Abstract
The 57-kb gonococcal genetic island (GGI) encodes a type IV secretion system (T4SS) that is found in most strains of N. gonorrhoeae. This T4SS functions to secrete single-stranded DNA that is active in natural transformation. The GGI has also been found in some strains of N. meningitidis. We screened 126 isolates of N. meningitidis and found the GGI in 17.5% of strains, with the prevalence varying widely among serogroups. The GGI is found in a significant number of serogroup C, W-135, and X strains but was not found in strains of serogroup A, B, or Y. Through detailed PCR mapping and DNA sequencing, we identified five distinct GGI types in meningococci. DNA sequencing and a genetic assay revealed that the GGI was likely integrated into the meningococcal chromosome by the site-specific recombinase XerCD and that the GGI can be excised and lost from the genome. Functional studies showed that in contrast with the gonococcal T4SS, the meningococcal T4SS does not secrete DNA, nor does it confer Ton-independent intracellular survival. Deletion of T4SS genes did not affect association with or invasion of host cells. These results demonstrate that the GGI is found in a significant proportion of meningococcal strains and that while some strains carry multiple insertions and deletions in the GGI, other strains carry intact T4SS genes and may produce functional secretion systems.
INTRODUCTION
Neisseria meningitidis (meningococcus [Mc]) is a symbiont of the human nasopharynx and can also cause meningitis and septicemia (30). Up to 10% of the healthy adult population is colonized transiently with Mc, although this percentage can be significantly higher in college residence halls and military barracks (5). The closely related species Neisseria gonorrhoeae (gonococcus [Gc]) is the etiological agent of the sexually transmitted disease gonorrhea. Untreated gonococcal infections can progress to pelvic inflammatory disease or disseminate further, causing arthritis, endocarditis, and meningitis (12, 38). One significant difference between Mc and Gc is that Mc are able to produce a polysaccharide capsule, while Gc do not produce a capsule due to the absence of some of the genes that are required for capsule biosynthesis (26). Approximately 80% of gonococcal strains possess the gonococcal genetic island (GGI), while a limited number of meningococci have been found to have the GGI (10, 37). In gonococci, the GGI is 57 kb and contains genes for a type IV secretion system (T4SS) (10, 14).
Several genomic islands have been identified in N. meningitidis. In 2000, Klee et al. (19) found eight genomic islands in strains of Mc, five of which were conserved across a panel of representative strains of meningococci or were homologous with known virulence factors. Another screen using strains of Mc with various pathogenic potentials found a prophage prevalent in the chromosomes of invasive disease-causing meningococci (1). By screening a pan-Neisseria microarray with meningococcal genomic DNA, Snyder et al. (37) found the GGI in six Mc strains. However, these were mostly from serogroups that are not frequently associated with disease. It was unclear from these results whether the GGI is common in disease isolates and whether GGI-carrying strains can produce a functional T4SS. In Gc, the GGI encodes a T4SS that is responsible for secretion of single-stranded DNA, which is active in natural transformation (14, 28, 32). Other T4SSs have been shown to secrete proteins necessary for intracellular infection, to be required for cell adhesion, and to secrete DNA and proteins directly into host cells (4, 6, 23, 34, 40).
Here we found that the GGI is present in approximately 17% of Mc strains and that the percentage varies by serogroup. Mapping of the GGIs in N. meningitidis strains using PCR identified five different GGI types that had regions where insertions or deletions occurred relative to the gonococcal GGIs. Insertions or deletions were found to disrupt genes for type IV secretion in several strains; however, two strains have GGI types that are nearly complete in their T4SS-encoding regions. Measurement of GGI loss frequencies suggested that changes in the difB sequences diminished GGI loss to a rate lower than that seen in N. gonorrhoeae. We also report that meningococci with a GGI do not exhibit a T4SS-dependent increase in DNA donation for natural transformation or differences in association with or invasion of ME180 host cells. Despite the occurrence of GGI types that have T4SS that are highly similar to the GGI in gonococci, the meningococcal T4SS does not function in the same manner and may secrete other effectors we have not detected.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All strains and plasmids used in this study are listed in Table 1 and in Table S1 in the supplemental material. Recent clinical isolates of meningococci were grown on GCB agar (Difco) plates supplemented with IsoVitaleX (Becton Dickinson Co.) at 37°C with 5% CO2 or grown with aeration in GC base liquid (GCBL) medium (1.5% Proteose peptone no. 3, 0.4% K2HPO4, 0.1% KH2PO4, 0.1% NaCl [pH 7.2]) containing IsoVitaleX and 0.042% NaHCO3. All other meningococci and all gonococci were grown on GCB agar plates with Kellogg's supplements at 37°C with 5% CO2 or in GCBL with Kellogg's supplements and 0.042% NaHCO3 with aeration (18, 24).
Table 1.
