Abstract
Infection with Salmonella enterica serovar Typhi in humans causes the life-threatening disease typhoid fever. In the laboratory, typhoid fever can be modeled through the inoculation of susceptible mice with Salmonella enterica serovar Typhimurium. Using this murine model, we previously characterized the interactions between Salmonella Typhimurium and host cells in the gallbladder and showed that this pathogen can successfully invade gallbladder epithelial cells and proliferate. Additionally, we showed that Salmonella Typhimurium can use bile phospholipids to grow at high rates. These abilities are likely important for quick colonization of the gallbladder during typhoid fever and further pathogen dissemination through fecal shedding. To further characterize the interactions between Salmonella and the gallbladder environment, we compared the transcriptomes of Salmonella cultures grown in LB broth or physiological murine bile. Our data showed that many genes involved in bacterial central metabolism are affected by bile, with the citric acid cycle being repressed and alternative respiratory systems being activated. Additionally, our study revealed a new aspect of Salmonella interactions with bile through the identification of the global regulator phoP as a bile-responsive gene. Repression of phoP expression could also be achieved using physiological, but not commercial, bovine bile. The biological activity does not involve PhoPQ sensing of a bile component and is not caused by bile acids, the most abundant organic components of bile. Bioactivity-guided purification allowed the identification of a subset of small molecules from bile that can elicit full activity; however, a single compound with phoP inhibitory activity could not be isolated, suggesting that multiple molecules may act in synergy to achieve this effect. Due to the critical role of phoP in Salmonella virulence, further studies in this area will likely reveal aspects of the interaction between Salmonella and bile that are relevant to disease.
INTRODUCTION
Salmonella enterica is a versatile bacterial pathogen equipped with the machinery required to infect a multitude of hosts (16). Multiple S. enterica serovars are able to infect humans, each causing specific clinical manifestations of disease (12, 16). Salmonella enterica serovar Typhimurium (Salmonella Typhimurium) causes localized, self-limiting gastroenteritis in humans, whereas S. enterica serovar Typhi (Salmonella Typhi) causes systemic, life-threatening typhoid fever (12, 26). Human infection with Salmonella Typhi can be modeled in the laboratory through the infection of susceptible mice with Salmonella Typhimurium (34). In this model, systemic infection ensues and Salmonella can infect multiple organs, such as the gastrointestinal tract, brain, liver, spleen, and gallbladder (19, 42). Many aspects of host colonization in this murine model are reminiscent of human Salmonella Typhi infection. In particular, gallbladder infection of patients is common during typhoid fever, and this can also occur in the absence of clinical symptoms; these patients are asymptomatic carriers that aid in the spread of the pathogen through fecal shedding (3, 23, 36, 39). Therefore, the interactions between pathogen and host in the gallbladder are important aspects that must be studied to better understand Salmonella infections in humans.
We have recently studied the interactions between Salmonella and its host using a gallbladder infection model (19). Salmonella can invade and infect the epithelial cells lining the lumen of the gallbladder in a murine host (19). Additionally, Salmonella can successfully colonize bile, where it can grow at rates comparable to those achieved in rich culture medium in the laboratory, presumably using phospholipids as carbon and energy sources (1, 19). Therefore, in order to further characterize the interactions between Salmonella and the host gallbladder environment, we used DNA microarrays to perform a transcriptome analysis of Salmonella growth in bile. We found that many genes are differentially regulated during growth in bile, with the central metabolism being deeply affected. Interestingly, we also found that phoP, a major regulator of virulence gene expression in Salmonella and many other bacteria (10, 13, 18), was strongly repressed by bile. Follow-up experiments determined that the repression of phoP expression does not occur through an effect on PhoPQ signaling; i.e., our evidence suggests that this phenotype is not due to direct sensing of a bile component by the PhoPQ signaling cascade. The repression of phoP expression could also be replicated using physiological (but not commercial) bovine bile, and a subset of small molecules from bile with full inhibitory activity was determined. Further studies in this area should reveal more details of the intricate interaction between Salmonella and bile and its impact on this pathogen's lifestyle.
MATERIALS AND METHODS
Chemicals.
Carbenicillin was purchased from EMD Chemicals (San Diego, CA). Streptomycin, dimethyl sulfoxide (DMSO), dehydrocholic acid, cholic acid, chenodeoxycholic acid, deoxycholic acid, glycolithocholic acid ethyl ester, taurocholic acid sodium salt, lithocholic acid, sodium taurodeoxycholate hydrate, sodium tauroursodeoxycholate, sodium taurochenodeoxycholate, methanol, and trifluoroacetic acid were purchased from Sigma-Aldrich (Oakville, Canada).
Commercial bile.
Commercial bovine bile was obtained from Sigma-Aldrich and is a crude bile extract from ox gallbladder. It is concentrated through boiling to approximately 50 to 60% total solids, vacuum dried, milled to a powder, and packaged. This powder is soluble in water at 50 mg/ml. For the purpose of comparison, we note that in order to approximate the natural composition of bile, this powder would have to be dissolved in water at a concentration of approximately 80 mg/ml.
Physiological bile.
We obtained fresh bile from C57BL/6 and 129Sv/ImJ Nramp1+/+ and Nramp1−/− mice from breeding colonies maintained at the Wesbrook Animal Unit at The University of British Columbia. Mice were fed a standard sterile chow diet (Laboratory Rodent Diet 5001; Purina Mills, St. Louis, MO) ad libitum throughout the experiments. Mice were sacrificed through CO2 asphyxiation, and bile was collected immediately. Bile samples were centrifuged for 5 min at 16,000 × g, and the supernatant was collected and frozen at −20°C until used. We also obtained fresh cow bile from a small slaughter plant as a kind gift from Neil Rawlyk, Sherry Tetland, Don Wilson, and Andrew Potter (Vaccine and Infectious Disease Organization, University of Saskatchewan). Bile was collected and saved at −20°C until used. In order to distinguish them from commercial bile, the fresh murine bile and bovine bile obtained and used in this study are referred to as “physiological bile.”
Microarray experiments.
Salmonella Typhimurium LT2 glass slide microarrays were obtained through the NIAID's Pathogen Functional Genomics Resource Center (PFGRC), managed and funded by Division of Microbiology and Infectious Diseases, NIAID, NIH, DHHS, and operated by the J. Craig Venter Institute. Two independent cultures of Salmonella Typhimurium SL1344 (43) were grown overnight in Luria-Bertani (LB) broth with streptomycin (100 μg/ml) at 37°C with shaking (225 rpm). Cells were pelleted by centrifugation and resuspended in sterile Dulbecco's phosphate-buffered saline (PBS; HyClone, Logan, UT). These suspensions were further diluted 1:50 in PBS and used to inoculate 70 μl of either LB broth or physiological murine bile (in 0.65-ml tubes) at a 1:100 dilution. Cultures were incubated for 24 h at 37°C with agitation (225 rpm). Then, samples were diluted in PBS, and serial dilutions were plated on LB plates containing 100 μg/ml of streptomycin and incubated overnight at 37°C for bacterial growth and enumeration by colony counting. The remaining cells were used in the subsequent steps, as described below. RNA isolation began with the addition of 2 volumes of RNAprotect bacterial reagent (Qiagen, Hilden, Germany) to the bacterial cultures and incubation at room temperature for approximately 5 min. Cells were pelleted by centrifugation, and RNA was isolated using the RNeasy minikit (Qiagen) with on-column DNase digestion, according to the manufacturer's recommendations. Synthesis of cDNA was performed as described herein. The entire RNA sample (≥3 μg) was mixed with 6 μg of random primers (Invitrogen, Burlington, Canada) and 40 U of RNaseOUT recombinant RNase inhibitor (Invitrogen) in a total volume of 18.5 μl and incubated at 70°C for 10 min. Samples were incubated on ice for 30 s, centrifuged to bring down condensation, and incubated with the following components (Invitrogen) at the final concentrations shown in parentheses: First Strand buffer (1×), dithiothreitol (10 mM), dNTPs (dATP, dCTP, dGTP, dTTP; 0.5 mM each) and 100 U of SuperScript II reverse transcriptase in a total volume of 30 μl. The mixture was incubated at 42°C overnight, and the reaction was stopped and the RNA degraded through the addition of 0.5 M EDTA and 1 M sodium hydroxide (10 μl each) and incubation at 65°C for 15 min. Then, 25 μl of 1 M Tris (pH 7) was added to neutralize the pH of the cDNA solution. cDNA was purified using the MinElute PCR purification kit (Qiagen) according to the manufacturer's recommendations, except that the Qiagen wash and elution buffers were substituted for by phosphate buffers, according to the PFGRC microarray protocol. cDNA was then precipitated using ammonium acetate and ethanol, as described elsewhere (17), and labeled with Cy3 (LB cultures) and Cy5 (bile cultures) using the ULS aRNA fluorescent labeling kit (Kreatech Biotechnology, Amsterdam, The Netherlands) according to the manufacturer's instructions. Samples were mixed, dried, and saved at −80°C until used. Prehybridization, hybridization, and washing steps were performed essentially as described in the PFGRC protocols, except that the hybridization buffer contained KREAblock blocking buffer (25%; Kreatech Biotechnology). After hybridization, slides were scanned using an Affymetrix 428 array scanner from Eurofins MWG operon (Huntsville, AL).
