Abstract
Within a given microbial population, a small subpopulation known as dormant persister cells exists. This persistence property ensures the survival of the population as a whole in the presence of lethal factors. Although persisters are highly important in antibiotic therapy, the mechanism for persistence is still not thoroughly understood. We show here that the cariogenic organism Streptococcus mutans forms persister cells showing noninherited multidrug tolerance. We demonstrated that the ectopic expression of the type II toxin-antitoxin systems, MazEF and RelBE, caused an increase in the number of persisters. In a search for additional persistence genes, an expression library was constructed, and several clones exhibiting a significant difference in persister formation after prolonged antibiotic treatment were selected. The candidate persister genes include genes involved in transcription/replication, sugar metabolism, cell wall synthesis, and energy metabolism, clearly pointing to redundant pathways for persister formation. We have previously reported that the S. mutans quorum-sensing peptide, CSP pheromone, was a stress-inducible alarmone capable of conveying sophisticated messages in the bacterial population. In this study, we demonstrate the involvement of the intraspecies quorum-sensing system during the formation of stress-induced multidrug-tolerant persisters. To the best of our knowledge, this is the first study reporting the induction of bacterial persistence using a quorum-sensing regulatory system.
INTRODUCTION
Numerous mechanisms of drug resistance have been described in bacteria; the main types of resistance are alteration of the antibiotic target site, antibiotic modification, and restricted antibiotic penetration (1, 10). Each of these mechanisms allows bacteria to grow in the presence of even high doses of the antibiotic. Bacterial populations also produce a small number of specialized cells that survive lethal concentrations of drugs without expressing an antibiotic resistance mechanism. These survivors are called persisters, cells that neither grow nor die in the presence of microbicidal antibiotics (3, 20, 31). It is thought that these persister cells enter into a state of dormancy, which allows them to prevent cell death due to antibiotics targeting bacterial cell growth (29). This persistence property ensures the survival of the population as a whole in the presence of lethal concentrations of antibiotics. For this reason, persister cells have been associated with the recalcitrant nature of chronic infections (18). It has also been suggested that persisters may play a significant role in biofilm tolerance (30). Indeed, biofilm infections show a surprising ability to resist killing by antibiotics, without having any obvious drug resistance mechanisms (8, 9, 12).
As opposed to the traditional view of bacterial cells expressing genetic resistance mechanisms for survival against antibiotics, persisters are not antibiotic-resistant mutants since they are genetically identical to the cells affected by the antibiotics. Instead, persisters are phenotypic variants of regular cells since, when recultured, they exhibit the same small survival fraction (53). Studies of persisters have identified multiple genes that have been implicated in bacterial persistence, and several of these are chromosomal toxin-antitoxin (TA) modules (18, 31, 33, 52). TA modules are typically composed of a pair of genes organized in operons and encoding a stable toxin and its labile antitoxin (54). Whereas toxins are always proteins, antitoxins are either RNAs (type I and type III) or proteins (type II) (19, 49). In Escherichia coli, multiple TA systems have been linked to the formation of persisters. It has been shown that the ectopic mild overexpression of TA systems induced bacterial persistence (44). Recently, Maisonneuve et al. (33) demonstrated that the successive deletion of the 10 mRNA interferase-encoding TA loci of E. coli progressively reduced the level of persisters, demonstrating quite convincingly that persistence was a phenotype common to TA loci. Persister formation can also be affected by other genes, including genes involved in phosphate metabolism, phospholipid synthesis, purine metabolism, folate biosynthesis, energy metabolism, DNA repair, and SOS response. An exhaustive overview of currently known persistence genes has been published recently (18).
It is largely believed that persisters are formed through a stochastic process, where fluctuations in the cellular, physical, and biochemical processes affecting gene expression can lead to persistence. Alternatively, persistence may be an inducible state in a population due to exposure to stress. A recent study revealed that DNA damage induced the formation of persisters through an SOS-dependent expression of the TisB toxin from the tisAB/istR TA locus in E. coli (13, 14). Bacteria are often subjected to a plethora of environmental stressors aside from DNA damage, and this opens the possibility that stress responses may ensure survival by inducing the formation of persisters.
Whereas most persister studies have largely been performed using E. coli, not much work has been performed using Gram-positive bacteria. Streptococcus mutans, the leading etiological agent of dental caries, colonizes the dental biofilm, where it is constantly exposed to a wide range of environmental conditions (e.g., constant cycles of “famine and feast,” fluctuations of pH, and high levels of salt from tooth demineralization) (35, 36). Work previously done in our lab has shown that S. mutans integrates its response to specific environmental stresses with its intraspecies bacterial quorum-sensing system, CSP-ComDE, via the CSP pheromone (41). The aim of the present study was to investigate the formation of persister cells in the oral pathogen S. mutans. The possible intimate connection between the intraspecies quorum-sensing regulatory pathway and the development of persisters was also investigated.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
A summary of the bacterial strains and plasmids used in the present study is provided in Table 1. Deletion mutants were constructed in S. mutans UA159 wild-type (WT) strain as described previously (27). S. mutans strains were grown in Todd-Hewitt medium supplemented with 0.3% yeast extract (THYE) and incubated statically at 37°C in air with 5% CO2. S. mutans biofilms were developed in polystyrene microtiter plates. The growth of the biofilm was initiated by diluting (1:30) an overnight culture into fresh 1/4-THYE broth supplemented with 20 mM glucose in the individual wells of the microtiter plate. The plates were then incubated at 37°C in air with 5% CO2 without agitation. Biofilms were grown to early (∼6 h), intermediate (ca. 20 to 24 h), or mature (∼72 h) phases. E. coli strains were cultivated aerobically in Luria-Bertani (LB) medium at 37°C. When needed, antibiotics were added as follows: chloramphenicol (20 μg/ml) for E. coli and chloramphenicol (10 μg/ml), spectinomycin (1 mg/ml), or erythromycin (10 μg/ml) for S. mutans. Cell growth was monitored by determining the optical density at 600 nm (OD600). Cell viability was assessed by counting CFU on replica agar plates. The MIC test was performed according to the broth microdilution method using THYE broth as described previously (46).
Table 1.
Bacterial strains and plasmids used in the study
| Strain or plasmid | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| Strains | ||
| S. mutans UA159 | Wild-type reference strain | Lab stock |
| E. coli DH10B | Host strain for cloning and plasmid production | Lab stock |
| ΔmazEF mutant | In-frame SMU.172-173 deletion mutant derived from S. mutans UA159; Emr | 47 |
| ΔrelBE mutant | In-frame SMU.895-896 deletion mutant derived from S. mutans UA159; Emr | This study |
| ΔmazEF ΔrelBE mutant | In-frame SMU.172-173 and SMU.895-896 double deletion mutant derived from S. mutans UA159; Emr Spr | This study |
| UA159(pIB166) | S. mutans UA159 harboring pIB166; Cmr | 47 |
| ΔmazEF(pMAS2) mutant | ΔmazEF harboring pMAS2; Emr Cmr | 47 |
| ΔrelBE(pVL1) mutant | ΔrelBE harboring pVL1; Emr Cmr | This study |
| ΔrnhB mutant | In-frame SMU.994 deletion mutant derived from S. mutans UA159; Emr | This study |
| ΔrnhC mutant | In-frame SMU.1873 deletion mutant derived from S. mutans UA159; Emr | This study |
| ΔfruA mutant | In-frame SMU.78 deletion mutant derived from S. mutans UA159; Emr | 28 |
| ΔscrR mutant | In-frame SMU.1844 deletion mutant derived from S. mutans UA159; Emr | This study |
| ΔclpP mutant | In-frame SMU.1672 deletion mutant derived from S. mutans UA159; Emr | This study |
| ΔcomC mutant | In-frame SMU.1915 deletion mutant derived from S. mutans UA159; Emr | 41 |
| ΔcomE mutant | In-frame SMU.1917 deletion mutant derived from S. mutans UA159; Emr | 41 |
| Plasmids | ||
| pIB166 | Shuttle plasmid containing the P23 lactococcal promoter; Cmr | 4 |
| pMAS2 | mazEF cloned into pIB166; Cmr | 47 |
| pVL1 | relBE cloned into pIB166; Cmr | This study |
Emr, erythromycin resistance; Cmr, chloramphenicol resistance; Spr, spectinomycin resistance.
