Abstract
Identification of Treponema pallidum rare outer membrane proteins (OMPs) has been a longstanding objective of syphilis researchers. We recently developed a consensus computational framework that employs a battery of cellular localization and topological prediction tools to generate ranked clusters of candidate rare OMPs (D. L. Cox et al., Infect. Immun. 78:5178–5194, 2010). TP0117/TP0131 (TprC/D), a member of the T. pallidum repeat (Tpr) family, was a highly ranked candidate. Circular dichroism, heat modifiability by SDS-PAGE, Triton X-114 phase partitioning, and liposome incorporation confirmed that full-length, recombinant TprC (TprCFl) forms a β-barrel capable of integrating into lipid bilayers. Moreover, TprCFl increased efflux of terbium-dipicolinic acid complex from large unilamellar vesicles and migrated as a trimer by blue-native PAGE. We found that in T. pallidum, TprC is heat modifiable, trimeric, expressed in low abundance, and, based on proteinase K accessibility and opsonophagocytosis assays, surface exposed. From these collective data, we conclude that TprC is a bona fide rare OMP as well as a functional ortholog of Escherichia coli OmpF. We also discovered that TprC has a bipartite architecture consisting of a soluble N-terminal portion (TprCN), presumably periplasmic and bound directly or indirectly to peptidoglycan, and a C-terminal β-barrel (TprCC). Syphilitic rabbits generate antibodies exclusively against TprCC, while secondary syphilis patients fail to mount a detectable antibody response against either domain. The syphilis spirochete appears to have resolved a fundamental dilemma arising from its extracellular lifestyle, namely, how to enhance OM permeability without increasing its vulnerability to the antibody-mediated defenses of its natural human host.
INTRODUCTION
Syphilis is a multistage, sexually transmitted illness renowned for its protean clinical manifestations and protracted natural history, both of which reflect the extraordinary invasiveness and immunoevasiveness of its etiologic agent, Treponema pallidum subsp. pallidum (34, 49). Although T. pallidum is often analogized to Gram-negative bacteria because it has both an inner and an outer membrane (IM and OM, respectively), the structure, composition, and physical properties of its cell envelope differ considerably from those of Gram-negative bacteria. For example, the OM of T. pallidum is devoid of lipopolysaccharide, contains phosphatidylcholine and phosphatidylserine as its major phospholipids, and is relatively permeable to lipophilic molecules (12, 22, 53). In Escherichia coli, lipoproteins are associated predominantly with the inner leaflet of the OM (60), while, with one exception, TP0453 (25), all lipoproteins localized to date in T. pallidum have been found to be tethered to the IM (7, 11, 32). In T. pallidum, the peptidoglycan (PG) resides approximately midway within the periplasmic space (30, 32), whereas in Gram-negative bacteria the murein layer is bound to the underside of the OM via covalent and noncovalent interactions with OM proteins (OMPs) and OM-associated lipoproteins (60). Most notably, the density of OMPs in T. pallidum is markedly lower than that in E. coli, approximately 1% based on freeze fracture electron microscopy (52, 64); the paucity of OM-spanning proteins and lack of surface-exposed lipoproteins are believed to be the ultrastructural basis for the spirochete's stealth pathogenicity (7, 48).
Rare OMPs presumably fulfill virulence-related and physiological functions in T. pallidum comparable to those of their much better characterized, nonorthologous Gram-negative counterparts (7, 16, 43, 60). Moreover, as with Gram-negative bacteria (58, 61), surface-exposed OMPs of T. pallidum represent potential targets for protective host defenses (15, 29, 50). Indeed, both rabbit and human syphilitic sera contain antibodies that label intact organisms by immunofluorescence (11) and promote opsonophagocytosis of spirochetes by monocytes/macrophages (13, 35, 36). These results suggest that at least some OMPs generate antibody responses that contribute to reduction of spirochete burdens and containment of disease. Over the years, a variety of factors have hindered efforts by investigators to identify T. pallidum OM-spanning proteins. Chief among these are their low abundance, their lack of sequence relatedness to known OMPs of Gram-negative organisms, and the fragility of the T. pallidum OM, all compounded by the spirochete's recalcitrance to in vitro cultivation and genetic manipulation (7, 22, 29, 31, 48).
Almost all known OM-spanning proteins adopt a β-barrel conformation (24, 56, 67). Three theoretical considerations derived from the vast literature on the topology and export of integral membrane proteins in bacteria support the assumption that T. pallidum rare OMPs likewise form β-barrels (11, 15, 48): (i) α-helical transmembrane domains act as stop-transfer sequences during translocation of polypeptides across the IM by the Sec export machinery (47); (ii) the presence of multiple short amphipathic segments circumvents the constraints to Sec-dependent export of integral membrane proteins across the IM (24, 60, 63); and (iii) even with the spirochete's remarkably low rate of multiplication (38) and comparatively permeable OM lipid bilayer (7, 12), fixed porin-like channels are needed to mediate nutrient uptake from the host milieu (43). Our recent demonstration that the spirochete contains an ortholog for BamA, TP0326, the central component of the molecular machine that chaperones newly exported precursors into the OM (24, 60, 63), constitutes prima facie evidence for the existence of unidentified β-barrels (19). Because the T. pallidum genome does not encode orthologs for known OMPs of proteobacteria other than TP0326/BamA (22), in silico identification of β-barrels rests upon structural predictions. Whereas the α-helical transmembrane domains of IM proteins are readily identifiable, such is not the case for the membrane-spanning elements (i.e., amphipathic β-strands) of OMPs (41, 56, 63).
We recently devised a consensus computational framework that employs a battery of cellular localization and topological prediction tools to generate ranked clusters of candidate rare OMPs, collectively referred to as the putative T. pallidum OMPeome (11). Among the most highly ranked candidates were the identical proteins TP0117/TP0131 (TprC/D, here referred to as TprC), members of the T. pallidum repeat (Tpr) family long suspected of harboring rare OMPs (7, 8, 22, 31, 33). Here we demonstrate that TprC does indeed meet criteria for designation as a bona fide rare OMP (48), namely, β-barrel structure, amphiphilicity, low abundance, and surface exposure. Moreover, similar to E. coli OmpF, an archetypal β-barrel (43, 60, 65), TprC is trimeric and forms channels in artificial membranes. We also found that TprC has a bipartite architecture consisting of a soluble N-terminal portion (TprCN), presumably periplasmic and bound directly or indirectly to peptidoglycan, and a C-terminal β-barrel (TprCC) solely responsible for membrane association and pore formation. Interestingly, T. pallidum-infected rabbits generate antibodies exclusively against TprCC, predicted to contain the surface-exposed determinants, while humans with secondary syphilis fail to mount a detectable antibody response against either domain. The syphilis spirochete appears to have resolved a fundamental dilemma arising from its extracellular lifestyle, namely, how to enhance OM permeability without increasing its vulnerability to the antibody-mediated defenses of its natural host.
MATERIALS AND METHODS
Propagation and harvesting of Treponema pallidum.
Animal protocols described in this work strictly follow the recommendations of the Guide for Care and Use of Laboratory Animals of the National Institutes of Health and were approved by the University of Connecticut Health Center Animal Care Committee under the auspices of Animal Welfare Assurance A347-01. The Nichols strain of T. pallidum subsp. pallidum was propagated by intratesticular inoculation of adult male New Zealand White rabbits with 1 × 108 treponemes per testis and harvested approximately 10 days later (25). To extract the treponemes, the testes were surgically removed, minced, and subsequently incubated on a rotator for 45 to 60 min in a 50-ml conical tube at room temperature (RT) in 5 ml of CMRL medium (Invitrogen, Carlsbad, CA) supplemented with 100 μl of protease inhibitor cocktail (PIC) (Sigma-Aldrich, St. Louis, MO). For the OMP surface accessibility and motility assays (see below), PIC was not added to the extraction medium. To remove testicular debris, the treponemal suspension was transferred to a sterile tube and centrifuged for 10 min at 500 × g. Treponemes were enumerated by dark-field microscopy using a Petroff-Hausser counting chamber (Hausser Scientific Company, Horsham, PA).
Bioinformatics analysis.
Multiple sequence alignments were performed in MacVector (Cary, NC; v.11.1.0) using sequences in the NCBI database and our in-house-derived sequence for Cali-77 TprC (see below). The conserved domains within TprC were identified using the NCBI conserved domain database (CDD) server (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi).
Cloning of tprC constructs.
DNAs encoding full-length TprC (TprCFl) and a portion of the TprC/D protein predicted by sequence alignment of the TprC/D/F/I family to be TprC/D specific (TprCSp; see Fig. S1A in the supplemental material) were PCR amplified from T. pallidum DNA using the primers listed in Table 1. The resulting amplicons were cloned into the BamHI and HindIII restriction sites of expression vector pET23b in frame with the C-terminal polyhistidine tag (Novagen, San Diego, CA). DNAs encoding the N-terminal and C-terminal portions of TprC (TprCN and TprCC, respectively) were amplified from the pET23b plasmid harboring tprCFl using primers listed in Table 1 and cloned into the EcoRI and HindIII restriction sites of pET23b in frame with the C-terminal polyhistidine tag (Novagen, San Diego, CA). All constructs were confirmed by nucleotide sequencing.
Table 1.