Microarray data analysis.
Scanned glass slide images were processed with Spotfinder and MIDAS version 2.22 (32). Each of the two slides was processed independently and contains two spots for each Salmonella gene, resulting in a total of 4 possible measurements of transcript abundance per gene. Genes that were detected in at least 3 of the 4 spots and showed average differences in signal intensity of 2.5-fold or more between the two growth conditions as well as individual differences of 2-fold or more in all spots with a detectable signal were selected and are reported.
Real-time PCR.
Salmonella was cultured in LB broth or physiological murine bile, essentially as described for microarray experiments. RNA was stabilized using RNAprotect bacterial reagent (Qiagen) and extracted using the RNeasy minikit (Qiagen) with the on-column DNase digestion, according to the manufacturer's recommendations. Synthesis of cDNA was performed using the QuantiTect reverse transcription (RT) kit (Qiagen). For RT-PCR, we used the QuantiTect SYBR green PCR kit (Qiagen) and the Applied Biosystems (Foster City, CA) 7500 system. Forward and reverse primers were added to reaction mixtures at a final concentration of 0.4 μM each. All results were normalized using the expression levels of the housekeeping gene gapA, encoding the glyceraldehyde-3-phosphate dehydrogenase enzyme (22), as the baseline. Averages of the data obtained with cultures grown in LB broth were normalized to 1, and the data from each sample (LB broth or bile) were normalized accordingly. Primers used for RT-PCR are shown on Table 1.
Table 1.
Primers used in this study
| Primer | Sequence (5′→3′)a |
|---|---|
| Real-time PCR | |
| PHOPF | CAATGAAGAGGTCATCAAACTCAC |
| PHOPR | GAGAACATCAATGGTATGACTTTCC |
| PMRAF | CATAATAACCAGGGTGAAAGTGAAC |
| PMRAR | CGTTATCCCAGTTGTAGATATCGTT |
| MGTAF | AATCCTTTCAACATCTTACTCACGA |
| MGTAR | ATTTTCATTAATAACCCGCAGTACG |
| MGTCF | AGGGAGAAAAACGTTATATCCTGAA |
| MGTCR | ATTTCTTTATAGCCCTGTTCCTGAG |
| PAGCF | ACATTTAAAGAACATTCCACTCAGG |
| PAGCR | AGCCGTTTATTTTTGTAGAGGAGAT |
| Cloning | |
| PPHOPF | ATTAGAGCTCTCGCGCTGTGACTCTGGTCG |
| PPHOPR | ATAAGGATCCATCCTCTACAACCAGTACGC |
| PHILAF | TCCTGAGCTCGATATAATGCCTGGAGCC |
| PHILAR | CCATGGATCCTATGAAATCATCAAAGACG |
Restriction enzyme sites are underlined.
Salmonella growth curves.
We obtained fresh, physiological bovine bile, adjusted its pH to match that of LB broth (around 7.3), and used it to perform Salmonella growth assays. LB broth and bile were tested either alone or in 1:1 mixtures with each other or PBS, as indicated. Streptomycin was present in all experiments at a final concentration of 100 μg/ml. Overnight cultures of Salmonella Typhimurium SL1344 were used to inoculate each culture medium at 1:200. Cultures were then incubated at 37°C with shaking (225 rpm), and growth was followed through measurements of the optical density at 600 nm.
GFP reporter assays.
In order to monitor the effect of bile or bile components on hilA or phoP expression, we amplified DNA fragments containing the promoters of hilA (−675 through +70, relative to the translational start codon) and phoP (−163 through +33, relative to the translational start codon) through PCR and independently cloned them upstream of the promoterless green fluorescent protein (GFP) gene gfp of pFPV25 (40) using the SacI and BamHI restriction sites and standard cloning procedures, as described elsewhere (2, 33). The primers used are shown on Table 1. The resulting plasmids were transformed into the appropriate Salmonella strains, and GFP production was analyzed through flow cytometry of bacterial cultures using a FACSCalibur (BD Biosciences, Franklin Lakes, NJ), as indicated. All cultures contained carbenicillin (100 μg/ml) and were incubated at 37°C with shaking (225 rpm). In each experiment, between 25,000 and 50,000 events were collected per sample.
Solid-phase extraction of the bioactive compound in physiological bovine bile.
In order to purify the active compound from physiological bovine bile, we used Sep-Pak C18 resin cartridges (Waters, Milford, MA). Columns were washed with 10 volumes of methanol (relative to the column bed weight) followed by a wash with the same volume of water. Ten volumes of physiological bovine bile was applied to the column, and the flowthrough was collected. Columns were then washed with 20 volumes of water, the molecules bound were eluted with 10 volumes of 50% methanol (in water) followed by 10 volumes of 100% methanol, and both fractions were collected.
RP-HPLC.
The fraction from the C18 cartridge purification eluted with 100% methanol described above was dried in a centrifuge equipped with a vacuum pump and resuspended in 0.1 volume of 25% methanol (in water). The suspension was filtered through a 0.22-μm pore-size membrane and used for further purification through reverse-phase high-performance liquid chromatography (RP-HPLC). RP-HPLC was performed using a Nucleosil C18 column (5-μm particle size, 25 by 0.46 cm; Sigma-Aldrich) with methanol-water mixtures containing 0.1% trifluoroacetic acid as the mobile phase, as indicated.
UPLC-MS.
For ultrahigh-performance liquid chromatography coupled with mass spectrometry (UPLC-MS), select fractions from RP-HPLC were dried and the residues were dissolved in 200 μl of methanol and diluted 1 to 500 with 75% methanol. Three microliters was injected into a 10- by 0.21-cm C18 UPLC column and run on the UPLC-quadrupole time of flight (QTOF) mass spectrometer in electrospray ionization [ESI(−)] mode.
Statistical analyses.
Data were analyzed by unpaired t tests with 95% confidence intervals using GraphPad Prism version 4.0 (GraphPad Software, Inc., San Diego, CA).
Microarray data accession number.
Raw microarray data were deposited in the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE33604.
RESULTS
Physiological murine bile alters the expression of multiple Salmonella genes.
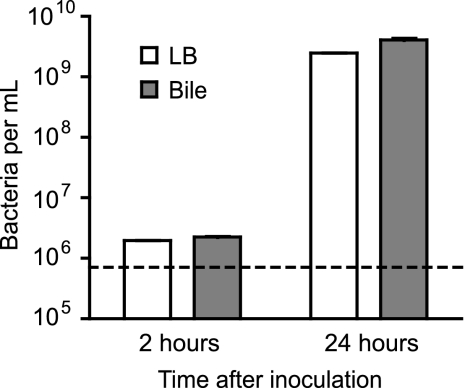
We have previously shown that Salmonella can successfully multiply in murine bile, despite the well-known antimicrobial properties of this physiological fluid (1, 19). In order to determine the impact of bile on Salmonella gene expression, we performed transcriptome comparisons between Salmonella grown in LB broth and Salmonella grown in physiological murine bile using DNA microarrays. As expected, following the inoculation of bile at an approximate concentration of 8 × 105 cells per ml, Salmonella showed signs of growth as early as 2 h postinoculation and displayed robust growth after 24 h of growth, achieving a concentration of 4 × 109 cells per ml (Fig. 1).
Fig 1.
Salmonella displays robust growth in physiological murine bile. Cultures were started by pelleting cells from an overnight Salmonella culture by centrifugation, resuspending them in one volume of PBS, diluting this solution 1:50 in PBS, and inoculating LB broth or bile with this suspension at a 1:100 dilution. This resulted in an initial bacterial concentration of approximately 8 × 105 cells per ml (indicated by the dashed line). Samples were incubated at 37°C with shaking for 24 h, and aliquots were removed, diluted, and spotted on LB broth plates for bacterial enumeration after 2 and 24 h of growth. Average results of three independent cultures with standard errors of means are shown.