Persistence assay.
Overnight cultures of S. mutans were diluted (1:20) into fresh THYE broth containing ofloxacin (20 μg/ml), oxacillin (2 μg/ml), cefotaxime (2 μg/ml), vancomycin (20 μg/ml), or rifampin (50 μg/ml), followed by incubation for 24 h at 37°C. Samples were withdrawn at the indicated times, serially diluted, and plated on THYE agar plates. The colonies were counted after 48 h of incubation. All assays were performed in triplicate from three independent cultures. The statistical significance was determined by using a Student t test and a P value of <0.01.
For the biofilm persistence assays, biofilms were developed in polystyrene microtiter plates as described above. After the incubation, the planktonic cells were carefully removed from the biofilms, harvested by centrifugation, washed with sterile phosphate-buffered saline (PBS), and resuspended in fresh THYE broth. Biofilm cells left intact in the wells were rinsed once with sterile PBS to remove loosely bound cells, and the biofilms were detached by physical scraping. The biofilm pellet collected by centrifugation was washed with sterile PBS and resuspended into fresh THYE broth. Both biofilm and planktonic cells were separately treated with ofloxacin (50 μg/ml) for 2 to 5 days. Samples were withdrawn at indicated time, serially diluted, and plated on THYE agar plates. Colonies were counted after 48 h of incubation. All assays were performed in triplicate from three independent cultures. Statistical significance was determined by using a Student t test and a P value of <0.01.
Heritability assay.
Overnight cultures of S. mutans cultivated in THYE broth without antibiotic were diluted (1:20) into fresh THYE broth containing ofloxacin (20 μg/ml) and incubated at 37°C for 24 h. Surviving cells were resuspended into fresh THYE broth without ofloxacin until the stationary phase was reached. The cells were then diluted (1:20) into fresh THYE broth containing ofloxacin (20 μg/ml) and incubated at 37°C for 24 h. This procedure was repeated three times. Samples were withdrawn at the indicated times during each passage, serially diluted, and plated on THYE agar plates. Colonies were counted after 48 h of incubation. All assays were performed in triplicate from three independent cultures. Statistical significance was determined by using a Student t test and a P value of <0.01.
Ectopic expression of type II TA modules.
The full-length coding regions of UA159 mazEF and relBE operons were PCR amplified using UA159 genomic DNA (gDNA) as a template and cloned under the control of the lactococcal promoter P23 in the shuttle plasmid pIB166. The recombinant plasmids pMAS2 (mazEF in pIB166) and pVL1 (relBE in pIB166) were transferred into S. mutans ΔmazEF and ΔrelBE mutants, respectively. Complemented strains were tested for persister cell formation using a persistence assay as described above.
Construction of a S. mutans genomic DNA library and selection of persister mutants.
Genomic DNA of UA159 WT strain was extracted and partially digested with the restriction enzyme Sau3AI for 1 h at 37°C. Fragments of ca. 0.25 to 1.5 kb were purified and cloned into the BamHI-digested expression plasmid pIB166. E. coli DH10B electrocompetent cells were transformed with the UA159 gDNA library for amplification. The expression library was then transferred to UA159 WT cells by natural transformation, and the clones were selected on THYE-chloramphenicol agar plates. A total of ∼2,000 clones were picked individually from transformation plates and screened for increased or decreased survival to ofloxacin. Briefly, overnight cultures of the picked clones were diluted (1:20) into fresh THYE broth containing ofloxacin (20 μg/ml) in the individual wells of a 96-well microtiter plate. Plates were incubated for 24 h at 37°C without agitation. At 5 and 24 h, ofloxacin-treated clones were diluted in sterile PBS and spot plated onto THYE agar plates. Colonies were counted after 48 h of incubation. Clones with an altered number of persister cells were selected. This selection comprised clones showing ≥2-fold increase or decrease in cell survival to ofloxacin versus WT control strain harboring the empty plasmid.
Environmental stresses and quorum-sensing peptide pheromone assays.
Overnight cultures of S. mutans were diluted (1:100) into fresh THYE broth and exposed for 2 h at 37°C (except for heat shock) to the following stresses: acid shock (THYE broth acidified to pH 5.0 by addition of HCl), amino acid starvation (100 μg of serine hydroxamate/ml), oxidative stress (0.5 mM H2O2), and heat shock (50°C). For the quorum-sensing peptide pheromone assay, diluted cells were incubated with 2 μM synthetic CSP (sCSP; Advanced Protein Technology Centre, Hospital for Sick Children, Toronto, Ontario, Canada) pheromone for 2 h at 37°C. Stressed cells and CSP-induced cells were then treated with ofloxacin at 20 μg/ml (stress and CSP assays), ciprofloxacin at 20 μg/ml (CSP assay), oxacillin at 10 μg/ml (CSP assay), or rifampin at 50 μg/ml (CSP assay) for 24 h at 37°C. Samples were withdrawn at the indicated times, serially diluted, and plated on THYE agar plates. Colonies were counted after 48 h of incubation. All assays were performed in triplicate from three independent cultures. Statistical significance was determined by using a Student t test and a P value of <0.01.
Gene expression analysis.
Transcriptional analysis was conducted by real-time quantitative PCR (qPCR). Total RNA was extracted using an RNeasy minikit (Qiagen). DNA-free RNA (10 μg) was reverse transcribed using a first-strand cDNA synthesis kit (MBI Fermentas). qPCRs were carried out using the SsoFast EvaGreen Supermix (Bio-Rad) and a CFX96 real-time PCR detection system (Bio-Rad). The 16S rRNA gene was used as an internal reference. qPCR assays were performed in triplicate with RNA isolated from three independent experiments. Statistical significance was determined by using a Student t test and a P value of <0.01.
RESULTS
S. mutans populations form multidrug-tolerant persister cells.
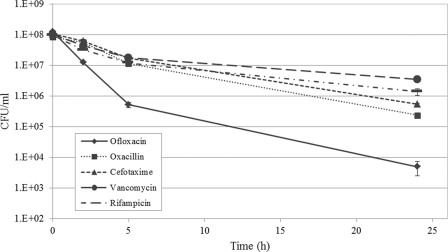
Persister cells are phenotypically tolerant to killing by antibiotics. At appropriate concentrations, bactericidal antibiotics should kill all normal growing cells, while dormant persisters remain in the population. If cell counts from these time-dependent killing assays are plotted out, it produces a biphasic killing pattern reflecting the rapid killing of the bulk of antibiotic-sensitive cells, and the presence of a surviving subpopulation of persister cells (18). In order to test whether S. mutans was able to develop persisters, several antibiotics exhibiting different mechanisms of action were tested. As shown in Fig. 1, treatments by all antibiotics displayed a typical biphasic killing curve, confirming the presence of multidrug-tolerant persisters in S. mutans cultures. The fluoroquinolone ofloxacin became the antibiotic of choice in the present study since it kills both normal growing cells and slowly growing or nongrowing stationary-phase cells, leaving dormant persisters intact (17).
Fig 1.