Strains and plasmids
| Strain or plasmid | Properties | Source or reference |
|---|---|---|
| N. meningitidis strains | ||
| 98/250521 | Clinical isolate, serogroup H | PHLS |
| 98/250521 Str | 98/250521 transformed with pNMD42 [rpsL(K43R)] | This study |
| 97/252675 | Clinical isolate, serogroup H | PHLS |
| 00/240868 | Clinical isolate, serogroup Z | PHLS |
| 01/241422 | Clinical isolate, serogroup Z | PHLS |
| 01/241471 | Clinical isolate, serogroup Z | PHLS |
| 01/241471 Str | 01/241471 transformed with pNMD42 [rpsL(K43R)] | This study |
| ATCC 13102 | Clinical isolate, serogroup C | ATCC |
| 13102 Str | ATCC 13102 transformed with pNMD42 [rpsL(K43R)] | This study |
| JD1628 | ATCC 13102 transformed with THH51 (pacA::ermC) | 8 |
| KL1011 | 01/241471 transformed with pHH35; traH deletion mutant | This study |
| KL1015 | 01/241471 transformed with ND515 (exp1::mTnCmPhoA) | This study |
| KL1017 | 01/241471 transformed with JD1628 (pacA::ermC) | This study |
| KL1018 | KL1017 transformed with pKL14 (pacA::ermC pilT::kan) | This study |
| KL1019 | KL1011 transformed with JD1628 (ΔtraH pacA::ermC) | This study |
| KL1020 | KL1019 transformed with pKL14 (ΔtraH pacA::ermC pilT::kan) | This study |
| KL1021 | 01/241471 transformed with KL1015 (exp1::mTnCmPhoA) | This study |
| KL1022 | 97/252675 transformed with KL1015 (exp1::mTnCmPhoA) | This study |
| KL1023 | 01/241471 transformed with KL1021 (exp1::mTnCmPhoA backcross) | This study |
| KL1024 | 97/252675 transformed with KL1022 (exp1::mTnCmPhoA backcross) | This study |
| KL1025 | KL1023 transformed with MS11 [rpsL(K43R) exp1::mTnCmPhoA] | This study |
| KL1028 | 98/250521 Str transformed with (98/250521XND515) (exp1::mTnCmPhoA backcross) | This study |
| KL1029 | KL1023 transformed with pNMD42 [rpsL(K43R)] | This study |
| KL1030 | KL1024 transformed with pNMD42 [rpsL(K43R)] | This study |
| KL1031 | KL1028 transformed with pKL15 [rpsL(K43R) exp1::mTnCmPhoA rpsL] | This study |
| KL1032 | KL1029 transformed with pKL15 [rpsL(K43R) exp1::mTnCmPhoA rpsL] | This study |
| KL1033 | KL1030 transfomed with pKL15 [rpsL(K43R) exp1::mTnCmPhoA rpsL] | This study |
| KL1034 | KL1031 transformed with pVD300recA4 [rpsL(K43R) exp1::mTnCmPhoA rpsL recA::tetM] | This study |
| KL1035 | KL1032 transformed with pVD300recA4 [rpsL(K43R) exp1::mTnCmPhoA rpsL recA::tetM] | This study |
| KL1036 | KL1033 transformed with pVD300recA4 [rpsL(K43R) exp1::mTnCmPhoA rpsL recA::tetM] | This study |
| KL1037 | 01/241471 Str transformed with pKL19 [rpsL(K43R) ΔtonB::ermC] | This study |
| KL1038 | 13102 Str transformed with pKL19 [rpsL(K43R) ΔtonB::ermC] | This study |
| KL1040 | 13102 Str transformed with KL1025 [rpsL(K43R) exp1::mTnCmPhoA] | This study |
| KL1041 | 13102 Str transformed with KL1040 [rpsL(K43R) exp1::mTnCmPhoA backcross] | This study |
| KL1044 | KL1041 transformed with pKL15 [rpsL(K43R) exp1::mTnCmPhoA rpsL] | This study |
| KL1045 | KL1044 transformed with pVD300recA4 [rpsL(K43R) exp1::mTnCmPhoA rpsL recA::tetM] | This study |
| KL1046 | KL1032 from loss assay [rpsL(K43R) ΔGGI] | This study |
| KL1047 | KL1046 transformed with pKL19 [rpsL(K43R) ΔGGI ΔtonB::ermC] | This study |
| KL1048 | JD1628 transformed with pKL14 (pacA::ermC pilT::kan) | This study |
| KL1049 | JD 1628 transformed with pHH35 (pacA::ermC ΔtraH) | This study |
| KL1050 | KL1049 transformed with pKL14 (pacA::ermC ΔtraH pilT::kan) | This study |
| KL1051 | KL1044 transformed with ND518 [rpsL(K43R) ΔT4SS] | This study |
| KL1052 | KL1051 transformed with pKL19 [rpsL(K43R) ΔT4SS ΔtonB::ermC] | This study |
| KL1053 | 13102 Str transformed with pSB2 (pSB2 insertion into 13102) | This study |
| KL1054 | 13102 Str transformed with pKL22 (pKL22 insertion into 13102) | This study |
| N. gonorrhoeae strains | ||
| MS11 | Wild-type N. gonorrhoeae | 39 |
| FA19 | Wild-type N. gonorrhoeae | H. S. Seifert |
| ND515 | MS11 transformed with HH522 (exp1::mTnCmPhoA) | 11 |