Primers used in this study
| Primer designation | Sequence (5′-3′)a | T. pallidum Nichols genome coordinates |
|---|---|---|
| TprCFL (5′) | TTTTGGATCCCATGGGCGTACTCACTCCG | 134982-134967 |
| TprCFL (3′) | GCCCAAGCTTCCATGTCACTTTCATTCCGCA | 136673-136697 |
| TprCSp (5′) | ACCGGATCCCATGAGAGCTATCCTCAAAGCA | 135789-135771 |
| TprCSp (3′) | GCGCTAAGCTTGACAACTCTTGGATCGGA | 136044-136062 |
| TprCNT (5′) | GATAGAATTCTATGGGGGTGTGGGCACAGCTGCAG | 135167-135147 |
| TprCNT (3′) | GATAAAGCTTACTCTGGTGTTGGTTACCGGCGTCGA | 135744-134770 |
| TprCCT (5′) | GATAGAATTCCATGCTCAACATAGACGCGCTCCTGC | 136114-136092 |
| TprCCT (3′) | GATCAAGCTTCCATGTCACTTTCATTCCGCAGA | 136671-136694 |
| TprC (5′) (Cali-77 amplification) | CAAGAGAAGGCGAGTGTGTAGGTGCTCATG | 134830-134801 |
| TprC (3′) (Cali-77 amplification) | GAAGAGGCAGCCCTCATCCGAGACAAA | 136769-136797 |
Underlined sequences indicate restriction sites.
Expression, purification, and folding of recombinant proteins.
326P1–4+, which contains polypeptide transport-associated (POTRA) domains 1 to 4 plus 14 residues from POTRA5 of TP0326, was expressed and purified as previously described (11). Expression, purification, and folding of the TP0326 β-barrel, 326βb (19); E. coli OmpG (19); and OmpF (37) were described previously. TprCFl, TprCN, and TprCC were expressed in the BL21(DE3) Rosetta-gami strain (Agilent Technologies, Inc., Santa Clara, CA). For batch purification, 1 liter of LB was inoculated with 50 ml of overnight culture grown at 37°C; IPTG (isopropyl-β-d-thiogalactopyranoside; final concentration, 0.1 mM) was added when the culture reached an optical density (600 nm) between 0.2 and 0.3. Cells were grown for an additional 3 h and then harvested by centrifugation at 6,000 × g for 15 min at 4°C. The pellets were resuspended with 20 ml of 50 mM Tris (pH 7.5), 100 μg of lysozyme, and 100 μl of protease inhibitor cocktail (PIC) and stored at −20°C. After thawing, the bacterial suspension was lysed by sonication for three 30-s pulses interspersed with 30 s of rest on ice. The pellet was recovered by centrifugation at 20,000 × g for 30 min at 4°C and then incubated in solubilization buffer (100 mM NaH2PO4 [pH 8.0], 10 mM Tris, 8 M urea) for 30 min at 4°C; the remaining insoluble material was removed by centrifugation at 20,000 × g for 30 min at 4°C. The supernatant was added to nickel-nitrilotriacetic acid (Ni-NTA) agarose matrix (Qiagen) that had been equilibrated in solubilization buffer and incubated with shaking at RT for 30 min. The matrix was washed with wash buffer (100 mM NaH2PO4 [pH 6.3], 10 mM Tris, 8 M urea) and subsequently eluted with elution buffer (100 mM NaH2PO4 [pH 4.5], 10 mM Tris, 8 M urea). SDS-PAGE and immunoblot analysis using a mouse monoclonal antibody directed against polyhistidine tag (Sigma-Aldrich) were employed to identify the protein during purification and assess its purity. Purified TprCFl and TprCC were incubated in folding buffer (1% n-dodecyl β-d-maltoside [DDM], 100 mM NaCl, 50 mM Tris [pH 7.5]) for 24 h at 4°C to ensure complete folding of the protein. The samples were centrifuged at 20,000 × g for 30 min at 4°C to remove misfolded aggregates. Purified TprCN was diluted 10-fold and further dialyzed to perform protein renaturation in different concentrations (6, 4, and 2 mM) of urea. Finally, TprCN was dialyzed against 50 mM Tris (pH 7.5) and 50 mM NaCl buffer.
TprCSp was expressed in the BL21(DE3) Rosetta-gami strain (Agilent Technologies, Inc., Santa Clara, CA). For purification, the harvested cell pellet was resuspended with 20 ml of 50 mM Tris (pH 7.5), 300 mM NaCl, 10 mM imidazole 10% glycerol, 100 μg of lysozyme, and 100 μl of PIC and stored at −20°C. After thawing, the bacterial suspension was lysed by sonication for three 30-s pulses interspersed with 30 s of rest on ice. The supernatants were then cleared of cellular debris by centrifugation at 18,000 × g for 20 min at 4°C and applied to a superflow Ni-NTA (Qiagen, Valencia, CA) immobilized metal-affinity chromatography (IMAC) column, which had been equilibrated with 50 mM Tris (pH 7.5), 300 mM NaCl, 10% glycerol (buffer A). The protein was eluted with buffer A supplemented with 250 mM imidazole. Fractions containing the protein were concentrated using an Amicon-Ultra concentrator (Millipore, Billerica, MA) with a nominal molecular mass cutoff of 10 kDa and dialyzed into phosphate-buffered saline (PBS). Protein concentrations were determined by measuring A280 in 20 mM sodium phosphate (pH 6.5) and 6 M guanidine hydrochloride (20). The ProtParam tool provided by the ExPASy proteomics server (23) was used to calculate the molar extinction coefficients (M−1 cm−1) of proteins.
Tryptophan fluorescence.
Spectra were obtained using a Hitachi F-2500 fluorescence spectrophotometer with samples placed in a 5-mm-path-length quartz cell at 25°C. The excitation wavelength was 295 nm, and the bandwidth of the excitation monochromator was 2.5 nm. The folding and denaturing buffers for TprCFl were 50 mM NaCl, 10 mM Tris (pH 7.5), 0.5% DDM (DDM buffer) and 100 mM NaH2PO4, 8 M urea (urea buffer), respectively. Tryptophan emission spectra were recorded between 300 and 400 nm. Background spectra without TprCFl were subtracted to obtain the final emission curves.
CD spectroscopy.
Circular dichroism (CD) analyses were performed using a Jasco J-715 spectropolarimeter (Jasco, Easton, MD). Far-UV CD spectra were acquired at 20°C in a 1-mm-path-length cuvette, with a 1-nm bandwidth, an 8-s response time, and a scan rate of 20 nm/min. Spectra of each sample, representing the average of nine scans, were baseline corrected by subtracting the spectral attributes of the buffer. The DICHROWEB server was utilized to assess the secondary structure contents of the proteins from their spectra (66).
Heat modifiability.
Recombinant proteins solubilized in SDS sample buffer (SB) were subjected to SDS-PAGE with or without boiling (10 min) followed by staining with GelCode Blue. To generate native lysates from freshly harvested T. pallidum, samples were incubated overnight at 4°C with 50 mM Tris (pH 7.0), 0.5% DDM, and 5% PIC (native lysis buffer) for solubilization. Insoluble material was removed from the sample lysed with native lysis buffer by centrifugation at 20,000 × g for 20 min at 4°C. To examine the heat modifiability of endogenous TprC, lysates solubilized in SB were split in half; one aliquot was boiled for 10 min. Subsequently, the lysates were resolved by SDS-PAGE and the gel including the stack was transferred to nitrocellulose membranes (0.45-μm pore size; GE Healthcare) at 25 V for 25 min using a semidry apparatus (Bio-Rad). Membranes were blocked for 1 h with PBS, 5% nonfat dry milk, 5% fetal bovine serum, and 0.1% Tween 20 and probed overnight at 4°C with primary antibodies directed against the TprC at a dilution of 1:5,000. After washing with PBS and 0.05% Tween 20 (PBST), the membranes were incubated for 1 h at 4°C with a horseradish peroxidase (HRP)-conjugated goat anti-rat antibody (Southern Biotech, Birmingham, AL) at a dilutions of 1:30,000. Following washes with PBST, the immunoblots were developed using the SuperSignal West Pico chemiluminescent substrate (Thermo Fisher Scientific).
Triton X-114 phase partitioning.
Phase partitioning of TprC in T. pallidum was performed using (i) standard (5, 11) and (ii) modified protocols. (i) Approximately 4 × 109 freshly harvested treponemes were pelleted by centrifugation at 10,000 × g for 20 min at 4°C; resuspended in 250 μl of PBS, 2% Triton X-114 (TX-114), and 0.005% PIC; and then diluted with 990 μl of PBS. After incubation for 1 h at 4°C, the solubilized T. pallidum organisms were dialyzed overnight in PBS at 4°C. Insoluble material was recovered by centrifugation at 20,000 × g for 20 min at 4°C, washed 4 times with PBS, and stored at −80°C. The supernatants were phase separated, and the resulting detergent and aqueous fractions were reextracted five additional times. Samples then were precipitated with 10 volumes of acetone overnight at −80°C. Each fraction (aqueous, detergent, and insoluble) was resuspended with 50 μl of 1× Laemmli sample buffer (Bio-Rad, Hercules, CA) and subsequently boiled for SDS-PAGE and immunoblotting analysis using TprC antiserum. (ii) In the modified protocol, the pelleted T. pallidum organisms were incubated in 100 μl of 2% DDM for 1 h at 4°C prior to phase partitioning according to the standard protocol described above. For recombinant proteins, 10 μg of each sample was phase partitioned using the standard protocol.
BN-PAGE analysis of endogenous and recombinant TprCFl.