To compare transcriptional profiles of cells grown in LB broth or bile, we isolated RNA from these cultures after 24 h of growth and analyzed transcript levels using Salmonella Typhimurium LT2 glass slide microarrays obtained through the Pathogen Functional Genomics Resource Center (PFGRC), operated by the J. Craig Venter Institute. By doing so, we found a total of 54 genes whose expression levels were affected 2.5-fold or more during growth in bile. Of these, 17 genes were activated, whereas 37 genes were repressed by bile (Table 2). Most of the genes differentially regulated were involved in bacterial central metabolism and respiration. We found that a significant portion of the genes repressed by bile were involved in the citric acid cycle and the generation of electron donors for aerobic respiration—e.g., succinate dehydrogenase, isocitrate dehydrogenase, aconitate hydratase, and others. Conversely, a number of the genes activated by bile were involved in alternative respiratory systems. For instance, we found that several hydrogenase-related genes (hydrogenase subunits, maturation endopeptidase, and chaperone) were activated by bile, suggesting a shift from the use of NAD(P)H and succinate to H2 as electron donors. Besides metabolic genes, we found that growth in bile affected the expression of genes involved in iron and copper metabolism and transport of sugars, amino acids, and oligopeptides, as well as virulence regulation.
Table 2.
Regulation of Salmonella gene expression during growth in physiological murine bile
| ORF no.a | Common name | Gene | Fold regulation |
|---|---|---|---|
| STM3146 | Hydrogenase 2 maturation endopeptidase | hybD | 10.36 |
| STM3506 | Ferrous iron transport protein B | feoB | 7.02 |
| STM3150 | Hydrogenase 2 small subunit | hypO | 4.55 |
| STM1907 | Copper homeostasis protein CutC | cutC | 4.46 |
| STM3511 | Putative DNA uptake protein | yhgI | 3.98 |
| STM3465 | Hypothetical protein | yhfA | 3.61 |
| STM3505 | Ferrous iron transport protein A | feoA | 3.56 |
| STM1324 | Putative cytoplasmic protein | 3.53 | |
| STM0629 | Cold shock protein CspE | cspE | 3.43 |
| STM3360 | Arginine repressor | argR | 3.17 |
| STM1200 | Thymidylate kinase | tmk | 3.16 |
| STM1121 | Putative cytoplasmic protein | ymdF | 3.15 |
| STM2856 | Hydrogenase assembly chaperone | hypC | 2.88 |
| STM3147 | Hydrogenase 2 large subunit | hybC | 2.87 |
| STM3302 | Pseudogene, yhhE | yhbE | 2.84 |
| STM0974 | Formate transporter | focA | 2.71 |
| STM2532 | Putative inner membrane lipoprotein | 2.55 | |
| STM0441 | Cytochrome o ubiquinol oxidase subunit III | cyoC | −10.71 |
| STM0439 | Protoheme IX farnesyltransferase | cyoE | −7.50 |
| STM3630 | Dipeptide transport protein | dppA | −6.35 |
| ORF01133 | Protein PhoH | phoH | −5.60 |
| STM0544 | Fimbrial protein | fimI | −5.33 |
| STM0154 | Dihydrolipoamide dehydrogenase | lpdA | −5.14 |
| STM0158 | Bifunctional aconitate hydratase 2/2-methylisocitrate dehydratase | acnB | −4.79 |
| STM2378 | 3-Oxoacyl-(acyl carrier protein) synthase I | fabB | −4.58 |
| STM4398 | d-Alanine/d-serine/glycine permease | cycA | −4.56 |
| STM0600 | Carbon starvation protein | cstA | −4.54 |
| STM3218 | Putrescine-2-oxoglutarate aminotransferase | oat | −4.43 |
| STM2389 | 3-ketoacyl-CoAb thiolase | fadI | −4.20 |
| PSLT052 | Plasmid partition protein A | parA | −4.18 |
| STM1231 | DNA-binding transcriptional regulator PhoP | phoP | −4.06 |
| STM4159 | Thiamine biosynthesis protein ThiH | thiH | −4.05 |
| STM3557 | Glycerol-3-phosphate transporter periplasmic binding protein | ugpB | −4.03 |
| STM0734 | Succinate dehydrogenase flavoprotein subunit | sdhA | −3.98 |
| STM0662 | Glutamate/aspartate transporter | gltL | −3.89 |
| ORF01131 | Bifunctional protein PutA | putA | −3.84 |
| STM1238 | Isocitrate dehydrogenase | icdA | −3.83 |
| STM1712 | Aconitate hydratase | acnA | −3.48 |
| STM0549 | Transcriptional regulator FimZ | fimZ | −3.45 |
| STM2141 | Fructose-bisphosphate aldolase | fbaB | −3.37 |
| STM0735 | Succinate dehydrogenase iron-sulfur subunit | sdhB | −3.25 |
| STM0738 | Succinyl-CoA synthetase subunit beta | sucC | −3.25 |
| STM1744 | Oligopeptide transport protein | oppC | −3.21 |
| PSLT046 | Putative carbonic anhydrase | −3.20 | |
| STM2190 | Galactose transport protein | mglB | −3.02 |
| STM2799 | DNA binding protein, nucleoid associated | stpA | −2.97 |
| STM1292 | Putative cytoplasmic protein | yeaC | −2.94 |
| STM0730 | Type II citrate synthase | gltA | −2.90 |
| STM3321 | Putative σ54 modulation protein | yhbH | −2.84 |
| STM1742 | Oligopeptide transport protein | oppF | −2.78 |
| STM3033 | Plasmid maintenance protein | −2.72 | |
| STM4274 | Putative inner membrane protein | yjcH | −2.65 |
| STM4126 | Soluble pyridine nucleotide transhydrogenase | udhA | −2.63 |
| ORF04918 | Virulence protein VsdF | −2.59 |
STM and pSLT designations refer to Salmonella enterica serovar Typhimurium LT2. Open reading frame (ORF) designations refer to Salmonella enterica serovar Typhimurium 14028.
CoA, coenzyme A.
phoP expression is strongly repressed by physiological murine bile.
Among the many genes whose expression was repressed by bile, we found one main regulator of virulence gene expression, phoP. Because both growth in bile and phoP expression are directly linked to Salmonella virulence, we chose to focus our studies on the regulation of phoP expression by bile, a previously unreported phenomenon with likely implications for Salmonella virulence and human disease. Our microarray experiments showed that phoP expression was repressed approximately 4-fold during growth in bile. In order to confirm this observation as well as to assess whether the effect of bile on phoP expression resulted in the regulation of phoP-controlled genes, we used real-time PCR to study transcript levels of phoP and several other phoP-regulated genes during growth in LB broth or physiological murine bile. Confirming our initial observation of phoP repression by bile through microarrays, our RT-PCR results indicated that phoP expression was repressed approximately 3.2-fold when Salmonella was grown in bile (Fig. 2). We also compared transcript levels of pmrA, mgtA, mgtC, and pagC, all of which are regulated by PhoP (14, 21, 37), during growth in LB broth or bile. Supporting our initial observations of phoP repression by bile, we found that transcript levels of both pmrA and pagC were significantly repressed by bile. Both pmrA and pagC are activated by PhoPQ (14, 21), and our results suggest that repression of phoP by bile causes the repression of these two PhoPQ target genes. Although the other genes tested did not reach statistically significant regulation by bile, mgtA displayed an obvious expression trend that suggests it may also be affected by bile through the regulation of phoP (Fig. 2).
Fig 2.
Salmonella growth in physiological murine bile modulates transcript levels of phoP and phoP-regulated genes. Salmonella was grown in LB broth or bile for 24 h, RNA was isolated, cDNA was synthesized, and RT-PCR was performed using primers specific for the genes indicated. Expression levels during growth in LB broth were normalized to 1, and expression in bile was adjusted accordingly. All results were normalized using the expression levels of gapA. Average expression levels of three independent cultures with standard errors of means are shown. P values were >0.05 (no asterisk), <0.03 (*), or <0.003 (**).
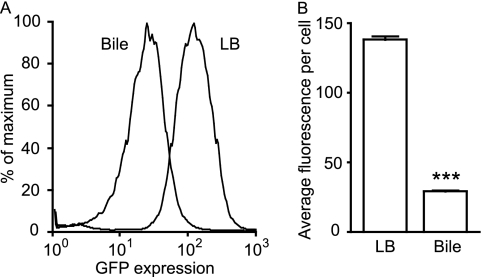
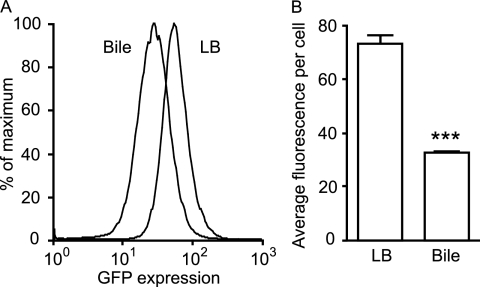
In order to further confirm the effect of bile on phoP expression and study this phenomenon in more detail, we constructed a phoP expression reporter strain by inserting the phoP promoter upstream of the promoterless gfp of pFPV25. The resulting plasmid was then inserted into wild-type Salmonella. Overnight cultures of this phoP reporter strain were used to inoculate 50 μl of LB broth or physiological murine bile (in 0.65-ml tubes) at a 1:100 dilution. The cultures were incubated for 4 h and then diluted 4-fold in PBS for flow cytometry analysis of GFP expression. Figure 3 shows that gfp expression is significantly reduced during Salmonella growth in bile compared to LB broth. In these experiments, the average fluorescence intensity per cell was approximately 4.7-fold lower in bile-grown cells than that in LB broth-grown cells.