| ND518 | ND500 transformed with pJS3D (ΔGGI Erm island) | This study |
| ND569 | ND519 transformed with pNMD38 (exp1::mTnCmPhoA difA+ difA+ rpsL) | 11 |
| Plasmids | ||
| pKL6 | traD complement in pKH37 | 31 |
| pKL13 | 01/24147171 77F-83R in pNMD37 at SpeI/SacI | This study |
| pKL14 | Kanr from pKH99 digested with EcoRV/Ecl136II in pHH42 digested with AgeI and blunted | This study |
| pKL15 | rpsL ermC from pKL13 into pKL6 at HindIII | This study |
| pKL16 | 3′ end of NGO1380-5' end of exbB from 13102 into pIDN1 at BamHI/PstI | This study |
| pKL17 | tonB deletion from pKL16, religated at XmaI | This study |
| pKL18 | ermC from pIDN1 inserted in tonB deletion from pKL17 at XmaI | This study |
| pKL19 | ΔtonB::ermC insert from pKL18 in pKH6 at SacI | This study |
| pKL22 | 13102 183F-144R in pIDN1 at PspOMI/KpnI | This study |
| pSB2 | 13102 195F-64R inserted in pIDN1 at SacI/SacII | This study |
| pSB3 | Chromosme walking product containing 3.5 kb of GGI from 13102 adjacent to yfeA | This study |
| pSB4 | Chromosme walking product containing 4.2 kb of GGI from 13102 adjacent to yecA | This study |
| pVD300recA4 | tetM inserted in recA | 35 |
| pHH35 | traH deletion, 990-bp in-frame deletion in pJD1181 | 11 |
| pHH42 | pilT mutant (K136Q) | 42 |
| pKH6 | cat from pGCC6del inserted at HindIII/ClaI of pIDN1, ermC cut out with SalI and XbaI | This study |
| pKH37 | Complementation vector | 20 |
| pKH99 | Kanr from pHSS6 into BamHI site of pIDN2 | This study |
| pNMD37 | PCR product of difB and flanking DNA outside GGI from IN522 into pNMD36 at SpeI/SacI | This study |
| pNMD42 | MS11 rpsL(K43R) into pIDN2 at KpnI/XhoI | This study |
| pHSS6 | Cloning vector | 36 |
| pIDN1 | Cloning vector | 15 |
| pIDN2 | Cloning vector | 15 |
GGI screening.
Recent clinical isolates were obtained from the Wisconsin State Laboratory of Hygiene (WSLH), Active Bacterial Core Surveillance Team and the Emerging Infection Programs Network of the U.S. Centers for Disease Control and Prevention (CDC), and L. H. Harrison (University of Pittsburgh). Colonies were resuspended in colony lysis solution (1% Triton X-100, 2 mM EDTA, 20 mM Tris HCl [pH 8.0]) and lysed at 95°C for 15 min (41). DNA from lysates was used as the template for PCR to screen for the GGI. We used primers 77F and hlhggiR (see Table S2 in the supplemental material for primer sequences) to amplify dif and flanking DNA to test for the absence of the GGI, and we used 137F and 118R to amplify a conserved fragment within the GGI. Due to the heterogeneous nature of sequences between dif and ung in meningococci, Southern blotting was used to confirm the absence of the GGI in several isolates. Chromosomal DNA isolation and purification was performed as described by Dillard (7). Probes were generated by PCR amplification of fragments from traI (53F-235R), oriT (247F-254R), ydhAB (193F-218R), and yea (197F-72R). Southern blotting was performed according to the method described by Sambrook et al. (33).
PCR walking and GGI typing.
Chromosomal DNA was isolated and purified from gonococcal strain MS11 and meningococcal strains 01/241471, 98/250521, 01/241422, 97/252675, 00/240868, A22, and ATCC 13102 using the DNeasy kit (Qiagen) according to manufacturer's instructions. Meningococcal strains were obtained from the Manchester Public Health Laboratory Service (PHLS), D. A. Caugant (Norwegian Institute of Public Health), and the American Type Culture Collection (ATCC). We performed successive PCRs beginning with DNA flanking the GGI. Each target was approximately 2 kb in N. gonorrhoeae strain MS11, and each product overlapped the next by about 500 bp (see Table S3 in the supplemental material for primers). The amplified DNA was run on a 0.8% agarose-TBE gel. Any fragments from meningococci that were a different size than the MS11 fragment were sequenced at the University of Wisconsin—Madison Biotechnology Center using the BigDye fluorescent sequencing method according to the manufacturer.