Freshly harvested treponemes were solubilized overnight at 4°C with 50 mM Tris (pH 7.0), 0.5 to 4.0% DDM, and 5% PIC. Prior to electrophoresis, lysates were cleared of detergent-insoluble material by centrifugation at 20,000 × g for 20 min at 4°C. Native lysates consisting of 5 × 108 to 1 × 109 spirochetes were resolved in a 4 to 12% Bis-Tris acrylamide gel (Bio-Rad) at 4°C using the blue-native polyacrylamide gel electrophoresis (BN-PAGE) method (68). The cathode buffer (50 mM Tricine [pH 7.0] and 15 mM Bis-Tris) contained 0.02% Coomassie brilliant blue G-250 (CBB-G250) for the first 1/3 of the run, after which the gel was run with fresh cathode buffer without CBB-G250. For the duration of the run, the anode buffer consisted of 50 mM Bis-Tris (pH 7.0). Resolved lysates were transferred to a nitrocellulose membrane in 50 mM Tricine (pH 7.0), followed by immunoblotting using TprCSp antiserum. Samples containing TprCFl were diluted to reduce the concentration of DDM to 0.5% and incubated on ice for 30 min prior to BN-PAGE. The resolved samples were transferred as described above to nitrocellulose followed by immunoblotting using TprCFl antiserum.
Electron microscopy.
The TX-114-insoluble material, washed extensively with PBS, was prepared for whole-mount electron microscopy by the single-droplet method (51) on Parlodion (Ted Pella, Inc., Tustin, CA) and carbon-coated 400-mesh copper grids (Ted Pella). Specimens were negatively stained with 1% uranyl acetate (Sigma) and examined at 60 kV of accelerating voltage on a Hitachi H-765 electron microscope.
Preparation of liposomes.
All phospholipids were purchased from Avanti Polar Lipids (Alabaster, AL). Large unilamellar vesicles (LUVs) were generated using an Avanti Mini-Extruder. A mixture of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 1-palmitoyl-2-oleoyl-sn-glycero-3-[phospho-l-serine] (sodium salt), 1-palmitoyl-2-oleoyl-sn-glycero-3-[phospho-(racemic)-glycerol] (sodium salt), and 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (69.3:17:13:0.7 mol%, respectively) in chloroform was dried at RT under argon and kept in vacuum for at least 4 h. This ratio simulates the phospholipid composition of the T. pallidum outer membrane (53). To hydrate the lipid mixture, 0.5 ml of 50 mM HEPES (pH 7.5), 100 mM NaCl (liposome buffer) was added to the dried phospholipids, and the sample was incubated for 30 min at 37°C. The lipids were resuspended by vortexing and then passed 21 times at 23°C through the extruder equipped with a 100-nm-pore-size polycarbonate filter. The filter dictates the final size of the vesicles (mean diameter, ∼110 nm) and reduces the chances of contamination with larger particles or foreign material. The resulting liposomes were stored at 4°C for up to 2 weeks. Liposomes containing terbium-dipicolinic acid complex [Tb(DPA)33−] were prepared as described above except that liposome buffer, including 3 mM terbium(III) chloride (TbCl3) and 9 mM 2,6-pyridinedicarboxylic acid (Sigma) neutralized to pH 7, was added to the lipid film. The suspended phospholipid mixtures were frozen in liquid N2 and thawed at 37°C a total of 6 times to reduce the number of multilamellar liposomes and to enhance the trapped volumes of the vesicles. Loaded liposomes were separated from nonencapsulated Tb(DPA)33− by gel filtration using Superdex TM 75 (GE Healthcare) in 50 mM HEPES (pH 7.5), 100 mM NaCl.
Liposome flotation assay.
TprCFl, 326P1-4+, and 326βb (300 nM final concentration) were incubated with 100 μg of LUVs for 1 h at 22°C in 50 mM Tris (pH 7.5), 100 mM NaCl buffer. Incorporation was terminated by adding sucrose to a final concentration of 50%, and the mixtures were transferred to an ultracentrifuge tube. Discontinuous sucrose gradients were made by gently adding layers of 40 and 6% sucrose to the ultracentrifuge tubes. After centrifugation at 90,000 × g for 1 h at 4°C, the liposome (top layer)- and non-liposome (middle and bottom layer)-containing fractions were carefully removed.
Pore formation assay.
Pore formation assays were performed using LUVs loaded with the fluorophore Tb(DPA)33− as previously described (28, 37). For endpoint measurements, LUVs loaded with Tb(DPA)33− were diluted to a final lipid concentration of 100 μM using 50 mM Tris (pH 7.5), 100 mM NaCl supplemented with 5 mM EDTA. The net initial emission intensity (F0) was determined after equilibration of the sample at 25°C for 5 min. Aliquots of 326P1-4+, TprCN, TprCFl, TprCC, and 326βb proteins were added to the liposome suspension at 100 nM final concentrations, and samples were incubated for 30 min at 37°C. After reequilibration to 25°C, the final net emission intensity (Ff) of the sample was determined (i.e., after blank subtraction and dilution correction) and the fraction of Tb(DPA)33− quenched was estimated using Ff/F0.
Immunologic reagents.
Rat polyclonal antisera directed against 326P1-4+ (11), TroA (TP0163) (1), TP0453 (25), and Tp47 (TP0574) (26) were described previously. Rat antisera directed against TprCFl and TprCSp were generated in 6-week-old Sprague-Dawley rats by intraperitoneal injection with 30 μg of purified protein in a 1:1 mixture of phosphate-buffered saline (PBS) (pH 7.4) and complete Freund's adjuvant; 1 month and 6 weeks later, the animals received 15-μg booster doses in a 1:1 mixture of PBS and incomplete Freund's adjuvant. Rabbit antiserum directed against TprCFl was generated by immunizing adult male New Zealand White rabbits subcutaneously with 100 μg of protein in a 1:1 mixture of PBS and TiterMax Gold; 1 month and 6 weeks later, the animals were boosted with 50 μg of protein in TiterMax Gold. The mouse monoclonal antibody (hybridoma clone HIS-1) specific for polyhistidine tags was purchased from Sigma-Aldrich. Immune rabbit serum (IRS) was described previously (11, 26); normal rabbit serum (NRS) was obtained from healthy, uninfected animals. Normal human serum (NHS) was obtained from a healthy volunteer without a history of syphilis and confirmed to be nonreactive by rapid plasma reagent testing. Sera from HIV-seronegative persons with secondary syphilis were obtained from individuals enrolled at a study site located in Cali, Colombia (14). Serum specimens were obtained following informed consent under protocols approved by the human subject boards at the Connecticut Children's Medical Center, the University of Connecticut Health Center (UCHC), and the Centro Internacional de Entrenamiento e Investigaciones Médicas (CIDEIM).
Quantitative immunoblot analysis.
The number of copies of TprC per T. pallidum cell was determined by quantitative immunoblotting as previously described (19). Lysates from 5 × 108 treponemes and graded amounts of recombinant TprCFl were electrophoresed on 10% SDS-polyacrylamide gels and immunoblotted with TprCSp antibodies. A standard curve, generated by densitometric analysis of the scanned immunoblots using ImageJ (NIH, v. 1.44c), was used to calculate the total amount of endogenous TprC in the resolved treponemal lysates. The amount of TprC per T. pallidum cell was determined by dividing the corresponding densitometrically derived value in the lysates by numbers of T. pallidum organisms. Copy numbers were determined by the molecular mass (MMTprC = 62,881 Da without signal sequence).
PCR amplification of tprC from a skin biopsy specimen obtained from a patient with secondary syphilis.
Following informed consent, a 4-mm punch biopsy specimen was obtained from affected skin of a patient with untreated secondary syphilis (designated Cali-77) seen at our Cali, Colombia, study site (14). DNA was extracted using the QIAamp DNA minikit (Qiagen Inc., Valencia, CA) according to procedures recommended by the manufacturer, eluted from the Qiagen columns in 100 μl of elution buffer at 70°C, and stored at −80°C. The concentration of DNA was determined spectrophotometrically by absorbance at 260/280 nm. The tprC gene (tp0117) plus flanking DNA was amplified using the primers listed in Table 1, cloned into the TOPO cloning vector (Invitrogen), and sequenced.
Accessibility of endogenous TprC to surface proteolysis.
The accessibility of TprC to proteolysis in intact treponemes was assessed as recently described for TP0326 (19). All steps were performed at RT unless stated otherwise. To determine the lowest concentration of proteinase K (PK) (Invitrogen) required to achieve surface proteolysis of TprC, freshly harvested organisms (5 × 108/ml in CMRL without PIC) were subjected to digestion with graded concentrations (0.1 to 10 μg) of PK for 1 h. For proteolysis of periplasmic controls, spirochetes were harvested by centrifugation at 10,000 × g for 20 min, resuspended with 200 μl of PK lysis buffer (50 mM Tris [pH 7.0], 0.5% Triton X-100, 0.1% β-mercaptoethanol, and 50 μg of lysozyme [Sigma-Aldrich]), incubated for 1 h, and then treated with 10 μg/ml of PK for 1 h. The activity of the protease was stopped by the addition of phenylmethylsulfonyl fluoride (PMSF) to 1 mg/ml. Intact treponemes were pelleted by centrifugation at 20,000 × g for 20 min and subsequently resuspended in SB. Immunoblotting was performed as described above. To assess motility, aliquots (10 μl) of each sample immediately following PK treatment (1 h of incubation) were transferred onto a glass microscope slide, gently overlaid with a cover glass, and then viewed by dark-field microscopy on an Olympus BX41 microscope (Center Valley, PA) using a 100× (1.4-numerical-aperture [NA]) oil immersion objective. The motility of the organisms was observed visually and recorded using a Retiga Exi charge-coupled device (CCD) camera (QImaging, Surrey, BC, Canada) and StreamPix (NorPix, Montreal, QC, Canada) software. ImageJ was used to adjust the brightness and contrast of the images. Images were converted into movies using QuickTime Pro (Apple Inc., Cupertino, CA; v.7.0).
