. 190:1658–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hiniker A, Bardwell JC. 2004. In vivo substrate specificity of periplasmic disulfide oxidoreductases. J. Biol. Chem. 279:12967–12973 [DOI] [PubMed] [Google Scholar]
- 28. Inaba K, et al. 2006. Crystal structure of the DsbB-DsbA complex reveals a mechanism of disulfide bond generation. Cell 127:789–801 [DOI] [PubMed] [Google Scholar]
- 29. Jacob-Dubuisson F, et al. 1994. PapD chaperone function in pilus biogenesis depends on oxidant and chaperone-like activities of DsbA. Proc. Natl. Acad. Sci. U. S. A. 91:11552–11556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jander G, Martin NL, Beckwith J. 1994. Two cysteines in each periplasmic domain of the membrane protein DsbB are required for its function in protein disulfide bond formation. EMBO J. 13:5121–5127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kang JW, Kwon AR, Kim DH, Choi EC. 2001. Cloning and sequencing of the astA gene encoding arylsulfate sulfotransferase from Salmonella Typhimurium. Biol. Pharm. Bull. 24:570–574 [DOI] [PubMed] [Google Scholar]
- 32. Kang JW, et al. 2001. Cloning, sequence analysis, and characterization of the astA gene encoding an arylsulfate sulfotransferase from Citrobacter freundii. Arch. Pharm. Res. 24:316–322 [DOI] [PubMed] [Google Scholar]
- 33. Kawaji H, Mizuno T, Mizushima S. 1979. Influence of molecular size and osmolarity of sugars and dextrans on the synthesis of outer membrane proteins O-8 and O-9 of Escherichia coli K-12. J. Bacteriol. 140:843–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kim DH, Konishi L, Kobashi K. 1986. Purification, characterization and reaction mechanism of novel arylsulfotransferase obtained from an anaerobic bacterium of human intestine. Biochim. Biophys. Acta 872:33–41 [DOI] [PubMed] [Google Scholar]
- 35. Kim DH, Kobashi K. 1986. The role of intestinal flora in metabolism of phenolic sulfate esters. Biochem. Pharmacol. 35:3507–3510 [DOI] [PubMed] [Google Scholar]
- 36. Klauck E, Böhringer J, Hengge-Aronis R. 1997. The LysR-like regulator LeuO in Escherichia coli is involved in the translational regulation of rpoS by affecting the expression of the small regulatory DsrA-RNA. Mol. Microbiol. 25:559–569 [DOI] [PubMed] [Google Scholar]
- 37. Kwon AR, Oh TG, Kim DH, Choi EC. 1999. Molecular cloning of the arylsulfate sulfotransferase gene and characterization of its product from Enterobacter amnigenus AR-37. Protein Expr. Purif. 17:366–372 [DOI] [PubMed] [Google Scholar]
- 38. Lawley TD, et al. 2006. Genome-wide screen for Salmonella genes required for long-term systemic infection of the mouse. PLoS Pathog. 2:87–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lawrenz MB, Miller VL. 2007. Comparative analysis of the regulation of rovA from the pathogenic yersiniae. J. Bacteriol. 189:5963–5975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lin D, Kim B, Slauch JM. 2009. DsbL and DsbI contribute to periplasmic disulfide bond formation in Salmonella enterica serovar Typhimurium. Microbiology 155:4014–4024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lloyd AL, Rasko DA, Mobley HL. 2007. Defining genomic islands and uropathogen-specific genes in uropathogenic Escherichia coli. J. Bacteriol. 189:3532–3546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lucchini S, et al. 2006. H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog. 2:746–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lum DH, Tan J, Rosen SD, Werb Z. 2007. Gene trap disruption of the mouse heparan sulfate 6-O-endosulfatase gene, Sulf2. Mol. Cell. Biol. 27:678–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Malojcić G, et al. 2008. A structural and biochemical basis for PAPS-independent sulfuryl transfer by aryl sulfotransferase from uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 105:19217–19222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mathew JA, Tan YP, Srinivasa Rao PS, Lim TM, Leung KY. 2001. Edwardsiella tarda mutants defective in siderophore production, motility, serum resistance and catalase activity. Microbiology 147:449–457 [DOI] [PubMed] [Google Scholar]