Recent clinical isolates with the GGI were analyzed for GGI type by colony lysis PCR (see Table S1 in the supplemental material). GGI+ N. meningitidis isolates were judged to carry GGI type 1 if there was a 3.5-kb fragment from a PCR using primers 155F and 106R, representing a deletion from yecB-parB (see Table S3 in the supplemental material for primer sequences and targets). To test for GGI type 2, insertions in yaa and traD were observed using primers 83F-82R and 137F-118R, respectively. GGI type 3 was identified by the presence of a 1.4-kb product from a PCR with 77F-76R (truncation of traD) and a 1.4-kb product from 45F-46R (IS1655 in traK). To distinguish GGI type 3 from GGI type 4, we looked for a 1.1-kb product (type 3) or a 1.2-kb product (type 4) from a reaction with 63F-184R. GGI type 5 was identified through amplification from 155F-106R and a 3.0-kb product from 113F-1105-6R, indicating a deletion from traE-ydbA.
Loss assay.
We constructed strains to select for GGI loss in four strains: 01/241471, 97/252675, 98/250521, and ATCC 13102. Each strain was transformed by spot transformation with pNMD42 to introduce an rpsL allele conferring streptomycin (Str) resistance at the chromosomal locus. This strain was transformed with chromosomal DNA from ND515, carrying exp1::mTnCmPhoA, in order to introduce a chloramphenicol (Cm) resistance marker and phoA into the GGI, allowing blue/white screening. The resulting strain was transformed with pKL15 to introduce an erythromycin (Erm) marker and an rpsL allele in the GGI. The rpsL allele in the GGI encodes for streptomycin sensitivity, which is dominant over streptomycin resistance (22). In order to prevent homologous recombination between the two rpsL genes, the strain was transformed with pVD300recA4, which has a tetracycline (Tet) resistance marker inserted into recA. Each of the final strains was resistant to Tet, Erm, and Cm, PhoA positive on XP indicator plates, and sensitive to Str.
Loss frequency was determined essentially as described by Domínguez et al. (11). Individual colonies were streaked onto GCB Cm plates (10-μg/ml final concentration) and grown overnight to ensure the presence of the GGI at the beginning of the assay. To allow for GGI loss, Cm-resistant colonies were grown overnight on GCB agar plates without selection. After 16 to 18 h of growth, bacteria were swabbed from plates into GCBL and plated in duplicate on GCB plates without selection to enumerate total CFU/ml and on GCB-Tris-XP (40 μg/ml) plates with Str (100 μg/ml) to select and screen for GGI loss. Loss frequency is the ratio of Str-resistant, PhoA− CFU to total CFU. Reported frequencies are the geometric means from three independent experiments.
Coculture transformation.
Donor strains for coculture transformation were constructed in ATCC 13102 and 01/241471 backgrounds, as these strains had the most complete T4SS-encoding regions. 01/241471 was transformed with chromosomal DNA from JD1628 (ATCC 13102 pacA::ermC) to introduce an Erm resistance marker. Test strains were then transformed with pKL14 to insert a kanamycin (Kan) resistance marker in pilT to ensure that the strain could act only as a DNA donor in natural transformation. Negative-control strains were constructed by transforming strains with pacA::ermC with pHH35 to generate an in-frame deletion of traH (11). traH is required for DNA secretion via the T4SS (15). The traH deletion strains were then transformed with pKL14 to introduce the pilT mutation. Recipients were constructed by transformation of ATCC 13102 or 01/241471 with pNMD42, introducing Str resistance.
We grew strains overnight on GCB plates without selection. Individual colonies were then restreaked onto appropriate selection, GCB with Erm (10 μg/ml) for donors and GCB with Str (100 μg/ml) for recipients. After 16 to 18 h growth, strains were inoculated in GCBL plus supplements at an optical density at 540 nm (OD540) of 0.19 to 0.21. After 2 h growth, strains were diluted 1:32 in fresh GCBL. At this point, donors and recipients were mixed together and grown in coculture for 3 h. At 0 h and 3 h, strains were plated in duplicate onto GCB without selection, GCB Erm, GCB Str, and GCB Erm (5 μg/ml) Str (100 μg/ml). Transformation frequency was calculated as the number of transformants (CFU/ml on GCB Erm Str) per donor (CFU/ml on GCB Erm). Reported frequencies are the geometric means from three independent experiments.
Intracellular survival assay.
We deleted tonB in ATCC 13102 and 01/241471 by spot transformation with pKL19 and selection on GCB plates containing Erm (10 μg/ml). We also constructed tonB deletions in strains lacking the GGI obtained by selection in the loss assay (01/241471 background) or by transformation with chromosomal DNA (ATCC 13102 background). Strains were grown overnight on GCB plates. Piliated colonies were selected, restreaked, and grown for 16 h on GCB plates.