Opsonophagocytosis assays.
Opsonophagocytosis assays were performed as described previously (8, 26, 35, 59). Rabbit peritoneal macrophages were elicited by intraperitoneal injection of 10 ml of 15% sterile Proteose Peptone no. 3 (Sigma-Aldrich). Cells were harvested 3 to 5 days later by peritoneal lavage with PBS containing 10 U of heparin (Sigma-Aldrich) per ml, centrifuged at 900 × g for 10 min, and resuspended in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (Mediatech), 100 U of penicillin/ml, and 100 μg of streptomycin/ml. The macrophages then were counted with a hemacytometer and plated in poly-d-lysine-treated culture slides (BioCoat; BD Biosciences) at a density of 5 × 105 cells/ml. After incubation for 2 h at 37°C, nonadherent cells were removed by washing the monolayers twice with DMEM. The cells were maintained overnight at 37°C in a reduced-oxygen atmosphere (3% O2 and 5% CO2). The following day, the adherent macrophages were washed twice with gassed DMEM. The cells then were incubated for 4 h at 37°C in a reduced-oxygen atmosphere (3% O2 and 5% CO2) with T. pallidum (10 organisms per macrophage) in the presence of CMRL medium containing 10% FBS plus 10% heat-inactivated NRS, IRS, rabbit anti-TprC, or rabbit anti-Tp47. After incubation, supernatants were removed to count the remaining T. pallidum organisms. Internalization of treponemes by macrophages was determined by indirect immunofluorescence assay (IFA). Slides were fixed with 4% (vol/vol) paraformaldehyde for 10 min at room temperature (RT), blocked with CMRL with 10% FCS for 30 min at RT, and incubated with rat anti-FlaA antiserum (1:75) for 1 h at RT. The slides then were washed with PBS-Tween and incubated with goat anti-rat–Alexa Fluor 488 (Invitrogen) for 30 min at RT. The slides were given one final wash and mounted in Vectashield antifade reagent (Invitrogen) containing 4′,6-diamidino-2-phenylindole (DAPI). Fluorescent images were acquired on an epifluorescence Olympus BX-41 microscope using a 40× (1.4-NA) oil immersion objective equipped with a Retiga Exi CCD camera (Q Imaging, Tucson, AZ) and the following Omega filter sets: DAPI, fluorescein isothiocyanate (FITC), and rhodamine. The percentages of rabbit macrophages containing degraded fluorescent spirochetes were systematically quantified for each of the conditions studied.
Nucleotide sequence accession number.
The tprC gene plus flanking DNA was sequenced and given the accession number JQ418492.
RESULTS
Recombinant TprC forms an amphiphilic β-barrel.
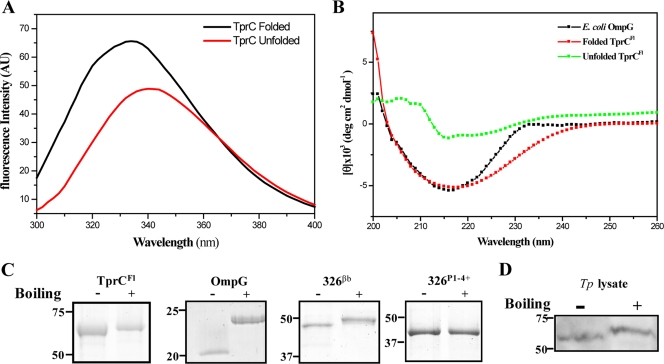
Experiments were conducted with recombinant protein to test the prediction that TprC possesses physical properties consistent with an amphiphilic β-barrel. Upon folding, the fluorescence emission maximum of tryptophan-containing integral membrane proteins blueshifts as the tryptophan residues move from an aqueous to a hydrophobic environment (27). We began, therefore, by using this method to monitor folding of full-length recombinant TprCFl, which contains 12 tryptophan residues. Figure 1A shows that unfolded TprCFl has an emission maximum at 342 nm compared to the maximum of 334 nm for the folded protein in a detergent buffer; the folded protein also displayed increased emission intensity. We next employed circular dichroism (CD) spectroscopy to assess its β-sheet content (Fig. 1B). In contrast to the unfolded protein, folded TprCFl displayed broad minima centering on 218 nm, indicating a predominance of β-structure, as did the E. coli OmpG control. β-Barrel-forming proteins characteristically retain a high degree of β-sheet content when solubilized in SDS at RT and, consequently, run with lower apparent molecular masses by SDS-PAGE than when denatured by boiling; this property is termed heat modifiability (2, 9). Along with OmpG and the β-barrel domain of TP0326 (326βb), folded TprCFl displayed heat modifiability, whereas boiling did not affect the SDS-PAGE mobility of 326P1-4+, which contains the periplasmic polypeptide transport-associated (POTRA) domains 1 to 4 plus 14 residues from POTRA5 (Fig. 1C) (19).
Fig 1.
TprC forms a β-barrel. (A) Tryptophan fluorescence emission spectra of unfolded and folded TprCFl in urea and DDM buffer, respectively. (B) CD spectra of unfolded and folded TprCFl (5 μM) in DDM buffer and OmpG (5 μM) in 50 mM NaCl, 10 mM Tris (pH 7.5), 0.2% n-octyl-β-d-glucopyranoside. (C) Heat modifiability of TprCFl, E. coli OmpG, 326βb, and 326P1-4+. Proteins were stained with GelCode Blue following SDS-PAGE with (+) or without (−) boiling in final sample buffer (SB). Molecular mass standards (kDa) for SDS-PAGE and immunoblot analyses are on the left of each panel. (D) T. pallidum samples (2 × 108 organisms) were dissolved in SB with (+) or without (−) boiling and immunoblotted using rat anti-TprCSp antiserum.
We next used TX-114 phase partitioning to determine whether TprCFl possesses the amphiphilic character expected of an OM-spanning β-barrel. As shown in Fig. 2A, TprCFl partitioned exclusively into the detergent-enriched phase, as did 326βb, but not 326P1-4+. To extend these findings, we examined the ability of TprCFl to incorporate into liposomes that simulate the phospholipid composition of the T. pallidum OM (53). Following separation on discontinuous sucrose gradients, TprC and 326βb were recovered from the liposome-containing top fractions, whereas 326P1-4+ was found only in the middle and bottom fractions containing unincorporated material (Fig. 2B).
Fig 2.
TprC is amphiphilic and trimeric and displays pore-forming activity. (A) Ten micrograms each of TprCFl, 326P1-4+, and folded 326βb was phase partitioned in TX-114 and stained with GelCode Blue following SDS-PAGE. Lanes: aqueous (Aq) and detergent-enriched (Det) phases. (B) Liposomes were reconstituted with 10 μg each of TprCFl, 326P1-4+, or 326βb followed by sucrose density gradient ultracentrifugation. Fractions were subjected to immunoblotting with antisera directed against TprCFl, 326P1-4+, or 326βb following SDS-PAGE. Lanes: top fractions (TF) contain liposome-incorporated material whereas middle and bottom fractions (MF and BF, respectively) contain unincorporated material. (C) BN-PAGE and immunoblot analysis of TprCFl and T. pallidum lysate solubilized in 0.5% DDM with 50 mM Tris (pH 7.0). Molecular mass standards (kDa) are shown on the left. (D) Quenching of LUVs encapsulating Tb(DPA)33− after incubation with 100 nM TprCFl, E. coli OmpF, 326βb, or 326P1-4+ in 50 mM Tris (pH 7.5), 100 mM NaCl supplemented with 5 mM EDTA. Each bar represents the mean ± standard error of the mean for three independent experiments.
TprC is trimeric and forms channels in large unilamellar vesicles (LUVs).
OMPs of Gram-negative bacteria often exist as multimers (40). We used BN-PAGE to examine the oligomeric state of TprCFl. As shown in Fig. 2C, TprCFl migrated exclusively as a trimer under nondenaturing conditions. The observations that (i) E. coli OmpF, an archetypal porin, is trimeric (43, 60, 65) and (ii) the major sheath protein (Msp) of Treponema denticola, the presumptive evolutionary ortholog of the Tpr family (8, 22), forms large pores in black lipid membranes (21) prompted us to ask if TprCFl can form channels in artificial membranes. Figure 2D shows that this was indeed the case. TprCFl promoted greater efflux of the small (10-Å, 650-Da) fluorophore Tb(DPA)33− from LUVs than did OmpF. Pore formation by BamA orthologs has been studied extensively and found to reside exclusively in the β-barrel portion of the molecule (4, 55, 62). To validate the results for TprCFl, we also assessed pore formation by 326βb and 326P1-4+. As anticipated, 326βb promoted efflux of fluorophore, though to a lesser extent than either OmpF or TprCFl, while 326P1-4+ was devoid of activity (Fig. 2D).
TprC expressed by T. pallidum is heat modifiable, trimeric, and amphiphilic but tethered to the peptidoglycan sacculus.