S. mutans multidrug tolerant persisters. During the antibiotic challenges, cells were removed at the indicated time points, serially diluted, and spot plated onto THYE agar plates for CFU determination. The S. mutans UA159 WT strain showed a biphasic killing curve for each of the antibiotics tested, where the majority of antibiotic-sensitive cells are rapidly killed, leaving intact a surviving subpopulation of persister cells.
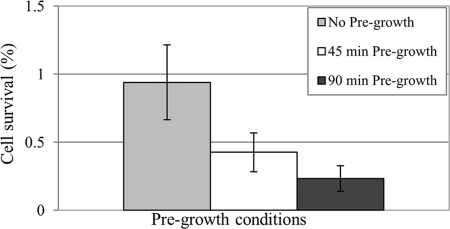
We next sought to determine whether S. mutans persisters were mainly formed during the logarithmic or stationary phase of growth. Overnight cultures of the UA159 WT strain were diluted into fresh medium to an OD600 of ∼0.05, and the cells were allowed to grow at 37°C for 45 or 90 min before ofloxacin treatment. The results showed that the percentage of cell survival decreased when cells exited from stationary phase and entered the lag phase (45 min of pregrowth) or early logarithmic phase (90 min of pregrowth) before being challenged with ofloxacin versus no pregrowth control (Fig. 2). These results strongly suggest that S. mutans persisters accumulate mainly during stationary phase and represent ∼1% of the stationary population. These results also suggest that pregrowth of diluted stationary-phase cultures caused some persisters to revert back to normal growing cells, thus lowering the number of persisters in the population.
Fig 2.
Effects of pregrowth period on cell survival after ofloxacin treatment. UA159 WT cultures were diluted into fresh THYE broth and pregrown for 45 or 90 min at 37°C before being treated with ofloxacin (20 μg/ml) for 5 h; no pregrowth was used as a control. Aliquots of cells were removed at time zero and 5 h after the introduction of the antibiotic. Cells were serially diluted and spot plated onto THYE agar, and the percentage of cell survival obtained after 5 h of ofloxacin treatment was determined from plate counts.
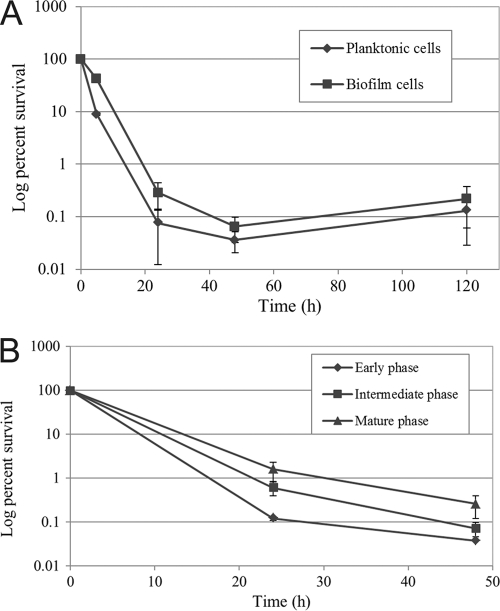
To determine whether slow-growing S. mutans biofilms produce substantial numbers of persisters, we also performed a persistence assay using 20-h-old biofilms. Over a period of 5 days, a distinctive biphasic killing curve can be observed for both the planktonic and the biofilm phases (Fig. 3A). Interestingly, the level of surviving persister cells was not statistically significantly greater in the biofilm phase compared to its free-floating planktonic phase at all time points tested. Subsequently, we wondered whether the age of the biofilm could affect the number of persisters formed. Biofilm cells obtained from different ages corresponding to early, intermediate, and mature biofilm phases were treated with ofloxacin for 2 days. The results showed that the number of persisters varied depending on the biofilm age, with older biofilms producing the highest number of persisters (Fig. 3B). These results suggest that S. mutans persisters accumulate as the biofilm matures, probably due to altered microenvironments within the biofilm.
Fig 3.
Formation of persisters in S. mutans biofilms. Static biofilms of UA159 WT strain were developed in polystyrene microtiter plates using 1/4-THYE broth supplemented with glucose. (A) After 20 h of growth, the planktonic phase was carefully removed, and the biofilm layer was gently washed with sterile PBS to remove loosely bound free-floating cells. The planktonic and biofilm phases were separately challenged with ofloxacin for 5 days. Paired t tests were performed on the results at 120, 48, and 24 h for both the planktonic and the biofilm persisters, and no statistically significant differences were found. (B) Early (∼6 h), intermediate (∼20 to 24 h), and mature (∼72 h) biofilms were treated with ofloxacin for 2 days. For both panels A and B, ofloxacin-treated cells were removed at the indicated time points, serially diluted, and spot plated onto THYE agar plates, and the percentage of cell survival was determined from plate counts.
S. mutans persister cells exhibit a nonheritable antibiotic tolerance phenotype.
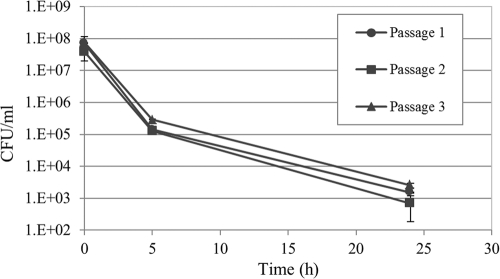
Unlike antimicrobial resistance, multidrug tolerance is a transient, nonheritable phenotype (38). To investigate the nonheritable nature of S. mutans persisters, we treated stationary-phase cells with ofloxacin for 24 h and performed three consecutive cycles of ofloxacin treatments on the surviving population derived from the persisters. As shown in Fig. 4, cultures grown up from persisters were as sensitive to ofloxacin as the parent culture from which the persisters were derived. Moreover, the killing curves maintained a biphasic pattern similar to that for the original population. The number of persisters remained approximately the same as repeated cycles of antibiotic exposure failed to enrich in persisters. These results confirmed that the formation of persisters in S. mutans populations is a nonheritable mechanism.
Fig 4.
Nonheritability nature of S. mutans persister phenotype. An overnight culture of UA159 WT strain was diluted to the early logarithmic phase and immediately treated with ofloxacin (20 μg/ml) for 24 h. The surviving cells were then regrown into fresh antibiotic-free THYE broth until the stationary phase was reached and then diluted to early logarithmic phase and immediately treated with ofloxacin (20 μg/ml) for 24 h. The procedure was repeated three times. The results showed that persisters are phenotypic variants of the parental strain that arise in a clonal population of genetically identical cells.
Ectopic expression of type II TA modules increases the number of persisters.
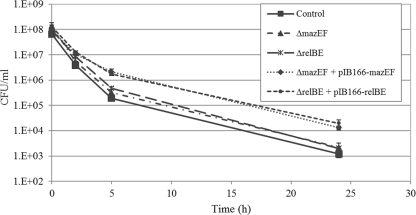
Toxin-antitoxin (TA) modules have been linked to persister formation primarily in E. coli and Mycobacterium tuberculosis, where certain TA modules were found to be upregulated in isolated persister cells (22, 44). To investigate the effects of ectopic overexpression of type II TAs on S. mutans persister formation, the mazEF and relBE operons were cloned under the control of a constitutive promoter into the shuttle vector pIB166. Our results showed that ectopic expression of MazEF and RelBE systems caused a log-fold increase in persister fraction under ofloxacin treatment, whereas deletion of the same TA systems had no discernible effect on the formation of persisters (Fig. 5). Similarly, WT cells overexpressing only the toxin displayed an ∼10-fold increase in cell survival (data not shown). As expected, a ΔmazEF ΔrelBE double deletion mutant did not affect persister production under the conditions tested, reinforcing the hypothesis that more than one single mechanism is responsible for persister formation. Although there is most likely an apparent redundancy of persister genes, our results suggested that both MazEF and RelBE type II TA systems function in contributing toward antibiotic tolerance through the formation of persister cells in S. mutans. In all studied type II TA systems, the antitoxin is less stable than the toxin and is degraded by a specific intracellular protease (6). The fact that an S. mutans ΔclpP mutant deficient in the ATP-dependent ClpP protease showed a reduced level of persisters (see Fig. S1 in the supplemental material) also suggests that higher levels of free toxin (MazF and/or RelE) could cause cell growth arrest, favoring bacterial persistence.