Fig 3.
phoP expression is strongly repressed by Salmonella growth in physiological murine bile. A Salmonella strain containing a phoP::gfp plasmid was grown overnight in LB broth with carbenicillin and subcultured 1:100 in LB broth or bile containing carbenicillin. After incubation at 37°C with shaking for 4 h, cultures were diluted in PBS, and gfp expression was analyzed through flow cytometry. (A) Histogram showing the relative proportions of cells (y axis) expressing gfp at various levels (x axis) during growth in LB broth or bile, as indicated. (B) Data from panel A expressed as average fluorescence intensity per cell. The data shown are the average from three independent cultures. Bars represent the standard errors of means. The P value was <0.0001 (***).
The effect of bile on phoP expression is independent of PhoPQ signaling.
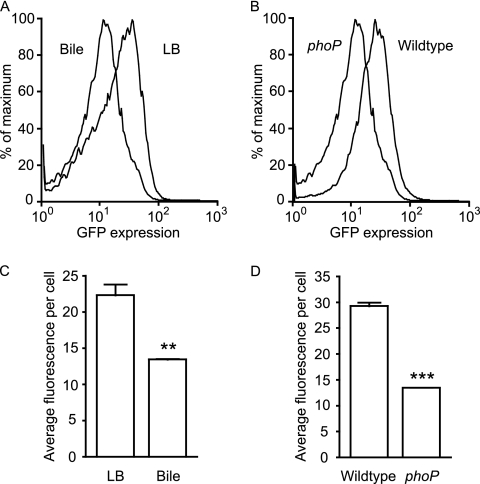
As is common with bacterial two-component regulatory systems, PhoPQ signaling controls phoPQ gene expression (35). Therefore, we decided to investigate the possibility that the effect of bile on phoP expression could be the result of a specific PhoP ligand being present in bile. If such a ligand existed and its binding repressed PhoPQ activity, one would expect that phoP expression would be repressed, resulting in a phenotype reminiscent of the one reported here. Therefore, we inserted the phoP::gfp fusion plasmid into a phoP knockout Salmonella strain constructed through P22-assisted transduction of a previously described phoP::Tn10 mutation into Salmonella Typhimurium SL1344 (9) and measured gfp expression. Overnight cultures of the resulting strain were used to inoculate aliquots of LB broth or physiological murine bile at a 1:100 dilution, as described above. The cultures were incubated for 4 h and then diluted 4-fold in PBS for flow cytometry analysis of GFP expression. If PhoPQ signaling were required for bile-mediated phoP repression, this repression would not occur in a phoP null mutant. Our results suggest that this is not the case. Figure 4 shows that bile can still repress phoP expression in a phoP::Tn10 background. Also, a comparison of phoP expression levels between wild-type and phoP::Tn10 Salmonella revealed that the introduction of a phoP mutation reduced phoP expression even during growth in bile. Altogether, our data show that bile does not repress phoP expression through repression of PhoPQ signaling.
Fig 4.
The repression of phoP expression by physiological murine bile is independent of PhoPQ signaling. The phoP::gfp reporter plasmid was inserted into wild-type Salmonella as well as a phoP::Tn10 mutant. Strains were grown overnight in LB broth with carbenicillin and subcultured 1:100 in LB broth or bile containing carbenicillin. After incubation at 37°C with shaking for 4 h, cultures were diluted in PBS, and GFP expression was analyzed through flow cytometry. (A) Histogram showing the relative proportions of cells (y axis) expressing gfp at various levels (x axis) during growth of the phoP::Tn10 Salmonella strain in LB broth or bile, as indicated. (B) Same as in panel A, except that levels of gfp production by the wild-type and phoP::Tn10 (phoP) Salmonella strains were analyzed during growth in physiological murine bile. (C) Data from panel A expressed as average fluorescence intensity per cell. (D) Data from panel B expressed as average fluorescence intensity per cell. The data shown are the average from three independent cultures. Bars represent the standard errors of means. P values were <0.005 (**) and <0.0001 (***).
Bile acids are not responsible for the bile-mediated repression of phoP expression.
Our data suggest that a molecule present in bile can repress the expression of the phoP gene without necessarily interfering with PhoPQ signaling. Because bile acids are the most abundant component of bile, accounting for approximately 67% of all of the organic content (8), we decided to study the effect of several bile acids on phoP expression. To this end, we prepared solutions of dehydrocholic acid, cholic acid, chenodeoxycholic acid, deoxycholic acid, glycolithocholic acid ethyl ester, taurocholic acid sodium salt, and lithocholic acid in DMSO and added these bile acids to LB broth at the final concentrations indicated. Controls containing only DMSO were also produced. The pH of the medium was not affected by DMSO or any of the bile acids tested. An overnight culture of the Salmonella phoP::gfp reporter strain was diluted 1:200 in LB broth containing each of the bile acids, and the cultures were incubated for 4 h. Cultures were diluted 1:20 in PBS and analyzed through flow cytometry. Table 3 shows that none of these bile acids, when added to bacterial cultures, repressed the expression of phoP. In fact, all bile acids tested caused a slight increase in phoP expression, although the change observed was minimal and was unlikely to be biologically relevant. Nevertheless, our data suggest that the molecule present in physiological murine bile that represses phoP expression is unlikely to be a bile acid.
Table 3.
Effect of individual bile acids on phoP expression
| Treatmenta | Relative fluorescence at concn shownb |
||
|---|---|---|---|
| 0.25 mM | 0.5 mM | 1 mM | |
| None | 100 (2.8) | 100 (2.7) | 100 (2.1) |
| Dehydrocholic acid | 101 (3.3) | 111 (1.2) | 101 (1.1) |
| Cholic acid | 101 (2.6) | 113 (2.3) | 96 (3.0) |
| Chenodeoxycholic acid | 99 (3.3) | 111 (0.8) | 99 (1.6) |
| Deoxycholic acid | 101 (2.0) | 115 (2.3) | 102 (0.5) |
| Glycolithocholic acid | 102 (2.0) | 113 (1.1) | 105 (0.4) |
| Taurocholic acid | 103 (1.7) | 116 (0.9) | 105 (0.5) |
| Lithocholic acid | 112 (3.7) | 121 (2.7) | 132 (5.4) |
Bile acids were suspended in DMSO and added to LB broth at the concentrations indicated. The controls contained the same concentration of DMSO, but without bile acids.
Arbitrary fluorescence values relative to control. Results shown are the average of three bacterial cultures, with standard errors of means shown within parentheses.
Commercial bovine bile does not produce the effects of physiological murine bile on phoP expression.
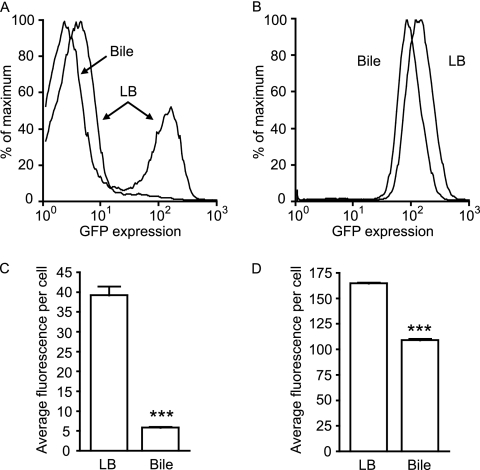
Commercial bovine bile has been previously shown to repress invasion gene expression in Salmonella (30, 31). To investigate whether the components of bile that affect invasion and phoP expression are the same, we fused the promoter of the major regulator of invasion gene expression in Salmonella, hilA, with gfp in pFVP25 and studied the production of GFP by Salmonella strains containing the hilA or phoP reporter plasmids during growth in LB broth with or without 3% (wt/vol) bovine bile. We prepared a solution of bovine bile in LB broth and adjusted the pH to match that of LB broth (around 7.3). An overnight culture of the Salmonella phoP::gfp reporter strain was used to inoculate LB broth, with or without bovine bile, at a 1:200 dilution. Cultures were incubated for 4 h, diluted 1:20 in PBS, and analyzed by flow cytometry. During growth in LB broth, the expression of hilA is bimodal, with most cells not producing GFP and a subpopulation of bright cells accounting for approximately 25% of the total number of cells (Fig. 5). In accordance with previous observations, hilA expression was strongly repressed by commercial bovine bile. During growth in bile-containing LB broth, the subpopulation of GFP-producing cells accounted for only 3.1% of the total number of cells. The repression of hilA by commercial bovine bile could also be clearly seen during the analysis of the average fluorescence values per cell in cells grown in LB broth with or without bile. In LB broth without bile, the average fluorescence per cell was 39.2, whereas in LB broth containing bile, this value was 5.9, accounting for an approximate fold change of 6.7 (Fig. 5). The effect of commercial bovine bile on phoP expression, however, was much less pronounced. Growth in LB broth containing bile caused a reduction in the average fluorescence per cell from 164.7 to 109, a 1.5-fold change. Although the effects of bile on both hilA and phoP were statistically significant, it is clear that the activity against hilA is distinct from that against phoP expression.