Intracellular growth in the absence of inorganic iron was assayed using a modification of the method described by Zola et al. (42). ME180 cervical epithelial cells were seeded at a density of 2.5 × 105 cells/ml to a 12 well plate and grown to 70 to 80% confluence at 37°C with 5% CO2 in McCoy's 5A medium (Gibco) supplemented with 10% heat-inactivated fetal bovine serum (HI-FBS) (Gibco). Meningococci were resuspended in McCoy's 5A supplemented with 10% HI-FBS and 24 μM Fe(NO3)3 and allowed to infect monolayers of ME180 cells at a multiplicity of infection (MOI) of 10 for 4 h at 37°C with 5% CO2. After initial infection, wells were washed twice with Chelex-treated PBS and then treated with McCoy's 5A with 10% HI-FBS and 25 μg/ml gentamicin (Sigma) for 1 h to kill any extracellular meningococci. After gentamicin treatment, cells from time zero were lysed by treatment with 0.5% saponin in Chelex-treated PBS for 1 min, harvested, and plated for meningococcal CFU counts onto GCB plates. The remaining cells had media replaced with McCoy's 5A plus 10% HI-FBS and were incubated at 37°C with 5% CO2 for 4 or 24 h. One hour before each time point, cells were treated with gentamicin, followed by lysis and harvest. Each experiment was performed in triplicate, and final CFU/ml values are the geometric means from three independent experiments.
Association and invasion assay.
ME180 cells were seeded into 24-well plates at a density of 4.0 × 105 cells/ml in a total volume of 0.5 ml of McCoy's 5A with 10% HI-FBS and grown to 80% confluence. The cells were infected with meningococcal strains ATCC 13102 and 01/241471 and isogenic T4SS− strains at an MOI of 100 for 3.5 h at 37°C with 5% CO2. Total CFU/ml was enumerated by plating supernatants and lysates to GCB plates. Lysis occurred by incubation with 0.5% saponin in PBS for 5 min. Wells to enumerate associated and invaded CFU/ml were washed six times with PBS. Contents of association wells were lysed and plated on GCB agar plates. Invaded wells were treated with 25 μg/ml gentamicin for 1 h and incubated at 37°C with 5% CO2. Wells were washed an additional six times with PBS, and lysates were plated onto GCB. Each experiment was carried out in triplicate. Association and invasion frequencies shown are the geometric means from three individual experiments.
RESULTS
Survey of N. meningitidis strains for the GGI.
When the GGI was initially discovered in N. gonorrhoeae, attempts were made to identify it in other Neisseria species using low-stringency Southern hybridization (10). It was not found in any symbiotic Neisseria species or any of 9 N. meningitidis strains. However, the two GGI genes used for the hybridization studies turned out to be variable in the GGI, and Snyder et al. (37) identified versions of the GGI in six Mc strains. Given the small number of Mc strains tested, it was not clear how common the GGI is in the meningococcal population. Therefore, we screened clinical isolates of N. meningitidis for the presence of the GGI using three panels of isolates. One panel contained N. meningitidis isolates from the Wisconsin State Laboratory of Hygiene (WSLH), which were collected from patients in Wisconsin with invasive meningococcal disease. The second was a panel from the University of Pittsburgh that also contained invasive disease isolates. The third panel was obtained from the U.S. Centers for Disease Control and Prevention (CDC) and contained invasive and noninvasive strains (see Table S1 in the supplemental material for a complete list of strains). To screen isolates by PCR, we used two sets of primers, one set internal to the GGI and a second set to amplify across the dif region that is the GGI insertion site in the chromosome (14). Results were considered conclusive if a product of the correct size was seen for only one reaction. Isolates that did not give conclusive results were screened by Southern blotting. Of the 126 isolates screened, we found that 22 isolates (17.5%) had the GGI. The percentage of GGI-positive strains varied by serogroup (Table 2).
Table 2.
Summary of results of GGI screening in N. meningitidis
| Serogroup | No. of GGI+ isolates/total | No. of isolates with island type |
||||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | ||
| All Mc | 22/126 | 2 | 1 | 6 | 1 | 12 |
| A | 0/21 | |||||
| B | 0/23 | |||||
| C | 6/22 | 1 | 5 | |||
| D | 0/1 | |||||
| H | 2/2 | 1 | 1 | |||
| W135 | 6/15 | 5 | 1a | |||
| X | 2/11 | 1 | 1 | |||
| Y | 0/23 | |||||
| Z | 6/8 | 1 | 5 | |||
GGI identified in draft genome sequence of strain α275 (GenBank accession number AM889138).
PCR walking, DNA sequencing, and GGI typing.
It was apparent from the results of Snyder et al. that some versions of the GGI in N. meningitidis carried deletions relative to the 57-kb Gc GGI (37). Therefore, we examined the GGIs of seven Mc strains for gene content using PCR, paying particular attention to the presence of genes required for type IV secretion. Six of the seven strains had been previously shown to have the GGI (37). The seventh strain we identified as possessing a distinct GGI type during the course of this study. The GGI-encoded T4SS in N. gonorrhoeae has been shown to secrete DNA active in natural transformation and to facilitate TonB-independent iron uptake during intracellular growth of gonococci (42). Thus, the presence of the T4SS genes in Mc might affect horizontal gene transfer or intracellular growth of N. meningitidis strains possessing the T4SS genes. In order to assess the content of the GGI in Mc, we performed overlapping PCR across the GGI and sequenced regions that gave PCR products different in size from that of N. gonorrhoeae strain MS11.