Fig 5.
Ectopic expression of type II TA modules in S. mutans. Overnight cultures of S. mutans strains were diluted to the early logarithmic phase and immediately treated with ofloxacin (20 μg/ml) for 24 h at 37°C. Cells were removed at the indicated time points, serially diluted, and spot plated onto THYE agar plates for CFU counting. UA159 WT strain harboring the empty plasmid was used as a control.
Identification of additional persistence genes.
We constructed an S. mutans gDNA library into an expression vector for gain of function since attempts by different groups to identify persister genes by screening transposon or insertion libraries for either increased or decreased survival to antibiotics were not successful (30). Approximately 2,000 clones were individually picked and tested for the production of altered number of persisters. For the initial screening, overnight cultures of S. mutans clones were diluted to early logarithmic phase into fresh THYE medium and immediately treated with ofloxacin for 24 h in 96-well plates (see Materials and Methods for details). Identified clones of interest displaying high- or low-level survival to antibiotic treatment in comparison to the control strain (WT harboring the empty plasmid) were selected for further analysis. Of the ∼2,000 clones tested, 20 clones of interest with unchanged growth parameters were initially identified and validated using our persistence assay: 16 clones exhibited a high persistence phenotype (increase in cell survival upon ofloxacin treatment versus control strain), and 4 clones displayed low persistence (decrease in cell survival upon ofloxacin treatment versus control strain) (Table 2). Plasmids were isolated from clones with the greatest differential survival (i.e., clones VL-1, VL-16, VL-39, VL-61, VL-390, VL-739, and VL-1176), sequenced to identify the DNA fragments expressed under the P23 promoter, and mapped to the UA159 genome. In order to confirm that the observed phenotypes (high or low persistence) were not the results of spontaneous mutations, we isolated the plasmid from each clone and transformed the WT strain with the respective plasmids. These new transformants were then tested using the persistence assay. The results demonstrated that similar phenotypes were observed between the original clones and the new transformants harboring the respective plasmids, confirming that the observed phenotypes were due to the expression plasmids.
Table 2.
Clones identified by high-throughput screening strategy
| Clonea | Phenotype (persistence) | % Survival at: |
Fold differenceb vs control at: |
Fragment(s) cloned (NCBI locus) | ||
|---|---|---|---|---|---|---|
| 5 h | 24 h | 5 h | 24 h | |||
| Control | Wild type | 0.25 | 1.6 × 10–3 | |||
| VL-1 | High | 0.66 | 9.8 × 10–3 | +2.6 | +6.1 | SMU.994 |
| VL-16 | High | 2.03 | 2.7 × 10–2 | +8.1 | +16.9 | SMU.402, SMU.1278 |
| VL-39 | Low | 0.17 | 4.5 × 10–4 | –1.5 | –3.5 | SMU.78 |
| VL-61 | High | 1.13 | 1.2 × 10–2 | +4.5 | +7.5 | SMU.1588-9 |
| VL-201 | High | 0.66 | 2.0 × 10–3 | +2.6 | +1.2 | NDc |
| VL-343 | High | 0.57 | 4.0 × 10–3 | +2.3 | +2.5 | ND |
| VL-363 | Low | 0.10 | 1.8 × 10–3 | –2.5 | +1.1 | ND |
| VL-390 | High | 2.04 | 3.3 × 10–2 | +8.2 | +20.6 | SMU.2154-6, SMU.1313 |
| VL-397 | Low | 0.13 | 1.5 × 10–3 | –1.9 | –1.1 | ND |
| VL-404 | High | 0.25 | 3.0 × 10–3 | 1.0 | +1.9 | ND |
| VL-418 | High | 0.47 | 4.9 × 10–3 | +1.9 | +3.1 | ND |
| VL-739 | High | 6.83 | 3.1 × 10–2 | +27.3 | +19.4 | IGR1445 |
| VL-766 | High | 0.37 | 3.4 × 10–3 | +1.5 | +2.1 | ND |
| VL-1176 | High | 0.81 | 8.3 × 10–3 | +3.2 | +5.2 | SMU.775 |
| VL-1306 | High | 0.35 | 3.7 × 10–3 | +1.4 | +2.3 | ND |
| VL-1341 | High | 0.91 | 5.6 × 10–3 | +3.6 | +3.5 | ND |
| VL-1357 | Low | 0.18 | 2.1 × 10–3 | –1.4 | +1.3 | ND |
| VL-1430 | High | 0.68 | 2.2 × 10–3 | +2.7 | +1.4 | ND |
| VL-1602 | High | 0.52 | 1.6 × 10–3 | +2.1 | 1.0 | ND |
| VL-1974 | High | 0.46 | 1.0 × 10–3 | +1.8 | –1.6 | ND |
Clones selected for sequencing and genomic mapping are indicated in boldface.
That is, the fold difference in cell survival toward ofloxacin compared to the wild-type strain carrying the control plasmid at 5 and 24 h. +, Increase in cell survival; –, decrease in cell survival.
ND, sequence not determined.
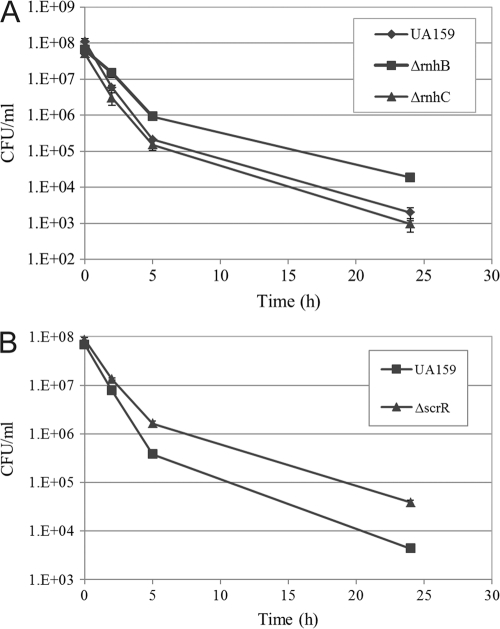
The clone VL-1 displaying a high persistence phenotype (∼2.6- and ∼6.1-fold increases in survival at 5 and 24 h, respectively; Table 2) had an ∼0.2-kb insert corresponding to a portion of the SMU.994 gene (also called rnhB) encoding a putative RNase H involved in DNA replication. An ΔrnhB deletion mutant was constructed in the WT strain and tested using a persistence assay. The results (Fig. 6A) showed that inactivation of rnhB caused an increased tolerance to ofloxacin (∼15 times more persisters than UA159 WT at 24 h). The MIC values of ofloxacin against the ΔrnhB mutant were not different from those for the WT strain, confirming that ΔrnhB was a true persistence mutant. Cultures of the ΔrnhB mutant exposed to rifampin or oxacillin also displayed an increase (≥2-fold) in survival (data not shown), suggesting that these persister cells are multidrug tolerant. The fact that both clone VL-1 and ΔrnhB mutant produced the same phenotype was surprising. We hypothesized that a hidden open reading frame(s) may be responsible for the high persistence phenotype observed for clone VL-1. In an attempt to verify this hypothesis, further analysis of the overexpressed gene fragment was done through bioinformatics. Interestingly, we found that clone VL-1 was encoding a putative peptide of 76 amino acid residues containing the conserved RNase HII motif. This result suggested to us that when overexpressed this peptide can compete with the native RnhB enzyme for its binding sites. If true, a gain-of-function mutation as observed for the knockout strain and overexpression clone would result in the same phenotype. Bioinformatic analysis of UA159 genome also revealed the presence of an additional RNase H, RnhC (SMU.1873), sharing 40% amino acid similarity with RnhB. Interestingly, while inactivation of rnhB increased the number of multidrug tolerant persister cells, rnhC does not seem to play a role in persister formation under the conditions tested since inactivation of rnhC did not alter the number of persisters (Fig. 6A).