Fig 5.
Commercial ox bile does not significantly repress phoP while still repressing hilA expression. Salmonella strains containing phoP::gfp or hilA::gfp reporter plasmids were grown overnight in LB broth with carbenicillin and subcultured 1:200 in LB broth containing carbenicillin with or without 3% (wt/vol) bovine bile (pH adjusted). After incubation at 37°C with shaking for 4 h, cultures were diluted in PBS, and gfp expression was analyzed by flow cytometry. (A) Histogram showing the relative proportions of cells (y axis) expressing gfp at various levels (x axis) during growth of the hilA::gfp Salmonella strain in LB broth with or without bovine bile, as indicated. (B) Same as in panel A, but with the phoP::gfp Salmonella reporter strain. (C) Data from panel A expressed as average fluorescence intensity per cell. (D) Data from panel B expressed as average fluorescence intensity per cell. The data shown are the average from three independent cultures. Bars represent the standard errors of means. P values were ≤0.0001 (***).
Physiological bovine bile alone does not support robust Salmonella growth in vitro.
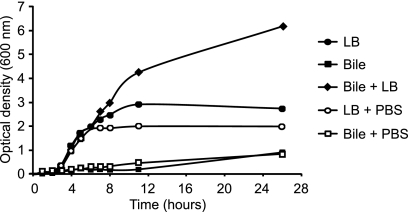
We have previously shown that physiological murine bile supports Salmonella growth at rates comparable to those seen in rich laboratory media (1). In order to begin characterizing the interactions between Salmonella and bile from other organisms, we obtained fresh, physiological bovine bile, adjusted its pH to match that of LB broth (around 7.3), and used it to perform Salmonella growth assays. Contrary to our findings using physiological murine bile, physiological bovine bile alone did not support Salmonella growth (Fig. 6). To determine if this was due to the absence of nutrients required for Salmonella growth in bovine bile or the antibacterial action of bile, we mixed LB broth or bile with PBS at a 1:1 ratio and analyzed Salmonella growth in these media. If the reason for the lack of Salmonella growth in bile were the presence of antibacterial molecules, we expected that diluting bile with PBS would decrease its antibacterial action and perhaps allow Salmonella to grow. As can be seen from Fig. 6, this was not the case. Diluting bile with PBS did not improve Salmonella growth. These results could be interpreted in two ways. First, it is possible that bile can still repress growth when diluted. The other possibility is that the lack of Salmonella growth in bile was solely due to the absence of nutrients in this fluid. To determine which of these was true, we mixed bile and LB broth at a 1:1 ratio and analyzed Salmonella growth. If the lack of growth were due to an antibacterial property that is still active in diluted bile, then Salmonella would not be able to grow in a 1:1 mixture of LB broth and bile. On the other hand, if the lack of growth were due to the absence of nutrients in bile, we expected Salmonella growth in the LB broth-bile mixture to be comparable to growth in the LB broth-PBS mixture (i.e., 50% LB broth in both cases). Although our results confirmed that the lack of growth in bile was not due to its antibacterial action, we found that the addition of bile to LB broth greatly increased Salmonella growth (Fig. 6). Altogether, our results indicate that bovine bile is rich in nutrients that can be used by Salmonella but lacks one or more nutrients that are required by Salmonella to grow; provided that these nutrients are added to the medium (by the addition of LB broth, for example), Salmonella can show extremely high growth in bovine bile.
Fig 6.
Physiological bovine bile alone does not support Salmonella growth. Bile or LB broth samples were kept undiluted, diluted with PBS, or mixed, as indicated, always at a 1:1 ratio. Streptomycin was added to all cultures at a final concentration of 100 μg/ml, and the pH of all solutions was adjusted to approximately 7.3. Overnight cultures of Salmonella Typhimurium SL1344 were then subcultured 1:200 in each of the culture media and incubated at 37°C with shaking; growth was monitored at the indicated time points by measuring the optical density of the cultures at 600 nm. The data shown are the average from three independent cultures. Bars represent the standard errors of means.
Physiological bovine bile represses phoP expression.
The experiments using physiological murine bile described above show that this fluid acts as a strong repressor of phoP expression, whereas commercial bovine bile fails to repress phoP. This could be due to one of two reasons. First, it is possible that the phoP inhibitory activity is a very specific phenomenon and is exclusive to murine bile. Alternatively, it is possible that commercial bovine bile is not fully representative of physiological bovine bile and that the inhibitory activity is lost during processing or packaging. To determine if the inhibitory activity observed in physiological murine bile is extended to other mammalian species, we used fresh, physiological bovine bile to perform Salmonella phoP expression assays. Because we determined that bovine bile alone does not support Salmonella growth, we compared levels of phoP expression during growth in 1:1 mixtures of LB broth and bile or LB broth and water. The phoP::gfp Salmonella reporter strain was grown overnight, subcultured 1:200 in the appropriate media, and allowed to grow for 4 h. Samples were diluted, and fluorescence was assayed through flow cytometry, as described above. Figure 7 shows that, like physiological murine bile, physiological bovine bile can also repress phoP expression, suggesting that this phenomenon is a conserved property of bile from multiple species. In order to determine if this was due solely to the fact that the pH of physiological bovine bile was adjusted to match that of LB broth, we repeated these experiments comparing phoP expression in bile without adjusting its pH (pH 8.5) against expression in LB broth after adjusting its pH to match that of bile (8.5). Our results showed that the pH of bile does not affect its capacity to repress phoP; i.e., bovine bile repressed phoP expression to a similar extent at pH 7.3 and 8.5 (data not shown).
Fig 7.
Physiological bovine bile represses phoP expression. LB broth was diluted 1:1 with either water or fresh bovine bile. Carbenicillin was added, and the pH of the solutions was adjusted. Overnight cultures of the Salmonella phoP::gfp reporter strain were then used to inoculate these solutions at a 1:200 dilution. Cultures were incubated at 37°C with shaking for 4 h, diluted in PBS, and analyzed by flow cytometry. (A) Histogram showing the relative proportions of cells (y axis) expressing GFP at various levels (x axis) during growth of the phoP::gfp Salmonella strain in LB broth-water (LB) or LB broth-bile (Bile) mixtures, as indicated. (B) Data from panel A expressed as average fluorescence intensity per cell. The data shown are the average from three independent cultures. Bars represent the standard errors of means. The P value was ≤0.0002 (***).
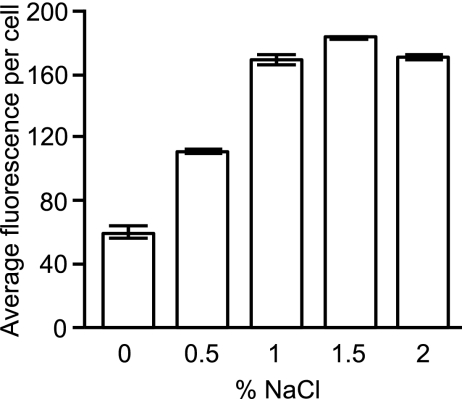
The repression of phoP expression by bile is not due to osmolarity.
After determining that both murine bile and bovine bile can repress phoP expression and that this is not due to the pH of the solution, we set out to investigate if this phenomenon was caused by a specific molecule present in bile or if it was a more general consequence of the chemical properties of this biological fluid. Bile is rich in organic and inorganic compounds, including salts, and therefore it was possible that differences in osmolarity between bile and the culture media used in our experiments could be responsible for the repression of phoP. To determine if this was the case, we measured phoP expression during Salmonella growth in LB media containing different concentrations of sodium chloride. The concentration of NaCl in LB broth is 1%, and our data show that increased concentrations of NaCl had no effect on phoP expression (Fig. 8). This shows that increased osmolarity cannot explain the phenomenon of phoP repression by bile. Nevertheless, although bile is rich in salts, it was still a formal possibility that the osmolarity of bile is lower than that of LB broth and that this, in turn, could be responsible for phoP repression. Indeed, our results showed that Salmonella growth in LB broth containing lower levels of NaCl caused significant repression of phoP expression (Fig. 8). However, when we compared phoP expression during Salmonella growth in a mixture of LB broth and water versus LB broth and bile, we found that expression was significantly lower in the mixture containing bile (Fig. 7), indicating that a potentially lower osmolarity of bile could not explain the phenomenon of phoP repression.