T. pallidum Nichols encodes two Tprs that have an extremely high degree of sequence relatedness to TprC: the truncated TprF and the full-length TprI (MMs of 39 and 66 kDa, respectively, with signal sequences; see alignment in Fig. S1A in the supplemental material). In order to assess the properties of TprC in T. pallidum, it was first necessary to generate an antiserum specific for the protein. We accomplished this by cloning a fragment of the tprC gene, designated tprCSp, encoding residues 290 to 381, a stretch of the polypeptide distal to the C terminus of TprF and highly divergent in sequence from TprI (see Fig. S1A). In contrast to the antiserum raised against TprCFl, which reacts with several polypeptides in T. pallidum lysates by standard immunoblotting, including one with the predicted molecular mass of mature TprC (64 kDa), the antiserum produced by animals immunized with TprCSp recognized only the 64-kDa protein (see Fig. S1B), which we now designate as endogenous TprC. Additional immunoblot experiments revealed that the antiserum against TprCSp was exquisitely sensitive, capable of detecting less than 1 ng of TprCFl (data not shown). Endogenous TprC displayed heat modifiability (Fig. 1D) and migrated as a trimer in DDM-solubilized lysates examined by BN-PAGE (Fig. 2C). TX-114 phase partitioning, however, yielded a surprising result. When treponemes were subjected to our standard phase-partitioning protocol, virtually all of the TprC was recovered with the TX-114-insoluble material (Fig. 3A, top panel). As previously reported (51), by transmission electron microscopy, this material consists of peptidoglycan-containing cell ghosts (sacculi) with attached flagellar filaments (Fig. 3B). Immunoblot analysis of the sacculi confirmed that this insoluble material contains TprC and, based on immunoreactivity with rabbit syphilitic serum, retained lipoproteins (Fig. 3C). In contrast, when phase partitioning was performed on DDM-solubilized lysates (see Materials and Methods), TprC partitioned exclusively in the detergent-enriched phase (Fig. 3A, bottom panel). The simplest interpretation of these collective results is that TprC expressed by T. pallidum is a trimeric OM β-barrel but also is linked directly or indirectly to the PG by noncovalent bonds which can be disrupted by DDM but not by TX-114.
Fig 3.
TprC expressed by T. pallidum associates with the peptidoglycan sacculus. (A) TX-114 phase partitioning of T. pallidum lysates (1 × 109 organisms) without (−) or with (+) preincubation with 2% DDM. Lanes: whole cells (WC), TX-114-insoluble material (Ins), aqueous phase (Aq), and detergent-enriched phase (Det). (B) Extensively washed TX-114-insoluble material visualized in negatively stained whole mounts by transmission electron microscopy. (C) Immunoblot analysis of TX-114-insoluble material with anti-TprCSp or immune rabbit serum (IRS). Molecular mass standards (in kilodaltons) are shown at left.
TprC is expressed in T. pallidum at low abundance and is surface exposed.
To further our characterization of TprC as a potential rare OM protein, we next examined its expression in T. pallidum. In our recent study of TP0326/BamA (19), we determined the mean copy number of tprC transcript to be 212 ± 35 per 10,000 copies of flaA. Although more than 3-fold greater than levels of tp0326 transcript (61 ± 16), this value is markedly lower than the more than 2,500 flaA-normalized copies of tp0547 message, which encodes the abundant lipoprotein carboxypeptidase Tp47 (17, 19). Expression of TprC on a per-cell basis was here determined by quantitative immunoblot analysis (Fig. 4A). The mean copy number (163 ± 42), calculated from three independent experiments, agrees extremely well with both the quantitative reverse transcription-PCR (qRT-PCR) result and our previous determination that treponemes express approximately 15,000 copies of FlaA protein per cell (45).
Fig 4.
TprC is expressed at low abundance and is accessible to proteinase K in motile treponemes. (A) Quantitative immunoblot analysis of TprC expressed in T. pallidum. T. pallidum lysates in amounts indicated were immunoblotted with anti-TprCSp antiserum followed by densitometric analysis; a standard curve generated from densitometric values obtained for graded amounts of TprCFl was used to determine the copy number of TprC per T. pallidum cell. Molecular mass standards (kDa) are indicated on the left. (B) Immunoblot analysis of TprC, detected by the anti-TprCSp antiserum, in motile treponemes (1.0 × 108 T. pallidum organisms/lane) treated for 1 h with graded concentrations of proteinase K (PK). (C) PK accessibility of TprC and two periplasmic controls in intact and detergent-lysozyme-treated organisms incubated with (+) or without (−) 10 μg/ml of PK. Each lane represents 1.0 × 108 T. pallidum organisms immunoblotted with antisera to TprCSp, TP0453, or TroA. (D) Live imaging of intact treponemes with and without incubation with 100 μg/ml of PK. Asterisks and arrowheads indicate flexing and wave form propagation, respectively. Time scale is 0 to 0.12 s.
In Gram-negative bacteria, OMPs often are noncovalently bound to peptidoglycan (60). Thus, the above results indicating that TprC appears to be tethered to the murein layer by no means excluded the possibility that the protein also has surface-exposed domains. In fact, surface exposure would be expected given the evidence that TprCFl contains a β-barrel. Consequently, we investigated the accessibility of TprC to PK treatment in freshly harvested, motile treponemes using immunoblot analysis with the TprCSp antiserum to monitor integrity of the protein. We first determined the lowest concentration of PK required for surface proteolysis so as to minimize inadvertent damage to the spirochetal OMs. As shown in Fig. 4B, with intact treponemes, TprC was partially degraded at 0.1 μg/ml of PK and fully degraded at 1 μg/ml. Figure 4C shows that a 10-fold excess of PK (10 μg/ml) degraded TprC but had no effect on the periplasmic controls TP0453 and TP0163 (TroA). TP0453 and TP0163 were degraded, however, in treponemes treated with Triton X-100 and lysozyme in addition to PK, thereby confirming that these two proteins are not intrinsically protease resistant. The lack of degradation of TP0453 in intact organisms is noteworthy because this polypeptide is tethered by its N-terminal lipids to the inner leaflet of the OM and is susceptible to proteolysis following minor perturbations of the bilayer (25). T. pallidum motility is exquisitely sensitive to disruption of the OM (25). As an additional means of confirming that the OMs of the treponemes remained intact during exposure to PK, we monitored their motility by dark-field microscopy and live imaging. Figure 4D shows that organisms incubated with 100 μg/ml of PK remained highly motile.
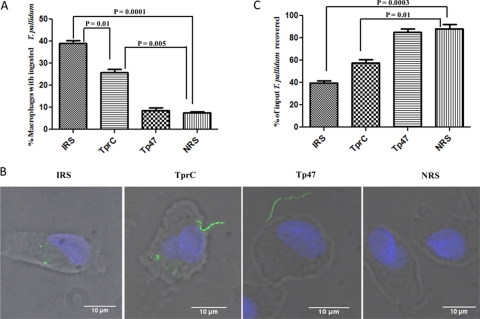
To complement the surface proteolysis results, we assessed the ability of rabbit antisera to TprCFl to promote internalization of live treponemes by rabbit peritoneal macrophages (8, 26, 35, 59). As shown in Fig. 5A and B, compared to NRS and antiserum directed against Tp47, TprCFl antiserum greatly increased uptake of organisms, although not to the same level as did IRS. As a qualitative measure of the degree of surface antibody binding by the population of treponemes, we enumerated organisms remaining in the supernatants following the 4-h incubation period with the macrophages. Even at the relatively low multiplicities of infection (MOIs) used (10:1), substantial percentages of spirochetes remained in the supernatants with each antiserum (Fig. 5C), although the percentages of organisms remaining with IRS and anti-TprCFl were significantly lower than those with NRS and Tp47. These results suggest that there are differences within treponemal populations with respect to levels of expression of surface-exposed opsonic targets.
Fig 5.
TprC antibodies have opsonic activity. (A) Percentages of rabbit peritoneal macrophages containing internalized treponemes. Results shown are means ± standard errors of the means for three independent experiments. (B) Representative micrographs of macrophages incubated with motile treponemes and the indicated sera. Each image is a composite of bright-field images, nuclei stained with DAPI, and FITC-labeled T. pallidum. (C) Percentages of treponemes recovered following incubation of rabbit peritoneal macrophages for 4 h with the indicated sera. Results shown are means ± standard errors of the means for three independent experiments. P values of <0.05 (Student's t test) were considered significant.
TprC has a bipartite architecture consisting of an N-terminal putative periplasmic domain and a C-terminal, pore-forming β-barrel.
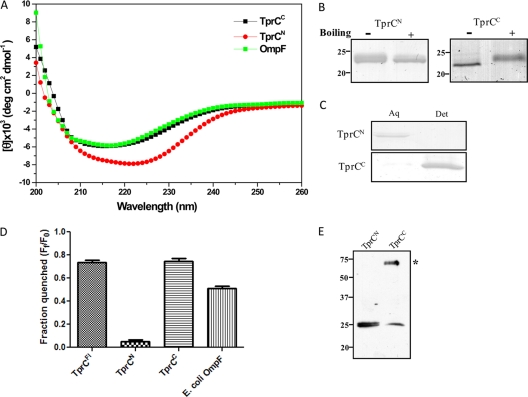
In T. pallidum, the PG is located approximately midway within the periplasmic space (30, 32), a distance of 10 to 20 nm from the inner leaflet of the OM (depending on the presence of flagellar filaments), rather than in proximity to the inner leaflet of the OM as in Gram-negative organisms (60). In order to be bound to the murein layer, an OMP would require a substantial periplasmic portion in addition to its OM-spanning β-barrel. In accord with this line of thinking, the NCBI's Conserved Domain Database server (39) predicted that TprC contains N- and C-terminal regions phylogenetically related to domains in Msp of T. denticola (see Fig. S1 in the supplemental material). Based upon the CDD analysis, we designed primer pairs (Table 1) to generate separate N- and C-terminal constructs, designated TprCN and TprCC, respectively, for biophysical characterization. The results, presented in Fig. 6, demonstrate that TprCN and TprCC have markedly disparate properties. Whereas TprCN is a mixture of α-helix and β-sheet (24.5% α-helix, 35.7% β-sheet, and 39.8% random coil) by CD spectroscopy, TprCC consists predominantly of β-sheet (5.2% α-helix, 57.5% β-sheet, and 37.3% random coil) (Fig. 6A). In contrast to TprCN, TprCC is heat modifiable (Fig. 6B) and partitions into the TX-114 detergent-enriched phase (Fig. 6C). Of particular importance, TprCC displayed channel-forming activity virtually identical to that of TprCFl as opposed to the near-negligible pore formation by TprCN (Fig. 6D). Lastly, unboiled TprCC forms an SDS-stable trimer, whereas TprCN is monomeric (Fig. 6E).