Fig 6.
Effect of inactivation of candidate persister genes on S. mutans persister formation. Overnight cultures of UA159 WT strain and its RNase (ΔrnhB and ΔrnhC) (A) and sucrose operon repressor (ΔscrR) (B) deletion mutants were diluted to the early logarithmic phase and immediately treated with ofloxacin (20 μg/ml) for 24 h at 37°C. Cells were removed at the indicated time points, serially diluted, and spot plated onto THYE agar plates for CFU counting.
S. mutans cells overexpressing the clone VL-739 displayed a high persistence phenotype (∼27.3- and ∼19.4-fold increases in survival at 5 and 24 h, respectively; Table 2). Sequencing of the insert revealed that clone VL-739 had an ∼0.5-kb insert corresponding to the intergenic region IGR1445. This intergenic region corresponds to the promoter region of both scrA and scrB genes. The scr regulon of S. mutans is composed of three genes—scrA, scrB, and scrR—coding for a sucrose-specific IIABC PTS component, a sucrose-6-phosphate hydrolase, and a sucrose operon repressor, respectively (51). Since cells lacking the ScrR repressor showed a significant change in the level of persister cells (∼8.8 times more persisters than UA159 WT at 24 h; Fig. 6B), our results suggest that a derepression of the scrA/scrB promoter region located in IGR1445 may directly or indirectly lead to an increase in persister formation. The MIC values of ofloxacin against ΔscrR mutant were not different from those for WT strain, confirming that the ΔscrR strain was a true persistence mutant.
Clone VL-39 displayed a low persistence phenotype (∼1.5- and ∼3.5-fold decreases in survival at 5 and 24 h, respectively; Table 2). Sequencing of the insert showed that it contained a fragment of ∼0.4 kb corresponding to a portion of the fruA gene encoding a β-d-fructosidase. This enzyme is responsible for the hydrolysis of fructans, a class of fructose-based dietary polysaccharides, enhancing S. mutans cell survival during periods of nutrient starvation (5). To test persister formation in cells lacking FruA, a fruA deletion mutant was tested for its ability to persist in the presence of ofloxacin. Our results showed that a ΔfruA mutant did not differ from the WT control strain in its ability to form persisters (data not shown). The role of fruA gene (or possibly a small peptide derived from the FruA protein) in S. mutans persister formation is currently unclear and warrants further investigation.
The clone VL-16 displaying a high persistence phenotype (∼8.1- and ∼16.9-fold increases in survival at 5 and 24 h, respectively; Table 2) contained two Sau3AI fragments. Sequence analysis of the cloned insert revealed that these fragments corresponded to portions of SMU.402 and SMU.1278 encoding a pyruvate formate-lyase (PFL) and a putative phosphoglycolate phosphatase, respectively. Although work previously done in S. mutans has shown that PFL was responsible for pyruvate heterofermentation under anaerobic conditions (25), it is still unclear how this may relate to persistence.
Cells overexpressing the clone VL-61 displayed a high persistence phenotype (∼4.5- and ∼7.5-fold increases in survival at 5 and 24 h, respectively; Table 2). Sequencing of the cloned fragment revealed that clone VL-61 had an ∼0.7-kb insert corresponding to portions of SMU.1588 and SMU.1589 encoding putative glycosyltransferases involved in the biosynthesis and degradation of peptidoglycan.
The nucleotide sequence of clone VL-390 displaying a high persistence phenotype (∼8.2- and ∼20.6-fold increases in survival at 5 and 24 h, respectively; Table 2) revealed that it contained an insert of ∼1.4 kb containing two fragments of different sizes. The first fragment corresponded to a region encompassing the SMU.2154, SMU.2155, and SMU.2156 genes and coding for a putative peptidase, a conserved hypothetical protein, and RecF recombinase, respectively. The second fragment corresponded to SMU.1313 encoding a putative ATP-dependent DNA helicase.
Finally, sequencing of the insert of clone VL-1176 displaying a high persistence phenotype (∼3.2- and ∼5.2-fold increases in survival at 5 and 24 h, respectively; Table 2) showed that it contained a fragment of ∼0.6 kb corresponding to a portion of SMU.775 encoding a conserved hypothetical protein.
Altogether, these results obtained through genetic screening suggest that S. mutans did not evolve a dedicated mechanism, allowing it to adopt a persistence phenotype. In contrast, the picture that emerges thus far from our results and from several studies is pointing to redundant pathways for bacterial persister formation.
Environmental stressors induce the formation of persister cells.
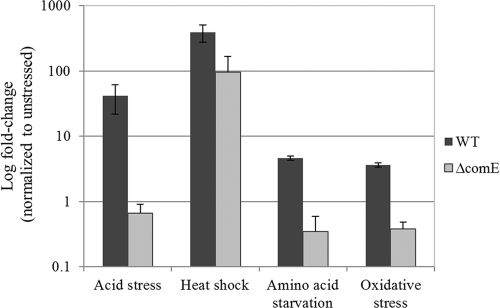
A clonal population may have two (bi-stability) or more (multi-stability) subpopulations with distinct phenotypic properties, such as persistence (15). Switching to generate alternate phenotypes can occur spontaneously (stochastic switching) or in response to environmental perturbations (responsive switching). Since S. mutans cells are constantly exposed to fluctuating and adverse environmental conditions in the oral cavity, ranging from variations in temperature, acidic pH, oxidative stress, and nutrient-limiting conditions (24, 36), we hypothesized that certain environmental stressors could positively influenced the number of persisters. To determine whether S. mutans persisters could be formed in response to different environmental stresses, S. mutans UA159 WT cultures were exposed to heat, acidic pH, oxidative stress, and serine hydroxamate (to mimic amino acid starvation) for 2 h prior to being challenged with ofloxacin for 24 h. As shown at Fig. 7, all tested stressors produced higher levels (ranging from 4- to >100-fold increases) of antibiotic tolerant persister cells in the WT strain, suggesting that the onset of environmental stress may induce the persistence phenotype in S. mutans populations.
Fig 7.
Effect of various environmental stressors on S. mutans persister formation. Overnight cultures of UA159 WT strain and its ΔcomE mutant were diluted (1:20) into fresh THYE broth and challenged with an environmental stress for 2 h before being treated with ofloxacin (20 μg/ml) for 24 h at 37°C. Aliquots of cells were removed at the introduction of the antibiotic ofloxacin and after the antibiotic treatment (24 h) to determine cell survival by spot plating onto THYE agar plates.
Formation of S. mutans persisters is positively influenced by the stress-inducible quorum-sensing peptide.