Fig 8.
The repression of phoP expression by bile is not due to osmolarity. Overnight cultures of the Salmonella phoP::gfp reporter strain were subcultured at 1:200 in LB broth containing various concentrations of NaCl, as indicated. Cultures were allowed to grow for 4 h at 37°C with shaking, diluted, and analyzed for gfp expression through flow cytometry. The data shown are the average from three independent cultures. Bars represent the standard errors of means.
The repression of phoP by bile is due to specific molecules with hydrophobic properties.
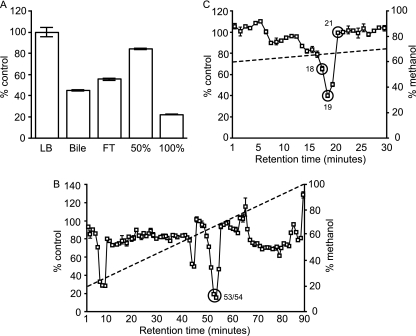
To further characterize the phoP inhibitory activity present in bile, we attempted to purify the bioactive molecule. Because many hydrophobic organic molecules are present in bile (steroids, bile acids, and phospholipids), we chose to utilize cartridges containing a C18 resin for initial purification (Sep-Pak; Waters). We applied bile samples to the cartridges, collected the flowthrough, and eluted the bound molecules with 50% and 100% methanol, sequentially. We then evaporated each fraction, added it to culture media, and analyzed phoP expression after Salmonella growth. To do so, overnight cultures of the Salmonella phoP::gfp reporter strain were subcultured 1:200 into the appropriate samples and allowed to grow for 4 h. After this period, samples were diluted in PBS, and gfp production was assayed through flow cytometry. Although a significant amount of bioactivity was recovered in the flowthrough, our data show that the bioactivity was mostly retained by the C18 resin and was eluted only with 100% methanol, suggesting that the bioactive molecule is of a hydrophobic nature (Fig. 9A). In order to further purify the bioactive molecule, we subjected the active fraction to reverse-phase high-performance liquid chromatography (RP-HPLC) using a linear gradient of 20% to 100% methanol over 90 min, at a flow rate of 1 ml per minute. Bioactivity of fractions was tested essentially as described above. This allowed us to identify three main fractions with phoP inhibitory activity, as shown in Fig. 9B. The fraction showing the highest level of activity (fractions 53 and 54) was evaporated and subjected to a second round of RP-HPLC, using a linear gradient of 60% to 70% methanol over 30 min, and the bioactivity of the resulting fractions was determined. This revealed a single fraction that concentrated most of the biological activity (fraction 19), and this fraction was selected for further studies (Fig. 9C). Altogether, these results show that the repression of phoP by bile is caused by one or more molecules with hydrophobic properties and is not a consequence of the general physicochemical properties of bile.
Fig 9.
The repression of phoP by bile is due to specific molecules with hydrophobic properties. Bovine bile was extracted as described below. Extracts were evaporated and resuspended in LB broth containing carbenicillin. Overnight cultures of the Salmonella phoP::gfp reporter strain were subcultured 1:200 in all solutions, which were then incubated for 4 h at 37°C with shaking, diluted, and analyzed for gfp production through flow cytometry. The expression level during growth in LB broth (negative control) was set to 100%, and the activity of test fractions was normalized accordingly. (A) Bovine bile was applied to C18 resin cartridges, and the flowthrough (FT) was collected. Cartridges were washed with water, and the bound fraction was eluted sequentially with 50% and 100% methanol. Fractions were dried and assayed for phoP inhibition as described above. (B) The 100% fraction from panel A was dried, resuspended in 25% methanol, and subjected to reverse-phase high-performance liquid chromatography (RP-HPLC) using a linear gradient of 20% to 100% methanol over 90 min (dashed line), at a flow rate of 1 ml per minute. Fractions were collected every minute, dried, and assayed for phoP inhibition as described above. (C) Fractions 53 and 54 from panel B were combined, dried, resuspended in 25% methanol, and subjected to RP-HPLC again using a linear gradient of 60% to 70% methanol over 30 min (dashed line), at a flow rate of 1 ml per minute. Fractions were collected every minute, dried, and assayed for phoP inhibition as described above. The data shown are the average from three (A) or two (B and C) independent cultures. Bars represent the standard errors of means.
The most abundant compound in the bioactive fraction is TDC.
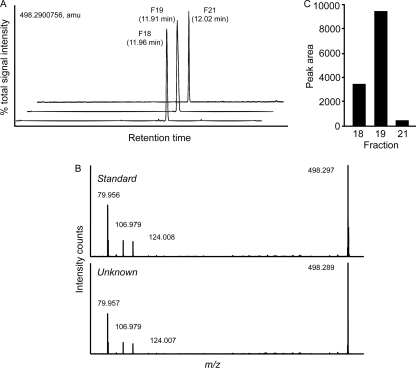
To try and identify the compound responsible for phoP inhibition, we subjected the HPLC fraction with the highest biological activity (fraction 19) to ultrahigh-performance liquid chromatography coupled with mass spectrometry (UPLC-MS). For these experiments, samples were run on a C18 column, and UPLC-QTOF MS was performed in ESI(−) mode. Our results showed that a single compound of m/z 498.2900756 accounted for over 98% of all signal present in fraction 19 (Fig. 10). We then queried METLIN, the Metabolite and Tandem MS Database (http://metlin.scripps.edu/), with this m/z to try and determine putative identities. The database search generated three main hits: taurochenodeoxycholic acid, tauroursodeoxycholic acid, and taurodeoxycholic acid (TDC). To identify which of these compounds was the most abundant compound present in fraction 19, we performed UPLC-MS on standard compounds and compared retention times with the unknown compound of fraction 19 (retention time of 11.91 min). Our results clearly showed that the unknown compound was not tauroursodeoxycholic acid (retention time of 8.37 min) or taurochenodeoxycholic acid (retention time of 11.28). On the other hand, the retention time of the TDC standard matched that of the unknown compound very closely (11.99 min). In order to further confirm that the most abundant compound of fraction 19 was TDC, we performed collision-induced dissociation tandem MS on the standard and unknown compounds. As can be seen in Fig. 10, the fragmentation patterns of both the TDC standard and the unknown compound were identical, indicating that TDC is indeed the most abundant compound in fraction 19.
Fig 10.
The most abundant compound of the bioactive fraction is taurodeoxycholic acid. Fractions 18, 19, and 21 were dried, dissolved in 200 μl of methanol, diluted 1:500 with 75% methanol, and injected (3 μl) into a 10- by 0.21-cm C18 UPLC column and run on UPLC-QTOF MS in ESI(−) mode. (A) Base peak ion chromatograms of fractions 18, 19, and 21. Retention times are shown. The main compound detected in all fractions was m/z 498.2900756 (m/z determined from fraction 19). amu, atomic mass units. (B) m/z 498.2900756 from fraction 19 was subjected to collision-induced fragmentation MS, and chromatograms showing daughter ions of this compound (bottom panel) as well as a taurodeoxycholic acid standard (top panel) are presented. (C) Peak area of m/z 498.2900756 from fractions 18, 19, and 21 as an example of the criteria used to select potential culprits of the phoP inhibitory activity in the active fractions.
TDC alone does not repress phoP expression.
In order to ascertain that TDC was responsible for the repression of phoP, we measured phoP expression during growth in LB broth containing various concentrations of this bile acid. To our surprise, TDC did not cause any appreciable repression of phoP, even at the highest concentration used (100 mM; data not shown). Therefore, TDC alone cannot account for the repression of phoP observed during Salmonella growth in bile. This suggests that (i) m/z 498.2900756 is not TDC, even though it shows the same retention time and fragmentation patterns; (ii) another minor component of the active fraction is responsible for the activity; or (iii) TDC requires a “cofactor” to repress phoP expression. To further confirm that the most abundant compound of fraction 19 is indeed TDC, we performed nuclear magnetic resonance (NMR) experiments with the TDC standard and fraction 19 and confirmed the identity of the main compound of fraction 19 as TDC (data not shown). Also, in order to determine if a minor component of fraction 19 was responsible for the bioactivity against phoP, we attempted to further purify the active molecule from the active fraction. However, additional rounds of HPLC produced no active fractions (data not shown). Therefore, our data suggest that a mixture of molecules may be necessary for the inhibitory activity of bile against phoP expression.
Defining a subset of small molecules responsible for the repression of phoP by bile.