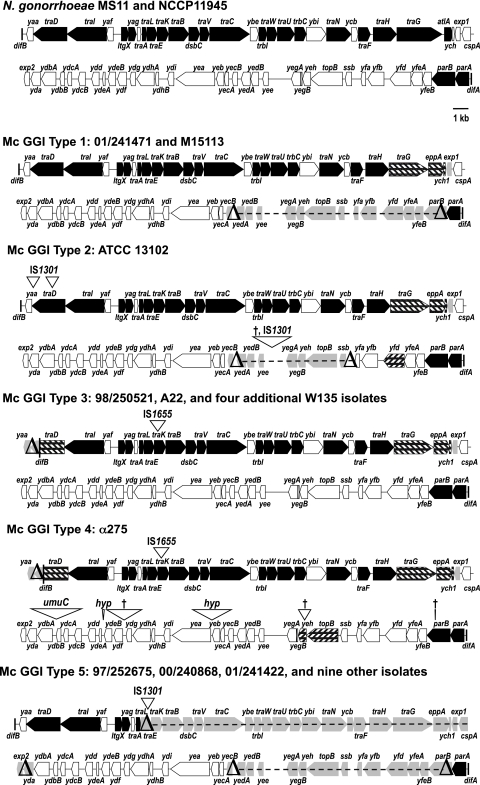
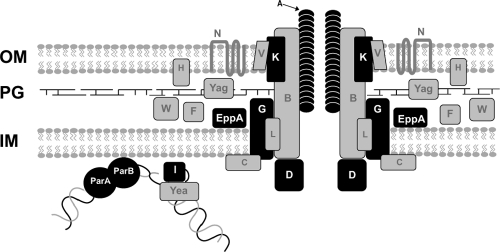
We found four different island types among seven strains by PCR walking and an additional GGI type in the draft genome of strain α275 (Fig. 1 and 2). The first island type, present in strain 01/241471, lacks the lytic transglycosylase gene, atlA, and instead encodes a peptidoglycan endopeptidase, EppA. This arrangement of genes (lacking atlA and carrying eppA) is also found in one-sixth of gonococcal strains (10, 20, 28). There is also a deletion of exp1 and an 11-kb deletion encompassing genes from yecB through parB. In N. gonorrhoeae, atlA and parB are both necessary for DNA secretion (10, 20, 25). However, all of the genes for structural components of the T4SS appear intact. This strain has the long allele of traA, and the signal sequence encoded by traA is different from that of gonococci (Fig. 3). Previous studies have shown that traA is not required for DNA secretion in Gc (16). In contrast with Snyder et al. (37), we were able to detect and sequence traE from 01/241471 and parA in all GGI+ meningococcal strains analyzed in the present study. These results, particularly the lack of atlA and parB, raise the question of whether this strain carries a functional T4SS and whether it could secrete DNA or possibly some other substrate. This GGI type is also found in a serogroup X strain, M15113 (Table 2). A model of the T4SS highlighting the components that differ between the meningococcal and gonococcal systems is shown in Fig. 2.
Fig 1.
Maps of GGIs in N. gonorrhoeae and N. meningitidis. Five GGI types have been identified in meningococci by overlapping PCR and bioinformatic analysis. Genes in black have been shown to be necessary for T4SS-dependent DNA secretion in gonococci. Genes in white are not necessary for DNA secretion. In meningococcal maps, genes filled with diagonal bars are similar to, yet distinct from, genes found in gonococcal strain MS11. Genes in light gray with deltas or dashes through them are deleted in meningococci. Inverted triangles represent insertions in meningococcal sequences, with the insertion sequence or homologous sequence indicated above (bases drawn to scale); a dagger indicates an insertion with no homologous sequence found.
Fig 2.
Schematic of the type IV secretion system in N. meningitidis. In gonococci, the partitioning protein homologs ParA and ParB may bind DNA and bring it to the T4SS apparatus. The relaxase, TraI, nicks DNA and it is unwound by a helicase, possibly Yea. The DNA-TraI complex may then interact with the putative coupling protein, TraD, and the DNA is secreted through the T4SS into the extracellular environment. TraK, the predicted secretin, may contribute to pore formation in the outer membrane. TraA is a predicted pilin. EppA is an endopeptidase that could potentially degrade peptidoglycan to allow for apparatus formation in the periplasm. Proteins depicted in black show differences between meningococci and gonococcal strain MS11, while proteins in gray are similar. OM, outer membrane; PG, peptidoglycan; IM, inner membrane.