- 46. Mendoza-Vargas A, et al. 2009. Genome-wide identification of transcription start sites, promoters and transcription factor binding sites in Escherichia coli. PLoS One 4:7526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Miki T, Okada N, Danbara H. 2004. Two periplasmic disulfide oxidoreductases, DsbA and SrgA, target outer membrane protein SpiA, a component of the Salmonella pathogenicity island 2 type III secretion system. J. Biol. Chem. 279:34631–34642 [DOI] [PubMed] [Google Scholar]
- 48. Miller SI, Kukral AM, Mekalanos JJ. 1989. A two-component regulatory system (phoP phoQ) controls Salmonella Typhimurium virulence. Proc. Natl. Acad. Sci. U. S. A. 86:5054–5058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Navarre WW, et al. 2006. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science 313:236–238 [DOI] [PubMed] [Google Scholar]
- 50. Okamoto K, Baba T, Yamanaka H, Akashi N, Fujii Y. 1995. Disulfide bond formation and secretion of Escherichia coli heat-stable enterotoxin II. J. Bacteriol. 177:4579–4586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Parkhill J, et al. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848–852 [DOI] [PubMed] [Google Scholar]
- 52. Puente J, Flores V, Fernandez M, Fuchs Y, Calva E. 1987. Isolation of an ompC-like outer membrane protein gene from Salmonella Typhi. Gene 61:75–83 [DOI] [PubMed] [Google Scholar]
- 53. Roy AB. 1981. Sulfotransferases, p 131–185. In Mulder GJ. (ed), Sulfation of drugs and related compounds. CRC Press, Inc, Boca Raton, FL [Google Scholar]
- 54. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 55. Sekura RD, Duffel MW, Jakoby WB. 1981. Aryl sulfotransferases. Methods Enzymol. 77:197–206 [DOI] [PubMed] [Google Scholar]
- 56. Sharma CM, et al. 2010. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature 464:250–255 [DOI] [PubMed] [Google Scholar]
- 57. Shi X, Bennett GN. 1995. Effects of multicopy LeuO on the expression of the acid-inducible lysine decarboxylase gene in Escherichia coli. J. Bacteriol. 177:810–814.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shimada T, Yamamoto K, Ishihama A. 2009. Involvement of the leucine response transcription factor LeuO in the regulation of the genes for sulfa drug efflux. J. Bacteriol. 191:4562–4571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Shimada T, Bridier A, Briandet R, Ishihama A. 2011. Novel roles of LeuO in transcription regulation of Escherichia coli genome: antagonistic interplay with the universal silencer H-NS. Mol. Microbiol. 82:378–397 [DOI] [PubMed] [Google Scholar]
- 60. Snyder JA, et al. 2004. Transcriptome of uropathogenic Escherichia coli during urinary tract infection. Infect. Immun. 72:6373–6381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Stratmann T, Madhusudan S, Schnetz K. 2008. Regulation of the yjjQ-bglJ operon, encoding LuxR-type transcription factors, and the divergent yjjP gene by H-NS and LeuO. J. Bacteriol. 190:926–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Totsika M, Heras B, Wurpel DJ, Schembri MA. 2009. Characterization of two homologous disulfide bond systems involved in virulence factor biogenesis in uropathogenic Escherichia coli CFT073. J. Bacteriol. 191:3901–3908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Uchimura K, et al. 2006. HSulf-2, an extracellular endoglucosamine-6-sulfatase, selectively mobilizes heparin-bound growth factors and chemokines: effects on VEGF, FGF-1, and SDF-1. BMC Biochem. 7:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ueguchi C, Ohta T, Seto C, Suziki T, Mizuno T. 1998. The leuO gene product has a latent ability to relieve bgl silencing in Escherichia coli. J. Bacteriol. 180:190–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wróbel M, Jurkowska H, Sliwa L, Srebro Z. 2004. Sulfurtransferases and cyanide detoxification in mouse liver, kidney, and brain. Toxicol. Mech. Methods 14:331–337 [DOI] [PubMed] [Google Scholar]
- 66. Yamanaka H, Kameyama M, Baba T, Fujii Y, Okamoto K. 1994. Maturation pathway of Escherichia coli heat-stable enterotoxin I: requirement of DsbA for disulfide bond formation. J. Bacteriol. 176:2906–29013 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.