In order to identify potential culprits of the phoP inhibitory activity displayed by bile, we performed UPLC-MS on fractions 18, 19, and 21 (from the last round of HPLC purifications). We chose these fractions because they showed various levels of bioactivity against phoP. Fraction 18 showed moderated activity, fraction 19 showed high activity, and fraction 21 showed no appreciable activity (Fig. 9C). This allowed us to compare the signal intensities (as peak areas) of the compounds detected in these fractions with the corresponding biological activity of each fraction, in order to determine the potential bioactive molecules in fraction 19. In fraction 19, the most active of all three, 10 different ions were found (data not shown). We then filtered this list to include only molecules that were also present on fraction 18 and that showed levels at least 1.5-fold higher in fraction 19 compared to those in fractions 18 and 21. This resulted in a list of 4 different ions (Table 4), one of which is m/z 498.2900756. Figure 10A and C show the abundance and signal intensity for this ion as an example of the filtering criteria utilized. Aside from m/z 498.2900756, none of the other m/z values produced any hits when the METLIN database was searched. Therefore, these may represent new molecules, and significant further studies will be required to determine their identities. Nevertheless, the experiments described above allowed us to identify a subset of small molecules, out of the several hundred present in bile (1), that are likely responsible for the biological activity observed.
Table 4.
Subset of molecules correlated with biological activity against phoP expression
| m/za | Peak area in fraction: |
||
|---|---|---|---|
| 18 | 19 | 21 | |
| 498.2900756 | 355,682 | 824,602 | 53,377 |
| 500.4995894 | 3,371 | 5,152 | 756 |
| 598.2227203 | 1,258 | 2,806 | NDb |
| 582.2482633 | 958 | 1,759 | ND |
Determined by UPLC-MS in ESI(−) mode.
ND, not detected.
DISCUSSION
A critical step for the success of microbes during the process of host colonization is the sensing of their surroundings and responding through the regulation of gene expression. This allows the microbe to turn on genes that will aid in host colonization and turn off genes that are not required and whose transcription would represent an unnecessary burden. This is true not only for microbial pathogens during the development of disease but also for symbiotic microorganisms that need to establish stable relationships with their hosts. Environmental sensing by microbes can occur through multiple mechanisms, the most widespread and important one of which involves the use of two-component regulatory systems (two-component systems [TCSs]) (4). TCSs are composed of a membrane-bound histidine kinase that acts as a sensor by binding a specific environmental signal and changing its phosphorylation state in response to it and a response regulator to which the histidine kinase can transfer its phosphate group. Changes in the phosphorylation state of response regulators cause conformational changes that alter their DNA binding capabilities, resulting in changes in gene expression (4).
Salmonella is one of many human pathogens that use TCSs to adapt to the conditions encountered in their host and regulate virulence gene expression in response (4, 28). A search of the Salmonella Typhimurium LT2 genome (http://cmr.jcvi.org/) using the terms “histidine kinase” and “response regulator” revealed a high number of TCSs, with 19 histidine kinases and 39 response regulators being found. One of the most important and well-studied TCSs in Salmonella is PhoPQ (13, 28). PhoQ is a sensor histidine kinase that responds to a variety of signals, including divalent cations, antimicrobial peptides, and pH (13, 18, 28). PhoP is the cognate response regulator, which translates PhoQ phosphorylation into regulation of gene expression. PhoPQ signaling is an intrinsic part of Salmonella virulence, as strains lacking this TCS show reduced virulence (20). Additionally, a strain containing a constitutively active PhoPQ TCS shows virulence defects, illustrating how a balance in PhoPQ activity and regulation of virulence gene expression is required for full virulence (21).
One of the environments that Salmonella has evolved to sense and adapt to is bile. Salmonella Typhi can colonize the gallbladders of individuals with typhoid fever and use this environment as a deposit of bacteria that constantly reseed the gastrointestinal tract and maintain fecal shedding (23, 36, 39). Infection of mice with Salmonella Typhimurium results in similar disease, with the gallbladder being heavily colonized (19). Due to the exposure to bile experienced by Salmonella during infection, the effect of bile on Salmonella has been studied in detail (6, 29–31). Prouty et al. have shown that bovine bile strongly represses host cell invasion (30, 31). Additionally, it was shown that bile induces Salmonella resistance to the antibacterial action of bile as well as antimicrobials through the regulation of transporters and PhoPQ-regulated genes (29, 41). However, in these studies, the authors state that PhoPQ signaling is not required for the induction of bile resistance by growth in bile and that bile does not affect phoPQ expression.
To increase our understanding of the interactions between Salmonella and bile, we performed transcriptome analyses of Salmonella grown in culture medium or physiological murine bile and showed that many genes are differentially regulated. Curiously, one of the genes repressed by bile was the response regulator phoP. We confirmed this observation using RT-PCR to show that the expression of phoP and other phoP-regulated genes is repressed by bile. However, neither commercial bovine bile nor individual bile acids were able to repress phoP expression to the same extent as physiological murine bile, explaining why the regulation of phoP by bile was missed in previous studies. Commercial bovine bile, however, was able to fully repress the expression of the invasion regulator hilA, suggesting that different components of bile may act to repress phoP and hilA. Regardless of this difference, Salmonella growth in bile seems to induce a program of gene expression focused on the repression of virulence genes. Although the reason for this is currently unknown, there are several plausible hypotheses, each focused on a different stage of gallbladder colonization. It is possible that the repression of hilA and phoP is simply a way to conserve energy when Salmonella is swimming in bile in the gallbladder lumen and has not yet reached the lining epithelium, where expression of these genes would be beneficial (the “entry hypothesis”). Support for this hypothesis comes from the fact that Salmonella cells can be readily observed inside the epithelial cells lining the gallbladder lumen (19). It is also possible that the repression of these genes is a consequence of adaptation to the gallbladder environment, where Salmonella could grow at high rates using bile as a carbon and energy source (the “growth hypothesis”). When encountering a nutritionally rich environment such as that of the gallbladder, Salmonella may suppress virulence gene expression to avoid damaging the host so that it can take full advantage of the nutritional status of the gallbladder environment to grow to high densities using bile components. The fact that many metabolic genes were affected by growth in bile may support this hypothesis and suggests that a metabolic rewiring is induced in bile. For instance, we found that many genes repressed by bile are involved in the citric acid cycle and the generation of electron donors for aerobic respiration, whereas genes activated by bile are involved in alternative respiratory systems. The last possibility is that when Salmonella encounters bile, it is ready to leave the host organism through excretion in the gastrointestinal tract and fecal shedding. Therefore, the activation of virulence genes would represent an unnecessary expense (the “exit hypothesis”).
Some of these hypotheses are not completely novel. Prouty and Gunn have proposed that during Salmonella infection of the gastrointestinal tract, an equivalent of our “entry hypothesis” takes place (31). The relatively high concentrations of bile present in the lumen of the small intestine may prompt Salmonella to repress invasion gene expression, until it reaches the intestinal epithelium, where the expression of these genes would be required for infection. Additionally, the utilization of bile as an environmental cue is not exclusive to Salmonella. For instance, bile salts have been previously shown to induce adherence to and invasion of cultured human cells by Shigella (27), a process mediated through the direct interaction of bile salts with the type-three secretion system involved in host cell invasion (7, 38). As with Salmonella and Shigella, species of Vibrio also link bile sensing with the regulation of virulence traits. Vibrio cholerae, for example, has been shown to repress the expression of its cholera toxin and toxin-coregulated pilus while upregulating motility in response to bile (15), and some of the molecules involved have been defined (5). Vibrio parahaemolyticus, on the other hand, can sense bile acids to induce the production of its thermostable direct hemolysin, capsule polysaccharide, and other virulence traits (11, 24, 25). In many cases, hypotheses for the evolutionary advantage of sensing bile and responding in specific manners can be made, although empirical evidence is lacking in most of these cases. Although for the most part the biological significance of gene regulation by bile in Salmonella remains unknown, our study contributes to the understanding of the interactions between Salmonella and bile and this pathogen's lifestyle during interactions with its host.
ACKNOWLEDGMENTS
We thank the anonymous reviewers of the manuscript for constructive criticism and insights. We are also greatly thankful to Neil Rawlyk, Sherry Tetland, Don Wilson, and Andrew Potter (Vaccine and Infectious Disease Organization, University of Saskatchewan) for the kind gift of fresh bovine bile, Jingxi Pan (University of Victoria Proteomics Centre) for help with UPLC-MS experiments, Vincent Chen (Michael Smith Laboratories, The University of British Columbia) for help with HPLC, and David Williams and Raymond Andersen (University of British Columbia Chemistry Department) for help with NMR experiments.
This work was funded by the Canadian Institutes of Health Research. L.C.M.A. and R.B.R.F. are supported by postdoctoral fellowships from the Canadian Institutes of Health Research. B.B.F. is the University of British Columbia Peter Wall Distinguished Professor.