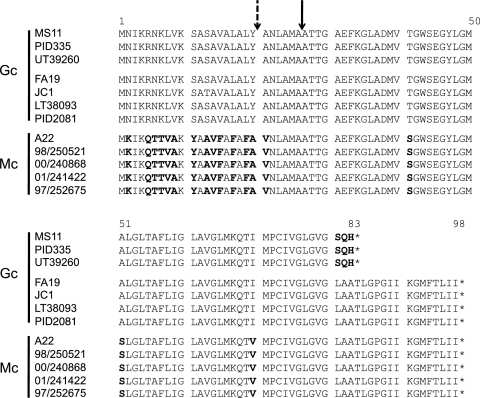
Fig 3.
Alignment of TraA from N. gonorrhoeae and N. meningitidis. There are three alleles of traA. Gonococci have a short or long allele, and meningococci carry a long allele with a different signal sequence. Amino acid differences are in bold. The signal sequence cleavage site is indicated by a solid arrow. An alternate potential cleavage site in meningococci is indicated by a dashed arrow.
The second island type is found in strain ATCC 13102. As with all the Mc strains that carry this region, atlA and exp1 are absent and eppA is present in this island type. IS1301 is inserted into traD downstream of the region encoding the first Walker A box. A second copy of IS1301 is inserted into yaa, a gene of unknown function. This strain also has the long allele of traA. There is an 8.2-kb deletion from yeb to ssb. It is replaced by 2.5 kb of sequence that contains an insertion of IS1301 preceded by two insertions of a 50-bp segment of GGI sequence found in the intergenic region between ydf and ydg. Between the two insertions of the ydf-ydg intergenic region is 1.3 kb of sequence that is not homologous to any known sequence. ATCC 13102 is the only strain we have found to carry this version of the GGI (Table 2).
The third island type is present in strains 98/250521 and A22. Four W135 recent clinical isolates we screened also have this island type. Many characteristics of this island type are also found in sequenced strain α275, carrying the fourth island type. Like the island types in 01/241471 and ATCC 13102, these strains have eppA and lack atlA and exp1. They also have a deletion in traD, resulting in the loss of 300 bp of traD coding sequence and the deletion of the yaa open reading frame. TraD is the predicted T4SS coupling protein, which would bind substrates for secretion by the T4SS apparatus, and it was recently shown that traD is necessary for DNA secretion (31), although a traD truncation mutant gives an intermediate level of DNA secretion (14). These GGIs also carry an insertion of IS1665 in traK immediately following the traK start. There was no consensus ribosome binding site or other in-frame start codon in the insertion, so it is unlikely that TraK is produced in these strains. TraK is predicted to be a secretin, an outer membrane protein that forms part of the channel for type IV secretion (14). Pachulec (25) recently found that traK is necessary for DNA secretion from N. gonorrhoeae. We found the genes traA and ybe to be present in strain A22 and yecB in both A22 and 98/250521, which differs from the results of Snyder et al. (37).
Recently a draft genome of strain α275 was made available (GenBank accession number AM889138). This strain contains a GGI that is 64 kb, 7 kb longer than the gonococcal strain MS11 GGI (28). This GGI represents a fourth island type in meningococci. It is highly similar to the GGI type 3 from difB through ydbA. The sequence from ydbB through parA contains numerous insertions (Fig. 1). The insertions range in size from 30 bp to 3.3 kb. The largest insertion contains sequence homologous to the umuC domain of DNA polymerase V. Two of the insertions have homology to hypothetical proteins, and three insertions have no known homology. There is a 56-bp deletion in yeh, an 80-bp deletion in topB, and a 30-bp deletion in parB. This island type is unique to α275.
The fifth island type is present in strains 01/241422, 00/240868, and 97/252675, as well as five serogroup C strains, three serogroup Z strains, and one serogroup X strain (Table 2). These strains have two large deletions. The 22-kb region from traE through yda is deleted and replaced by one or two copies of IS1301. The deletion removes a majority of the T4SS genes. The 11-kb deletion from yecB through parB seen in GGI type 1 is also present in these strains. Most genes in this region have no homologues or are homologous to other genes of unknown function. There is a single-stranded binding protein, SsbB, and a homolog of topoisomerase IV, TopB, encoded in this region as well. Neither is required for DNA secretion (25).
We used PCR to assign a GGI type to the 22 recent clinical isolates that carry the GGI (Table 2; also, see Table S1 in the supplemental material). All isolates gave results consistent with one of the five island types we established above. All GGI+ serogroup C isolates, except for ATCC 13102, carry island type 5. GGI type 3 was found only in serogroup W135 strains, and α275 was the only W135 strain with GGI type 4. Types 3 and 4 are identical from difB to ydbA; after ydbA, GGI type 4 has numerous insertions and deletions. Nearly all serogroup Z strains carry island type 5; 01/241471 is the only group Z strain with island type 1. We did not observe a correlation between invasive disease and presence or absence of the GGI in meningococci.