Work previously done in our lab has shown that S. mutans integrates its response to specific environmental stresses with its intraspecies bacterial quorum-sensing system, the CSP-ComDE regulatory circuit (41). We first monitored the expression of the CSP pheromone gene (comC) under the environmental stresses (heat, acidic pH, oxidative stress, and amino acid starvation) known to trigger the formation of S. mutans persisters in the WT strain. The levels of comC transcripts were significantly upregulated by heat at 50°C [(8.0 ± 3.5)-fold], acidic conditions at pH 5.0 [(40.4 ± 30.5)-fold], and in the presence of hydrogen peroxide [(21.5 ± 12.7)-fold] and serine hydroxamate [(38.5 ± 31.0)-fold], confirming the link between the stress response in S. mutans and its intraspecies quorum-sensing system.
In streptococci, the ComDE two-component system conveys the quorum-sensing CSP signal by phosphotransfer from the ComD membrane-bound receptor to ComE response regulator. Once phosphorylated, activated ComE alters gene expression, allowing cells to launch coordinated responses to their environments (39). We tested the direct impact of the CSP-ComDE quorum-sensing system in the stress-induced development of persisters using cells of a ΔcomE mutant, which is unable to respond to the CSP signaling pheromone, exposed to the same environmental stresses as those described above. Our results showed that whereas the WT strain produced an increased number of persisters under these stresses, the stress-inducible persistence phenotype was abolished for the ΔcomE mutant subjected to acidic pH, oxidative stress, and amino acid starvation (Fig. 7). Surprisingly, ΔcomE mutant cells exposed to heat stress at 50°C were still able to increase the numbers of ofloxacin-tolerant persisters but to a lesser degree than that observed for WT (95-fold increase for the ΔcomE mutant versus 389-fold increase for the WT), suggesting that heat stress also induces the formation of persisters through a quorum-sensing-independent pathway.
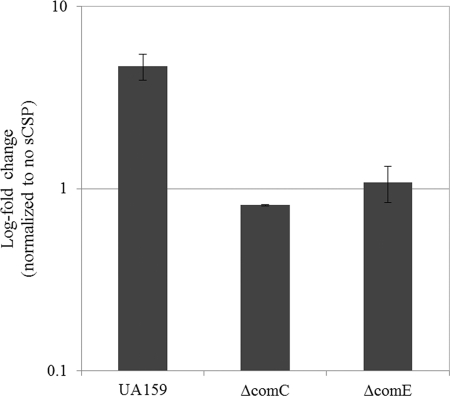
Finally, the intimate connection between the S. mutans CSP-ComDE intraspecies quorum-sensing pathway and the development of persisters was tested using UA159 WT strain and its quorum-sensing deficient mutants, the ΔcomC mutant, which is unable to produce the CSP signaling pheromone, and the ΔcomE mutant. Since environmental stresses increased the number of persisters and considering that stresses induced the expression of the CSP pheromone, we used exogenously added synthetic CSP (sCSP) to study the effect of the CSP pheromone on formation of S. mutans persisters. The WT strain and its ΔcomC and ΔcomE mutants were exposed to sCSP before being treated with ofloxacin for 24 h. Our data showed that the CSP pheromone significantly increased (∼5-fold) the number of ofloxacin-tolerant persister cells for the WT strain (Fig. 8). In contrast, the number of persisters did not change when the ΔcomC and ΔcomE mutant cells were preincubated with sCSP before the antibiotic treatment (Fig. 8). The same results were obtained with oxacillin (∼2-fold increase) and vancomycin (∼3-fold increase) for the WT strain (see Fig. S2 in the supplemental material). The fact that the addition of synthetic CSP to a ΔcomC mutant could not restore the WT phenotype strongly suggests that the production of the CSP pheromone is subject to positive-feedback regulation, as frequently observed in Gram-negative quorum-sensing systems, and that this feedback loop may play a role in signal integration and/or transmission. Altogether, these results clearly demonstrate that the intraspecies quorum-sensing system of S. mutans plays a role in the formation of multidrug-tolerant persisters.
Fig 8.
Effect of CSP pheromone on the development of S. mutans persisters. Cells of UA159 WT strain and its quorum-sensing mutants (ΔcomC and ΔcomE) were preincubated with synthetic CSP (2 μM sCSP) for 2 h at 37°C before being challenged with ofloxacin at 20 μg/ml. Aliquots of cells were removed at the introduction of the antibiotic and after the antibiotic treatment (24 h) to determine cell survival by spot plating onto THYE agar plates.
DISCUSSION
Resistant pathogens play a major role in acute infections, and a variety of acquired mechanisms lead to multidrug resistance. Bacterial populations also produce persister cells that are tolerant to killing by all antibiotics currently in use, a phenomenon known as multidrug tolerance (21, 30, 45, 53). In contrast to inherited antibiotic resistance resulting from mutations in existing genes or the acquisition of external resistance-encoding genes, multidrug tolerance conferred by persister cells is noninherited and is purely phenotypic. Persistence is a characteristic of a heterogeneous population of cells. It may therefore present an advantage at the level of the population and not at the level of single cells—a bet-hedging strategy to cope with a fluctuating stressful environment ensuring survival of the species during catastrophe (50). Although persisters are very important in antibiotic therapy, the identification of the phenotypic switch responsible for persistence remains largely unknown. Given the low frequency of persisters in a clonal population, it is conceivable that stochastic variations of gene expression would lead to toxic levels of a variety of proteins in a small fraction of cells, increasing their chance to become persisters, particularly when in a dense biofilm environment.
The natural habitat of S. mutans is the oral biofilm, one of the most complex human microbiota (55). The number of persisters in a growing population of bacteria can reach ca. 1% at the stationary phase (21). Similarly, S. mutans produced substantial numbers of persisters when growing in a biofilm. Interestingly, we demonstrated that older (72-h-old) S. mutans biofilms produced more persister cells than biofilms grown to the early (6-h-old) or intermediate (24-h-old) phases, suggesting that more persisters could be formed due to gradually limiting conditions prevailing in the biofilm (e.g., nutrient limitation and altered microenvironments). Biofilms can thus promote the survival of bacteria by generating persister cells and by creating a protective niche where bacteria (normal growing and dormant) can evade the immune system. Assuming that persistence can be governed by both stochastic and deterministic mechanisms (20, 22, 31), the clinical implications of persisters in the context of biofilm infections are highly significant.
Our results showed that fluctuations in the levels of a small number of genes triggered the formation of persisters in S. mutans cultures. Both S. mutans MazEF and RelBE type II TA modules function in contributing to antibiotic tolerance through the formation of persister cells. Overproduction of these TA modules did not cause cell lysis of S. mutans (verified experimentally for MazEF [47]; we assume it is the same for RelBE) but rather exerted an increase in persister cells that were more tolerant toward antibiotic killing. Not surprisingly, the removal of both TA loci did not affect antibiotic susceptibility in S. mutans, suggesting that additional type II TA systems could be required for persistence. For instance, a total of 45 putative toxins and antitoxins have been identified in the genome of S. mutans UA159 reference strain on the basis of protein sequence and comparative genomic analysis (34, 47); however, only four TA pairs (MazE/MazF, RelB/RelE, Xre/COG2856, and PIN/AbrB) possessed all of the features and could be predicted to function as bona fide type II TA systems. Further experiments will be required to explore the possibility that TA loci contribute cumulatively to the development of bacterial persistence in S. mutans.
The screening of an expression library has led us to identify additional genes involved in persister formation. One of these genes, rnhB, encodes a putative RNase H and caused a significant increase in the number of persisters when inactivated. Interestingly, inactivation of rnhC encoding another putative RNase H did not affect the levels of persisters, albeit the lack of this RNase led to noticeable growth defects, including significantly delayed colony formation and delayed time to reach the stationary phase. Although further studies would be required to elucidate the role of RnhB RNase in persister formation, our results reinforce the notion that the increase in persisters cannot be solely due to reduced bacterial growth (2). The genetic screening also produced candidate persister genes involved in transcription/replication, sugar metabolism, cell wall synthesis, and energy metabolism, confirming that S. mutans, like many other human pathogens (18, 20, 31), possesses redundant pathways of persister formation. The fact that there is more than one way to produce persisters strongly suggests that each bacterial population contains a diversity of persisters based on fluctuations and variability in cellular processes. This diversity hypothesis suggests that bacterial persistence may be a result of multiple distinct cellular physiologies within a population (2).