Footnotes
Published ahead of print 24 February 2012
REFERENCES
- 1. Antunes LC, et al. 2011. Metabolomics reveals phospholipids as important nutrient sources during Salmonella growth in bile in vitro and in vivo. J. Bacteriol. 193:4719–4725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Antunes LC, et al. 2010. Inhibition of Salmonella host cell invasion by dimethyl sulfide. Appl. Environ. Microbiol. 76:5300–5304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bakken K, Vogelsang TM. 1950. The pathogenesis of Salmonella typhimurium infection in mice. Acta Pathol. Microbiol. Scand. 27:41–50 [DOI] [PubMed] [Google Scholar]
- 4. Beier D, Gross R. 2006. Regulation of bacterial virulence by two-component systems. Curr. Opin. Microbiol. 9:143–152 [DOI] [PubMed] [Google Scholar]
- 5. Chatterjee A, Dutta PK, Chowdhury R. 2007. Effect of fatty acids and cholesterol present in bile on expression of virulence factors and motility of Vibrio cholerae. Infect. Immun. 75:1946–1953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Crawford RW, Gibson DL, Kay WW, Gunn JS. 2008. Identification of a bile-induced exopolysaccharide required for Salmonella biofilm formation on gallstone surfaces. Infect. Immun. 76:5341–5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dickenson NE, et al. 15 December 2010, posting date. Conformational changes in IpaD from Shigella flexneri upon binding bile salts provide insight into the second step of type III secretion. Biochemistry [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Esteller A. 2008. Physiology of bile secretion. World J. Gastroenterol. 14:5641–5649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fields PI, Swanson RV, Haidaris CG, Heffron F. 1986. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc. Nat. Acad. Sci. U. S. A. 83:5189–5193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gooderham WJ, Hancock RE. 2009. Regulation of virulence and antibiotic resistance by two-component regulatory systems in Pseudomonas aeruginosa. FEMS Microbiol. Rev. 33:279–294 [DOI] [PubMed] [Google Scholar]
- 11. Gotoh K, et al. 2010. Bile acid-induced virulence gene expression of Vibrio parahaemolyticus reveals a novel therapeutic potential for bile acid sequestrants. PLoS One 5:e13365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Grassl GA, Finlay BB. 2008. Pathogenesis of enteric Salmonella infections. Curr. Opin. Gastroenterol. 24:22–26 [DOI] [PubMed] [Google Scholar]
- 13. Groisman EA. 2001. The pleiotropic two-component regulatory system PhoP-PhoQ. J. Bacteriol. 183:1835–1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gunn JS, Miller SI. 1996. PhoP-PhoQ activates transcription of pmrAB, encoding a two-component regulatory system involved in Salmonella typhimurium antimicrobial peptide resistance. J. Bacteriol. 178:6857–6864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gupta S, Chowdhury R. 1997. Bile affects production of virulence factors and motility of Vibrio cholerae. Infect. Immun. 65:1131–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Haraga A, Ohlson MB, Miller SI. 2008. Salmonellae interplay with host cells. Nat. Rev. Microbiol. 6:53–66 [DOI] [PubMed] [Google Scholar]
- 17. Kappelhoff R, Overall C. 2007. The CLIP-CHIP oligonucleotide microarray: dedicated array for analysis of all protease, nonproteolytic homolog, and inhibitor gene transcripts in human and mouse. Curr. Protoc. Protein Sci. Chapter 21:Unit 21.19 [DOI] [PubMed] [Google Scholar]
- 18. Kato A, Groisman EA. 2008. The PhoQ/PhoP regulatory network of Salmonella enterica. Adv. Exp. Med. Biol. 631:7–21 [DOI] [PubMed] [Google Scholar]
- 19. Menendez A, et al. 2009. Salmonella infection of gallbladder epithelial cells drives local inflammation and injury in a model of acute typhoid fever. J. Infect. Dis. 200:1703–1713 [DOI] [PubMed] [Google Scholar]
- 20. Miller SI, Kukral AM, Mekalanos JJ. 1989. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc. Natl. Acad. Sci. U. S. A. 86:5054–5058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Miller SI, Mekalanos JJ. 1990. Constitutive expression of the PhoP regulon attenuates Salmonella virulence and survival within macrophages. J. Bacteriol. 172:2485–2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nelson K, Whittam TS, Selander RK. 1991. Nucleotide polymorphism and evolution in the glyceraldehyde-3-phosphate dehydrogenase gene (gapA) in natural populations of Salmonella and Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 88:6667–6671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nichols HJ. 1916. Experimental observations on the pathogenesis of gall-bladder infections in typhoid, cholera, and dysentery. J. Exp. Med. 24:497–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Osawa R, Arakawa E, Okitsu T, Yamai S, Watanabe H. 2002. Levels of thermostable direct hemolysin produced by Vibrio parahaemolyticus O3:K6 and other serovars grown anaerobically with the presence of a bile acid. Curr. Microbiol. 44:302–305 [DOI] [PubMed] [Google Scholar]
- 25. Pace JL, Chai TJ, Rossi HA, Jiang X. 1997. Effect of bile on Vibrio parahaemolyticus. Appl. Environ. Microbiol. 63:2372–2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Parry CM, Hien TT, Dougan G, White NJ, Farrar JJ. 2002. Typhoid fever. N. Engl. J. Med. 347:1770–1782 [DOI] [PubMed] [Google Scholar]
- 27. Pope LM, Reed KE, Payne SM. 1995. Increased protein secretion and adherence to HeLa cells by Shigella spp. following growth in the presence of bile salts. Infect. Immun. 63:3642–3648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Prost LR, Miller SI. 2008. The salmonellae PhoQ sensor: mechanisms of detection of phagosome signals. Cell. Microbiol. 10:576–582 [DOI] [PubMed] [Google Scholar]
- 29. Prouty AM, Brodsky IE, Falkow S, Gunn JS. 2004. Bile-salt-mediated induction of antimicrobial and bile resistance in Salmonella typhimurium. Microbiology 150:775–783 [DOI] [PubMed] [Google Scholar]
- 30. Prouty AM, et al. 2004. Transcriptional regulation of Salmonella enterica serovar Typhimurium genes by bile. FEMS Immunol. Med. Microbiol. 41:177–185 [DOI] [PubMed] [Google Scholar]
- 31. Prouty AM, Gunn JS. 2000. Salmonella enterica serovar Typhimurium invasion is repressed in the presence of bile. Infect. Immun. 68:6763–6769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Saeed AI, et al. 2003. TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34:374–378 [DOI] [PubMed] [Google Scholar]
- 33. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 34. Santos RL, et al. 2001. Animal models of Salmonella infections: enteritis versus typhoid fever. Microbes Infect. 3:1335–1344 [DOI] [PubMed] [Google Scholar]
- 35. Shin D, Lee EJ, Huang H, Groisman EA. 2006. A positive feedback loop promotes transcription surge that jump-starts Salmonella virulence circuit. Science 314:1607–1609 [DOI] [PubMed] [Google Scholar]
- 36. Sinnott CR, Teall AJ. 1987. Persistent gallbladder carriage of Salmonella typhi. Lancet i:976. [DOI] [PubMed] [Google Scholar]
- 37. Soncini FC, Garcia Vescovi E, Solomon F, Groisman EA. 1996. Molecular basis of the magnesium deprivation response in Salmonella typhimurium: identification of PhoP-regulated genes. J. Bacteriol. 178:5092–5099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stensrud KF, et al. 2008. Deoxycholate interacts with IpaD of Shigella flexneri in inducing the recruitment of IpaB to the type III secretion apparatus needle tip. J. Biol. Chem. 283:18646–18654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vaishnavi C, et al. 2005. Prevalence of Salmonella enterica serovar Typhi in bile and stool of patients with biliary diseases and those requiring biliary drainage for other purposes. Jpn. J. Infect. Dis. 58:363–365 [PubMed] [Google Scholar]
- 40. Valdivia RH, Falkow S. 1996. Bacterial genetics by flow cytometry: rapid isolation of Salmonella typhimurium acid-inducible promoters by differential fluorescence induction. Mol. Microbiol. 22:367–378 [DOI] [PubMed] [Google Scholar]
- 41. van Velkinburgh JC, Gunn JS. 1999. PhoP-PhoQ-regulated loci are required for enhanced bile resistance in Salmonella spp. Infect. Immun. 67:1614–1622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wickham ME, Brown NF, Provias J, Finlay BB, Coombes BK. 2007. Oral infection of mice with Salmonella enterica serovar Typhimurium causes meningitis and infection of the brain. BMC Infect. Dis. 7:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wray C, Sojka WJ. 1978. Experimental Salmonella typhimurium infection in calves. Res. Vet. Sci. 25:139–143 [PubMed] [Google Scholar]