Fig 6.
TprCC contains the β-barrel of TprC and forms a trimer. (A to C) CD spectroscopy, heat modifiability, and TX-114 phase partitioning of TprCN and TprCC. Aq and Det, aqueous and detergent-enriched phases, respectively. (D) Quenching of Tb(DPA)33− encapsulated within LUVs following incubation with TprCFl, TprCN, TprCC, and E. coli OmpF. (E) SDS-PAGE of unboiled TprCN and TprCC (asterisk indicates TprCC trimer). Numbers at left in panels B and E are molecular masses in kilodaltons.
T. pallidum-infected rabbits, but not humans, develop an antibody response against TprC.
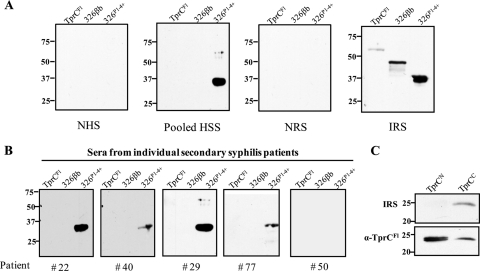
As an initial step toward elucidating the contribution of TprC antibodies to the opsonic activity of syphilitic sera, we sought to determine whether the protein induces an antibody response during syphilitic infection. The immunoblot analyses performed to address this question employed folded, unboiled TprC to enhance detection of antibodies against conformational determinants. As shown in Fig. 7A and B, antibodies directed against TprC were not detected either in pooled human secondary syphilitic sera or in the individual serum specimens used to form the pool, whereas rabbits mounted a detectable, albeit weak, response to the protein. Heterogeneity of surface-exposed epitopes could explain the lack of reactivity of human syphilitic sera with the Nichols strain-derived TprC used for the immunoblot analyses. This is unlikely, however, given the extremely high degree of sequence conservation of TprC sequences, including the β-barrels (see Fig. S2 in the supplemental material). Note that the Cali-77 sequence in the alignment was derived from a tprC gene that was PCR amplified from the skin of a patient seen at our Cali, Colombia, enrollment site (14); serum from this patient also was used for immunoblotting (Fig. 7B). Interestingly, antibody reactivities to 326P1–4+ and folded, unboiled 326βb differed strikingly from those for TprC. As recently reported (19), and also shown in Fig. 7A and B, humans often responded well to the POTRA domain but not to the β-barrel, while infected rabbits recognized both POTRA and β-barrel domains. Lastly, we wished to confirm that the opsonic rabbit sera used for the experiment shown in Fig. 5 contained antibodies directed against the β-barrel. This was found to be the case for both rabbit anti-TprCFl serum and IRS, although their reactivities differed considerably; the former reacted with both TprCN and TprCC while the latter recognized only TprCC (Fig. 7C).
Fig 7.
T. pallidum-infected rabbits, but not humans, mount an antibody response against TprC. (A) Reactivities of normal rabbit serum (NRS), immune rabbit serum (IRS; representative of 3 different animals), normal human serum (NHS), and pooled human secondary syphilitic sera (HSS) against folded, unboiled TprCFl; 326P1-4+; and folded, unboiled 326βb. (B) Immunoblot reactivities of individual patient sera. (C) Immunoblot reactivities of the anti-TprCFl antiserum and IRS used for the opsonophagocytosis assays in Fig. 5 against TprCN and folded, unboiled TprCC. Each immunoblot assay was performed using 100 ng of recombinant protein. Numbers at left are molecular masses in kilodaltons.
DISCUSSION
Certain general features of early human syphilis, such as its waxing and waning course and the tendency for disease progression to occur in the face of a robust immune response (34, 50), have long perplexed clinicians and syphilis researchers. In recent years, investigators have begun to devise ex vivo and translational strategies to relate these enigmatic disease manifestations to the syphilis spirochete's unusual cell envelope architecture (14, 42), most notably its paucity of surface antigenic targets (7, 10, 11). Critical to this effort is the need to molecularly characterize the bacterium's rare OMPs, entities which were originally observed by freeze fracture electron microscopy more than 20 years ago (52, 64) but only just beginning to be definitively identified more than a decade after the publication of the bacterium's genomic sequence (7, 22, 29). Based on theoretical considerations, we postulated that OMPs in T. pallidum would, like their nonorthologous Gram-negative counterparts, adopt a β-barrel conformation (11, 48). Our limited success in pursuing our quest for T. pallidum OMPs at the protein level prompted us to adopt an in silico-based approach in which we used a battery of computational tools to mine the T. pallidum genome for proteins predicted to form β-barrels; TprC, the focus of the present study, emerged from this analysis as one of our strongest candidates (11).
Strategies for characterizing OMP candidates must, by necessity, rely heavily upon the use of recombinant proteins but with an eye toward confirming key in vitro findings with the endogenous protein and establishing surface exposure in live treponemes. Along these lines, we used TprCFl to establish that this recombinant protein is rich in β-sheet content, is heat modifiable, forms trimers, and is capable of inserting into lipid bilayers whose phospholipid composition mimics that of the T. pallidum OM (53). While it is not possible to replicate all of these results with endogenous protein, we did find that the latter is heat modifiable, amphiphilic, and trimeric, results consistent with formation of β-barrel multimers in the spirochetal OM. Furthermore, we demonstrated surface exposure by two very different methods, protease accessibility and opsonophagocytosis, employing live organisms under conditions in which the integrity of the OM was maintained. The two approaches yielded an interesting counterpoint; one showed that all of the TprC in the population of treponemes is accessible to PK digestion, while the other revealed that not all treponemes in a population express sufficient TprC for opsonization (a result mirrored with IRS, which presumably contains antibodies against opsonic targets in addition to TprC). We also confirmed that sera with opsonic activity contained antibodies directed against the portion of TprC expected to harbor surface-exposed determinants. These results, coupled with our observation that tprC/TprC is expressed at exceedingly low copy numbers, leave little doubt as to its authenticity as a rare OMP.
In comparison with the extensively characterized porin E. coli OmpF, the recombinant protein also readily promoted efflux of the fluorophore Tb(DPA)33− from LUVs. Based on the overall similarity in biophysical characteristics of the recombinant and endogenous proteins, we propose that TprC does indeed function to enhance the permeability of the T. pallidum OM and, as such, is the first bona fide OM-spanning protein in the syphilis spirochete shown to do so. Importantly, a channel large enough to permit rapid efflux of Tb(DPA)33− (MM, 650 Da) should be large enough to allow passive influx of glucose, amino acids, and numerous other nutrients which T. pallidum must parasitize from the body fluids of its obligate human host. Additional support for this idea comes from Msp of T. denticola, reported to form extremely large ungated channels in black lipid membranes (21). Efforts to understand the mechanisms of spirochetal persistence have generally focused on the bacterium's ability to evade host defenses (7, 31, 49). In order to establish chronic infection, T. pallidum also must acquire essential nutrients from every microenvironment in which it takes up residence. Our findings provide a small physiological piece of the persistence puzzle which emerged from the discovery of the bacterium's unique, protein-scarce OM (48). We hypothesize that the copy number of TprC and the size of its channel have been calibrated during the reductive evolutionary process which gave rise to pathogenic treponemes (57) to meet the metabolic demands of this very slowly replicating organism (38) while maintaining the lowest possible immunologic profile.
With most Gram-negative OMPs, the entire polypeptide forms a cylinder consisting of an even number of β-strands framed by large external loops and small periplasmic turns (56, 67). The 22-stranded β-barrel predicted for TprC by TMBpro (54) conforms to this generalization (11). The data generated here using the full-length recombinant TprCFl gave no reason to question the assumption that the entire polypeptide forms the barrel. Two empirical findings, both obtained from T. pallidum, however, prompted the experiments which now lead to a substantially revised working model for TprC (Fig. 8), in addition to underscoring the uncertainties of in silico β-barrel structural predictions. One was the association of endogenous TprC with the TX-114-insoluble material, which consists of cell ghosts with remnant, unextracted proteins (51). The other came from cryo-electron tomographic analysis of T. pallidum, which localized the spirochete's thin PG layer to approximately midway within the periplasmic space (30, 32). The two findings together suggested that TprC contains a periplasmic domain that tethers the OM-inserted barrel to the PG sacculus. We confirmed this deduction by demonstrating that the β-barrel and channel-forming activity reside entirely within the C-terminal 202 residues; this portion of TprC also contains the predicted sequence for recognition of TprC precursors by the POTRA domain of BamA (19), a prerequisite for OM localization (3, 24, 55, 63) (see Fig. S1 in the supplemental material). We note that our data do not directly address the location of the stretch of polypeptide containing TprCSp, which we place in the periplasm (Fig. 8). Because we did not consistently detect a PK degradation product reactive with our TprCSp antiserum (data not shown), the possibility that this portion of the protein is surface exposed and, therefore, part of the β-barrel also needs to be considered. Arguing strongly against this topological assignment is our demonstration that TprCC alone, which does not include the contiguous TprCSp, is sufficient to circularize as a stable (i.e., heat-modifiable), trimeric β-barrel with the same properties as that of the full-length polypeptide. Given the weight of evidence that the outer membranes of the treponemes remained intact throughout incubation with PK, we attribute the inability to detect a PK degradation product reactive with our TprCSp antiserum to the induction of a cell envelope stress response that results in degradation of this fragment. The water solubility of TprCSp (data not shown) further supports its assignment to the periplasmic compartment.