Switching to generate alternate phenotypes can also occur in response to environmental fluctuations (20). In E. coli, persisters can develop through the upregulation of the TisB toxin via the SOS response to damaged DNA caused by ciprofloxacin (13, 14). Streptococci lack the classical SOS response, and the induction of the competence regulon has been proposed to act as a general stress response in Gram-positive bacteria (7, 43). Early work done in S. mutans suggested an intimate link between the competence cascade and the organism's response to acid stress (32). In S. mutans and Streptococcus pneumoniae, the intraspecies quorum-sensing CSP signal responsible for competence induction has been shown to act as a stress pheromone or “alarmone” in response to certain stressful environments (7, 41). It is well known that bacterial quorum-sensing signaling mechanisms can go well beyond the counting of cell numbers (40, 42). There is growing evidence that quorum sensing constitutes a global regulatory system in many different bacterial species (40). In streptococci, the CSP pheromone is sensed outside the cell by the histidine kinase of a two-component system. Binding of extracellular CSP pheromone to ComD sensor initiates the phosphorelay cascade (ComD∼P → ComE∼P) that leads to transcriptional activation of the competence regulon controlled by the alternate sigma factor ComX (39). Our results suggest that when a portion of the population is challenged with an incoming stress, the production of CSP pheromone allows communication to the other siblings within the population to prepare for a “stress-response” state via quorum sensing, in order to survive. It is possible that by mimicking this “incoming stress” signal through the treatment with exogenous CSP, cells within the population are able to undergo a different transcriptional profile, where some cells are induced into a persister state through an indirect downstream transcriptional regulation of the ComDE pathway. To our knowledge, this is the first time where bacterial persistence has been shown induced via a quorum-sensing regulatory system. While the recent study by Möker et al. (37) showed that cultures of Pseudomonas aeruginosa increased their persister numbers in response to quorum-sensing-related signaling molecules (phenazine pyocin and acyl-homoserine lactone), these authors stopped short of explaining the signaling pathway that leads to the increased formation in response to these signaling molecules. In S. mutans, several class II bacteriocins are expressed through the CSP-ComDE quorum-sensing system (16, 26, 48). Class II bacteriocins are antimicrobial peptides that function to kill competitors by dissipating the proton motive force via pore formation in the cytoplasmic membrane of target cells (23). Interestingly, preliminary work done in our lab demonstrated that inactivation of mutacin V (CipB), but not mutacin IV (NlmAB), completely abolished the increase in persister numbers observed for the WT strain following pretreatment with the CSP pheromone. It can be speculated that the CSP-induced mutacin V could induce the formation of persisters by insertion into the cytoplasmic membrane, thus decreasing the proton motive force and the ATP levels. Alternatively, mutacin V could act as a peptide regulator for the transcriptional control of persister genes, as was recently demonstrated for the development of genetic competence under CSP-induced conditions (16).
Bacteria are often subjected to a myriad of environmental stresses, and this opens the possibility that quorum sensing can serve to prime the cellular response to stress and ensure survival of the population by inducing the formation of antibiotic-tolerant persisters. Therefore, the prevention of persisters formed under challenging conditions by interfering with the intraspecies quorum-sensing system of the pathogen suggests different possible drug targets for the development of effective antipersister strategies. For instance, several algae and terrestrial plants produce compounds able to interfere with bacterial quorum sensing (11). The new data presented in the present study definitely benefits a field that is still in its scientific infancy.
Supplementary Material
ACKNOWLEDGMENTS
We thank Stephanie Koyanagi for technical assistance and Delphine Dufour for careful reading of the manuscript. We thank Indranil Biswas for the generous gift of plasmid pIB166.
This study was supported by Canadian Institutes of Health Research grant MOP-93555 to C.M.L. and Natural Sciences and Engineering Research Council of Canada grant RGPIN 355968 to C.M.L. C.M.L. is a recipient of a Canada Research Chair. V.L. is supported by a Harron Scholarship.
Footnotes
Published ahead of print 24 February 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Alekshun MN, Levy SB. 2007. Molecular mechanisms of antibacterial multidrug resistance. Cell 128:1037–1050 [DOI] [PubMed] [Google Scholar]
- 2. Allison KR, Brynildsen MP, Colins JJ. 2011. Heterogenous bacterial persisters and engineering approaches to eliminate them. Curr. Opin. Microbiol. 14:593–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bigger JW. 1944. Treatment of staphylococcal infections with penicillin. Lancet 244:497–500 [Google Scholar]
- 4. Biswas I, Jha JK, Fromm N. 2008. Shuttle expression plasmids for genetic studies in Streptococcus mutans. Microbiology 154:2275–2282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Burne RA, Schilling K, Bowen WH, Yasbin RE. 1987. Expression, purification, and characterization of an exo-β-d-fructosidase of Streptococcus mutans. J. Bacteriol. 169:4507–4517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Buts L, Lah J, Dao-Thi MH, Wyns L, Loris R. 2005. Toxin-antitoxin modules as bacterial metabolic stress managers. Trends Biochem. Sci. 30:672–679 [DOI] [PubMed] [Google Scholar]
- 7. Claverys JP, Prudhomme M, Martin B. 2006. Induction of competence regulons as a general response to stress in Gram-positive bacteria. Annu. Rev. Microbiol. 60:451–475 [DOI] [PubMed] [Google Scholar]
- 8. Costerton JW, Stewart PS, Greenberg EP. 2007. Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322 [DOI] [PubMed] [Google Scholar]
- 9. Davies D. 2003. Understanding biofilm resistance to antimicrobial agents. Nat. Rev. Drug Discov. 2:114–122 [DOI] [PubMed] [Google Scholar]
- 10. Davies J, Davies D. 2010. Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 74:417–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Defoirdt T, Boon N, Bossier P. 2010. Can. bacteria evolve resistance to quorum sensing disruption? PLoS Pathog. 6:e1000989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. del Pozo JL, Patel R. 2007. The challenge of treating biofilm-associated bacterial infections. Clin. Pharmacol. Ther. 82:204–209 [DOI] [PubMed] [Google Scholar]
- 13. Dörr T, Lewis K, Vulic M. 2009. SOS response induces persistence to fluoroquinolones in Escherichia coli. PLoS Genet. 5:e1000760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dörr T, Vulic M, Lewis K. 2010. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol. 8:e1000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dubnau D, Losick R. 2006. Bistability in bacteria. Mol. Microbiol. 61:564–572 [DOI] [PubMed] [Google Scholar]
- 16. Dufour D, Cordova M, Cvitkovitch DG, Lévesque CM. 2011. Regulation of the competence pathway as a novel role associated with a streptococcal bacteriocin. J. Bacteriol. 193:6552–6559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Eng RH, Padberg FT, Smith SM, Tan EN, Cherubin CE. 1991. Bactericidal effects of antibiotics on slowly growing and nongrowing bacteria. Antimicrob. Agents Chemother. 35:1824–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fauvart M, De Groote VN, Michiels J. 2011. Role of persister cells in chronic infections: clinical relevance and perspectives on anti-persister therapies. J. Med. Microbiol. 60:699–709 [DOI] [PubMed] [Google Scholar]