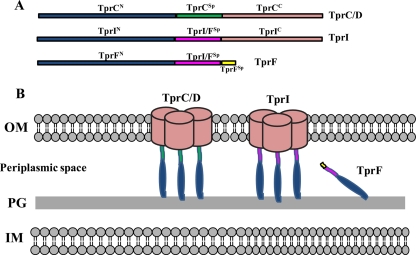
Fig 8.
Topological models of TprC, TprI, and TprF. (A) Comparison of domain architectures. (B) TprC/D and TprI are proposed to consist of trimers with identical β-barrels and N-terminal periplasmic domains that directly or indirectly tether the barrels to the peptidoglycan (PG) sacculus. TprF is entirely periplasmic. TprCSp is depicted as periplasmic based on its solubility in aqueous buffer. TprI shares with TprF a putative periplasmic fragment not present in TprC.
While we have not yet directly proven that TprCC and TprCN reside in the OM and periplasm, respectively, these are the only conceivable compartmental assignments. We do note, however, that TprCN lacks a recognizable OmpA-like peptidoglycan-binding motif (18, 44), an observation that we do not find surprising given the reverse bipartite topologies of TprC and OmpA (61) and the phylogenetic distance separating spirochetes from diderms which do possess OmpA orthologs (46). Furthermore, it has not escaped our attention that the topologic model proposed for TprC can be extended readily to TprI, which differs from TprC mainly within the periplasmic domain, and TprF, which is predicted to be entirely periplasmic (Fig. 8B). Inasmuch as the domain predictions for TprC were based on T. denticola Msp, it is tempting to speculate that this molecular architecture applies to more distantly related Tpr family members as well as the parental ortholog (8, 11, 22). Our finding a number of years ago that Msp is both surface exposed and periplasmic based on immunofluorescence analysis (6) is consistent with the concept of a broadly conserved bipartite domain structure for this phylogenetically related group of OMPs.
Our concept of stealth pathogenicity, dating from the pregenomic era, was based on the premise that rare OMPs would be poorly immunogenic because they are nonlipidated and expressed at extremely low copy numbers (48). The availability of properly folded bona fide OMPs has enabled us to assess this generalization, which has turned out to be less straightforward and far more intriguing than the “one size fits all” hypothesis originally envisioned. As reported elsewhere (19) and reproduced here, TP0326 often induces an antibody response in humans but the antibodies appear to be directed exclusively toward the periplasmic POTRA domains and away from the potentially lethal surface-exposed determinants. In contrast, neither domain of TprC appears to be immunogenic in humans. The higher copy number of TprC would suggest that expression levels alone cannot explain this dichotomy, although one must bear in mind the critical proviso that copy numbers were assessed in spirochetes harvested from one tissue in a nonnatural host at a single time point during infection. Nothing is known yet about the expression levels of these proteins during the period of acute disease when the immune system actually encounters them. In the case of TP0326, one must further postulate a dissociation in the response to the periplasmic and OM-spanning domains, also surprising because β-barrels containing large extracellular loops, as predicted for TP0326 (19), are expected to be antigenic (58).
Collectively, these results support our longstanding contention, buttressed by recent clinical studies (14), that poor antibody binding is a prime virulence determinant of the pathogen that finds clinical expression during the spirochetemia of secondary syphilis. They also represent something of a paradox since human syphilitic sera do contain antibodies capable of surface labeling and promoting opsonophagocytosis of subpopulations of treponemes (11, 13, 42). Either these antibodies are directed against yet-to-be-identified OMPs or the immunoblots are not giving a fully accurate readout of the responses to the proteins, possibilities that we intend to aggressively explore in the near future. The enigma of the immune response to rare OMPs ostensibly deepens when one considers that rabbits infected with T. pallidum via the intratesticular route make antibodies against the β-barrels of both TP0326 and TprC. We believe that this observation reflects the unique evolutionary relationship between the syphilis spirochete and its natural host. The syphilis field may finally have advanced to the point where the antibody response in rabbits can be dissected and exploited as an immunologic road map to the long-sought syphilis vaccine for humans.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH grants AI-26756 (J.D.R.), K23 AI62439 (J.C.S.), 5R03TW008023-3 (J.C.S. and A.R.C.), and 5R03TW008023 (J.C.S.) and by the Connecticut Children's Medical Center (CCMC) Arrison and Burr Curtis Research Funds (J.C.S.).
Footnotes
Published ahead of print 2 March 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Akins DR, et al. 1997. Tromp1, a putative rare outer membrane protein, is anchored by an uncleaved signal sequence to the Treponema pallidum cytoplasmic membrane. J. Bacteriol. 179:5076–5086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Beher MG, Schnaitman CA, Pugsley AP. 1980. Major heat-modifiable outer membrane protein in gram-negative bacteria: comparison with the ompA protein of Escherichia coli. J. Bacteriol. 143:906–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bennion D, Charlson ES, Coon E, Misra R. 2010. Dissection of β-barrel outer membrane protein assembly pathways through characterizing BamA POTRA 1 mutants of Escherichia coli. Mol. Microbiol. 77:1153–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bredemeier R, et al. 2007. Functional and phylogenetic properties of the pore-forming beta-barrel transporters of the Omp85 family. J. Biol. Chem. 282:1882–1890 [DOI] [PubMed] [Google Scholar]
- 5. Brusca JS, Radolf JD. 1994. Isolation of integral membrane proteins by phase partitioning with Triton X-114. Methods Enzymol. 228:182–193 [DOI] [PubMed] [Google Scholar]
- 6. Caimano MJ, Bourell KW, Bannister TD, Cox DL, Radolf JD. 1999. The Treponema denticola major sheath protein is predominantly periplasmic and has only limited surface exposure. Infect. Immun. 67:4072–4083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cameron CE. 2006. The T. pallidum outer membrane and outer membrane proteins, p 237–266 In Radolf JD, Lukehart SA. (ed), Pathogenic Treponema: molecular and cellular biology. Caister Academic Press, Norwich, United Kingdom [Google Scholar]
- 8. Centurion-Lara A, et al. 1999. Treponema pallidum major sheath protein homologue TprK is a target of opsonic antibody and the protective immune response. J. Exp. Med. 189:647–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Conlan S, Bayley H. 2003. Folding of a monomeric porin, OmpG, in detergent solution. Biochemistry 42:9453–9465 [DOI] [PubMed] [Google Scholar]
- 10. Cox DL, Akins DR, Porcella SF, Norgard MV, Radolf JD. 1995. Treponema pallidum in gel microdroplets: a novel strategy for investigation of treponemal molecular architecture. Mol. Microbiol. 15:1151–1164 [DOI] [PubMed] [Google Scholar]
- 11. Cox DL, et al. 2010. Surface immunolabeling and consensus computational framework to identify candidate rare outer membrane proteins of Treponema pallidum. Infect. Immun. 78:5178–5194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cox DL, Radolf JD. 2001. Insertion of fluorescent fatty acid probes into the outer membranes of the pathogenic spirochaetes Treponema pallidum and Borrelia burgdorferi. Microbiology 147:1161–1169 [DOI] [PubMed] [Google Scholar]
- 13. Cruz AR, et al. 2008. Phagocytosis of Borrelia burgdorferi, the Lyme disease spirochete, potentiates innate immune activation and induces apoptosis in human monocytes. Infect. Immun. 76:56–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cruz AR, et al. 2010. Secondary syphilis in Cali, Colombia: new concepts in disease pathogenesis. PLoS Negl. Trop. Dis. 4:e690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cullen PA, Cameron CE. 2006. Progress towards an effective syphilis vaccine: the past, present and future. Expert Rev. Vaccines 5:67–80 [DOI] [PubMed] [Google Scholar]
- 16. Cullen PA, Haake DA, Adler B. 2004. Outer membrane proteins of pathogenic spirochetes. FEMS Microbiol. Rev. 28:291–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Deka RK, Machius M, Norgard MV, Tomchick DR. 2002. Crystal structure of the 47-kilodalton lipoprotein of Treponema pallidum reveals a novel penicillin-binding protein. J. Biol. Chem. 277:41857–41864 [DOI] [PubMed] [Google Scholar]
- 18. De Mot R, Vanderleyden J. 1994. The C-terminal sequence conservation between OmpA-related outer membrane proteins and MotB suggests a common function in both gram-positive and gram-negative bacteria, possibly in the interaction of these domains with peptidoglycan. Mol. Microbiol. 12:333–334 [DOI] [PubMed] [Google Scholar]
- 19. Desrosiers DC, et al. 2011. TP0326, a Treponema pallidum β-barrel assembly machinery A (BamA) orthologue and rare outer membrane protein. Mol. Microbiol. 80:1496–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Edelhoch H. 1967. Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry 6:1948–1954 [DOI] [PubMed] [Google Scholar]
- 21. Egli C, Leung WK, Muller KH, Hancock RE, McBride BC. 1993. Pore-forming properties of the major 53-kilodalton surface antigen from the outer sheath of Treponema denticola. Infect. Immun. 61:1694–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fraser CM, et al. 1998. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science 281:375–388 [DOI] [PubMed] [Google Scholar]
- 23. Gasteiger E, et al. 2003. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31:3784–3788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hagan CL, Silhavy TJ, Kahne D. 2011. Β-barrel membrane protein assembly by the Bam complex. Annu. Rev. Biochem. 80:189–210 [DOI] [PubMed] [Google Scholar]
- 25. Hazlett KR, et al. 2005. TP0453, a concealed outer membrane protein of Treponema pallidum, enhances membrane permeability. J. Bacteriol. 187:6499–6508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hazlett KR, et al. 2001. The TprK protein of Treponema pallidum is periplasmic and is not a target of opsonic antibody or protective immunity. J. Exp. Med. 193:1015–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Heuck AP, Johnson AE. 2002. Pore-forming protein structure analysis in membranes using multiple independent fluorescence techniques. Cell Biochem. Biophys. 36:89–101 [DOI] [PubMed] [Google Scholar]
- 28. Heuck AP, Tweten RK, Johnson AE. 2003. Assembly and topography of the prepore complex in cholesterol-dependent cytolysins. J. Biol. Chem. 278:31218–31225 [DOI] [PubMed] [Google Scholar]
- 29. Ho EL, Lukehart SA. 2011. Syphilis: using modern approaches to understand an old disease. J. Clin. Invest. 121:4584–4592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Izard J, et al. 2009. Cryo-electron tomography elucidates the molecular architecture of Treponema pallidum, the syphilis spirochete. J. Bacteriol. 191:7566–7580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lafond RE, Lukehart SA. 2006. Biological basis for syphilis. Clin. Microbiol. Rev. 19:29–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu J, et al. 2010. Cellular architecture of Treponema pallidum: novel flagellum, periplasmic cone, and cell envelope as revealed by cryo electron tomography. J. Mol. Biol. 403:546–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lukehart SA. 2008. Scientific monogamy: thirty years dancing with the same bug: 2007 Thomas Parran Award Lecture. Sex. Transm. Dis. 35:2–7 [DOI] [PubMed] [Google Scholar]
- 34. Lukehart SA. 2008. Syphilis, p 1038–1046 In Fauci AS, et al. (ed), Harrison's principles of internal medicine, vol 17 McGraw-Hill, New York, NY [Google Scholar]
- 35. Lukehart SA, Miller JN. 1978. Demonstration of the in vitro phagocytosis of Treponema pallidum by rabbit peritoneal macrophages. J. Immunol. 121:2014–2024 [PubMed] [Google Scholar]
- 36. Lukehart SA, Shaffer JM, Baker-Zander SA. 1992. A subpopulation of Treponema pallidum is resistant to phagocytosis: possible mechanism of persistence. J. Infect. Dis. 166:1449–1453 [DOI] [PubMed] [Google Scholar]
- 37. Luthra A, et al. 2011. The transition from closed to open conformation of Treponema pallidum outer membrane-associated lipoprotein TP0453 involves membrane sensing and integration by two amphipathic helices. J. Biol. Chem. 286:41656–41668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Magnuson HJ, Eagle H, Fleischman R. 1948. The minimal infectious inoculum of Spirochaeta pallida (Nichols strain), and a consideration of its rate of multiplication in vivo. Am. J. Syph. Gonorrhea Vener. Dis. 32:1–19 [PubMed] [Google Scholar]
- 39. Marchler-Bauer A, et al. 2011. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 39:D225–D229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Meng G, Fronzes R, Chandran V, Remaut H, Waksman G. 2009. Protein oligomerization in the bacterial outer membrane (review). Mol. Membr. Biol. 26:136–145 [DOI] [PubMed] [Google Scholar]
- 41. Mizianty MJ, Kurgan L. 2011. Improved identification of outer membrane beta barrel proteins using primary sequence, predicted secondary structure, and evolutionary information. Proteins 79:294–303 [DOI] [PubMed] [Google Scholar]
- 42. Moore MW, et al. 2007. Phagocytosis of Borrelia burgdorferi and Treponema pallidum potentiates innate immune activation and induces gamma interferon production. Infect. Immun. 75:2046–2062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nikaido H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67:593–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Park JS, et al. 2012. Mechanism of anchoring of OmpA protein to the cell wall peptidoglycan of the gram-negative bacterial outer membrane. FASEB J. 26:219–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Parsonage D, et al. 2010. Broad specificity AhpC-like peroxiredoxin and its thioredoxin reductant in the sparse antioxidant defense system of Treponema pallidum. Proc. Natl. Acad. Sci. U. S. A. 107:6240–6245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Paster BJ, Dewhirst FE. 2000. Phylogenetic foundation of spirochetes. J. Mol. Microbiol. Biotechnol. 2:341–344 [PubMed] [Google Scholar]
- 47. Pugsley AP. 1993. The complete general secretory pathway in gram-negative bacteria. Microbiol. Rev. 57:50–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Radolf JD. 1995. Treponema pallidum and the quest for outer membrane proteins. Mol. Microbiol. 16:1067–1073 [DOI] [PubMed] [Google Scholar]
- 49. Radolf JD, Hazlett KRO, Lukehart SA. 2006. Pathogenesis of syphilis, p 197–236 In Radolf JD, Lukehart SA. (ed), Pathogenic treponema: cellular and molecular biology. Caister Academic Press, Norwich, United Kingdom [Google Scholar]
- 50. Radolf JD, Lukehart SA. 2006. Immunology of syphilis, p 285–322 In Radolf JD, Lukehart SA. (ed), Pathogenic Treponema: cellular and molecular biology. Caister Academic Press, Norwich, United Kingdom [Google Scholar]
- 51. Radolf JD, Moomaw C, Slaughter CA, Norgard MV. 1989. Penicillin-binding proteins and peptidoglycan of Treponema pallidum subsp. pallidum. Infect. Immun. 57:1248–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Radolf JD, Norgard MV, Schulz WW. 1989. Outer membrane ultrastructure explains the limited antigenicity of virulent Treponema pallidum. Proc. Natl. Acad. Sci. U. S. A. 86:2051–2055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Radolf JD, et al. 1995. Characterization of outer membranes isolated from Treponema pallidum, the syphilis spirochete. Infect. Immun. 63:4244–4252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Randall A, Cheng J, Sweredoski M, Baldi P. 2008. TMBpro: secondary structure, Β-contact and tertiary structure prediction of transmembrane Β-barrel proteins. Bioinformatics 24:513–520 [DOI] [PubMed] [Google Scholar]
- 55. Robert V, et al. 2006. Assembly factor Omp85 recognizes its outer membrane protein substrates by a species-specific C-terminal motif. PLoS Biol. 4:e377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Schulz GE. 2002. The structure of bacterial outer membrane proteins. Biochim. Biophys. Acta 1565:308–317 [DOI] [PubMed] [Google Scholar]
- 57. Seshadri R, et al. 2004. Comparison of the genome of the oral pathogen Treponema denticola with other spirochete genomes. Proc. Natl. Acad. Sci. U. S. A. 101:5646–5651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sette A, Rappuoli R. 2010. Reverse vaccinology: developing vaccines in the era of genomics. Immunity 33:530–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Shevchenko DV, et al. 1999. Membrane topology and cellular location of the Treponema pallidum glycerophosphodiester phosphodiesterase (GlpQ) ortholog. Infect. Immun. 67:2266–2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Silhavy TJ, Kahne D, Walker S. 2010. The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2:a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Smith SG, Mahon V, Lambert MA, Fagan RP. 2007. A molecular Swiss army knife: OmpA structure, function and expression. FEMS Microbiol. Lett. 273:1–11 [DOI] [PubMed] [Google Scholar]
- 62. Stegmeier JF, Andersen C. 2006. Characterization of pores formed by YaeT (Omp85) from Escherichia coli. J. Biochem. 140:275–283 [DOI] [PubMed] [Google Scholar]
- 63. Tommassen J. 2010. Assembly of outer-membrane proteins in bacteria and mitochondria. Microbiology 156:2587–2596 [DOI] [PubMed] [Google Scholar]
- 64. Walker EM, Zampighi GA, Blanco DR, Miller JN, Lovett MA. 1989. Demonstration of rare protein in the outer membrane of Treponema pallidum subsp. pallidum by freeze-fracture analysis. J. Bacteriol. 171:5005–5011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Watanabe Y, Inoko Y. 2009. Reassembly of an integral oligomeric membrane protein OmpF porin in n-octyl beta-D:-glucopyranoside-lipids mixtures. Protein J. 28:66–73 [DOI] [PubMed] [Google Scholar]
- 66. Whitmore L, Wallace BA. 2004. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 32:W668–W673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wimley WC. 2003. The versatile β-barrel membrane protein. Curr. Opin. Struct. Biol. 13:404–411 [DOI] [PubMed] [Google Scholar]
- 68. Wittig I, Braun HP, Schagger H. 2006. Blue native PAGE. Nat. Protoc. 1:418–428 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.