- 19. Fineran PC, et al. 2009. The phage abortive infection system, ToxIN, functions as a protein-RNA toxin-antitoxin pair. Proc. Natl. Acad. Sci. U. S. A. 106:894–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jayaraman R. 2008. Bacterial persistence: some insights into an old phenomenon. J. Biosci. 33:795–805 [DOI] [PubMed] [Google Scholar]
- 21. Keren I, Kaldalu N, Spoering A, Wang Y, Lewis K. 2004. Persister cells and and tolerance to antimicrobials. FEMS Microbiol. Lett. 230:13–18 [DOI] [PubMed] [Google Scholar]
- 22. Keren I, Minami S, Rubin E, Lewis K. 2011. Characterization and transcriptome analysis of Mycobacterium tuberculosis persisters. mBio 2(3):e00100–11 doi:10.1128/mBio.00100-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kjos M, et al. 2011. Target recognition, resistance, immunity and genome mining of class II bacteriocins from Gram-positive bacteria. Microbiology 157:3256–3267 [DOI] [PubMed] [Google Scholar]
- 24. Kolenbrander PE. 2000. Oral microbial communities: biofilms, interactions, and genetic systems. Annu. Rev. Microbiol. 54:413–437 [DOI] [PubMed] [Google Scholar]
- 25. Korithoski B, Lévesque CM, Cvitkovitch DG. 2008. The involvement of the pyruvate dehydrogenase E1α subunit in Streptococcus mutans acid tolerance. FEMS Microbiol. Lett. 289:13–19 [DOI] [PubMed] [Google Scholar]
- 26. Kreth J, Merritt J, Zhu J, Shi W, Qi F. 2006. Cell-density- and ComE-dependent expression of a group of mutacin and mutacin-like genes in Streptococcus mutans. FEMS Microbiol. Lett. 265:11–17 [DOI] [PubMed] [Google Scholar]
- 27. Lau PCY, Sung CK, Lee JH, Morrison DA, Cvitkovitch DG. 2002. PCR ligation mutagenesis in transformable streptococci: application and efficiency. J. Microbiol. Methods 49:193–205 [DOI] [PubMed] [Google Scholar]
- 28. Lévesque CM, et al. 2005. Involvement of sortase anchoring of cell wall proteins in biofilm formation by Streptococcus mutans. Infect. Immun. 73:3773–3777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lewis K. 2007. Persister cells, dormancy and infectious disease. Nat. Rev. Microbiol. 5:48–56 [DOI] [PubMed] [Google Scholar]
- 30. Lewis K. 2008. Multidrug tolerance of biofilms and persister cell. Curr. Top. Microbiol. Immunol. 322:107–131 [DOI] [PubMed] [Google Scholar]
- 31. Lewis K. 2010. Persister cells. Annu. Rev. Microbiol. 64:357–372 [DOI] [PubMed] [Google Scholar]
- 32. Li YH, Hanna MN, Svensäter G, Ellen RP, Cvitkovitch DG. 2001. Cell density modulates acid adaptation in Streptococcus mutans: implications for survival in biofilms. J. Bacteriol. 183:6875–6884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Maisonneuve E, Shakespeare LJ, Jørgensen MG, Gerdes K. 2011. Bacterial persistence by RNA endonucleases. Proc. Natl. Acad. Sci. U. S. A. 108:13206–13211 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34. Makarova KS, Wolf YI, Koonin EV. 2009. Comprehensive comparative-genomic analysis of type 2 toxin-antitoxin systems and related mobile stress response systems in prokaryotes. Biol. Direct. 4:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Marsh PD. 2005. Dental plaque: biological significance of a biofilm and community lifestyle. J. Clin. Periodontol. 32:7–15 [DOI] [PubMed] [Google Scholar]
- 36. Marsh PD. 2006. Dental plaque as a biofilm and a microbial community: implications for health and disease. BMC Oral Health 6:S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Möker N, Dean CR, Tao J. 2010. Pseudomonas aeruginosa increases formation of multidrug tolerance persister cells in response to quorum sensing signaling molecules. J. Bacteriol. 192:1946–1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Moyed HS, Bertrand KP. 1983. hipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J. Bacteriol. 155:768–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Oggioni MR, Morrison DA. 2008. Cooperative regulation of competence development in Streptococcus pneumoniae: cell-to-cell signaling via a peptide pheromone and an alternative sigma factor, p 345–362 In Winans SC, Bassler BL. (ed), Chemical communication among bacteria. ASM Press, Washington, DC [Google Scholar]
- 40. Parsek MR, Greenberg EP. 2005. Sociomicrobiology: the connection between quorum sensing and biofilms. Trends Microbiol. 13:27–33 [DOI] [PubMed] [Google Scholar]
- 41. Perry JA, Jones MB, Peterson SN, Cvitkovitch DG, Lévesque CM. 2009. Peptide alarmone signaling triggers an auto-active bacteriocin necessary for genetic competence. Mol. Microbiol. 72:905–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Platt TG, Fuqua C. 2010. What's in a name? The semantics of quorum sensing. Trends Microbiol. 18:383–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Prudhomme M, Attaiech L, Sanchez G, Martin B, Claverys JP. 2006. Antibiotic stress induces genetic transformability in the human pathogen Streptococcus pneumoniae. Science 313:89–92 [DOI] [PubMed] [Google Scholar]
- 44. Shah D, Zhang Z, Khodursky A, Kaldalu N, Kurg K, et al. 2006. Persisters: a distinct physiological state of Escherichia coli. BMC Microbiol. 6:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Spoering AL, Vulic M, Lewis K. 2006. GlpD and PlsB participate in persister cell formation in Escherichia coli. J. Bacteriol. 188:5136–5144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Suntharalingam P, Senadheera MD, Mair RW, Lévesque CM, Cvitkovitch DG. 2009. The LiasFSR system regulates the cell envelope stress response in Streptococcus mutans. J. Bacteriol. 191:2973–2984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Syed MA, et al. 2011. The chromosomal mazEF locus of Streptococcus mutans encodes a functional type II toxin-antitoxin addiction system. J. Bacteriol. 193:1122–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Van der Ploeg JR. 2005. Regulation of bacteriocin production in Streptococcus mutans by the quorum-sensing system required for development of genetic competence. J. Bacteriol. 187:3980–3989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Van Melderen L. 2010. Toxin-antitoxin systems: why so many, what for? Curr. Opin. Microbiol. 13:781–785 [DOI] [PubMed] [Google Scholar]
- 50. Veening JW, Smits WK, Kuipers OP. 2008. Bistability, epigenetics, and bet-hedging in bacteria. Annu. Rev. Microbiol. 62:193–210 [DOI] [PubMed] [Google Scholar]
- 51. Wang B, Kuramitsu HK. 2003. Control of enzyme IIscr and sucrose-6-phosphate hydrolase activities in Streptococcus mutans by transcriptional repressor ScrR binding to the cis-active determinants of the scr regulon. J. Bacteriol. 185:5791–5799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang X, Wood TK. 2011. Toxin-antitoxin systems influence biofilm and persister cell formation and the general stress response. Appl. Environ. Microbiol. 77:5577–5583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wiuff C, et al. 2005. Phenotypic tolerance: antibiotic enrichment on noninherited resistance in bacterial populations. Antimicrob. Agents Chemother. 49:1483–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yamaguchi Y, Park JH, Inouye M. 2011. Toxin-antitoxin systems in bacteria and archae. Annu. Rev. Genet. 45:61–79 [DOI] [PubMed] [Google Scholar]
- 55. Zaura E, Keijser BJF, Huse SM, Crielaard W. 2009. Defining the healthy “core microbiome” of oral microbial communities. BMC Microbiol. 9:259. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.