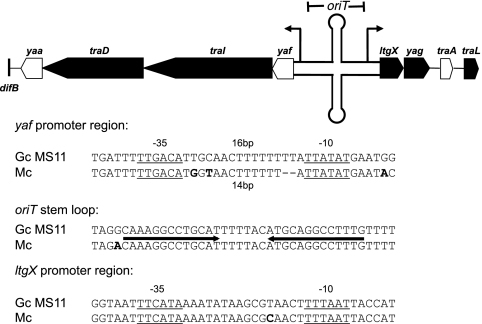
Sequences of the oriT and traI promoter regions.
Since several of the Mc GGIs contain deletions in T4SS genes, we hypothesized that these strains might carry mutations in the origin of transfer, oriT, or promoters for the T4SS operons. We sequenced the region between yaf and ltgX, which contains the oriT and promoters for the putative yaf traI traD yaa operon and the putative operon starting with ltgX, which contains most of the genes required for type IV secretion in N. gonorrhoeae, from seven meningococcal strains (14, 31, 32). The oriT is found in the 156-bp fragment within the PsiI and NgoMIV sites in the yaf-ltgX noncoding region (our unpublished results). When aligned with the gonococcal sequence, all meningococcal sequences have an identical oriT stem-loop sequence (Fig. 4). There are six separate base pair differences between gonococci and meningococci within the entire oriT. Meningococci also have a 2-bp deletion and a 2-bp insertion within 20 bp of each other. The proposed −10 and −35 sequences for the ltgX operon promoter are also intact, and spacing is conserved. However, the spacing between the presumed −10 and −35 sites for the promoter of the apparent yaf operon is altered in meningococci. In gonococci, there is 16 bp between the −10 and −35 hexamers; however, in meningococci, a 2-bp deletion results in a 14-bp spacer between the sites (Fig. 4).
Fig 4.
Analysis of the yaf-ltgX intergenic region. The intergenic region between yaf and ltgX contains the oriT and likely promoters for the transcript starting with yaf and the transcript starting with ltgX. The oriT is drawn as a stem-loop, and promoters are drawn as elevated arrows on the map. The yaf promoter is likely defective in meningococci due to a 2-bp deletion in the spacer region between the putative −10 and −35 sites. The oriT and the ltgX promoter are conserved between gonococci and meningococci. The meningococcal sequence shown is 100% identical in seven strains we sequenced.
traA sequencing.
The gonococcal genetic island in N. gonorrhoeae contains traA, encoding the putative T4SS pilin. Sequencing in gonococci revealed two alleles of traA, a long form and a short form (Fig. 3). A single nucleotide insertion results in a frameshift, and early termination of the short allele. Despite traA being required for F-plasmid conjugation, traA is not required for DNA secretion via the T4SS in gonococci (16). As type IV secretion of DNA occurs in a contact-independent manner in Gc, it is not unreasonable that a pilus would not be required for DNA secretion but would be necessary for contact-dependent conjugation in other systems. Jain et al. showed that the long allele of traA encodes a protein that when coexpressed in E. coli with trbC is processed and circularized (17). That study found that the signal sequence was cleaved after A26, which was the predicted cleavage site.
Meningococci contain a third allele of traA. To date, all traA+ meningococci have been found to have a 98-amino-acid TraA, but the signal sequence is different from that in gonococci (Fig. 3). There are also 3 amino acid changes in the predicted mature form of the protein, T41S, A51S, and I70V, from gonococci to meningococci (Fig. 3). Signal sequence prediction showed that the most likely cleavage site was after A26, as in gonococci. Another possible cleavage site, after A20, was also identified; however, the percent likelihood of cleavage at that site as predicted by SignalP 4.0 (27) was much lower than the value for a site after A26.
GGI loss.
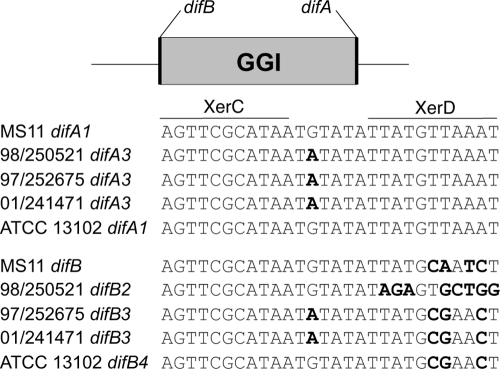
The GGI is flanked by a direct repeat. One copy of this repeat constitutes the dif site, a sequence recognized by the site-specific recombinases XerCD to separate chromosome dimers during replication (2, 3, 14). In Gc, XerCD action at the dif site difA and a second, partial dif site, difB, leads to excision of the GGI and its loss from the population at low frequency (11). We sequenced the dif site closest to traD, difB, in several Mc strains and found three different sequences (Fig. 5). The first sequence, difB2 (found in strains A22, 98/250521, and α275), has multiple changes in the XerD-binding site compared to the consensus sequence. The second sequence, difB3 (found in strains 01/241471 and 97/252675), has three base pair changes within the XerD-binding site and one base pair change within the spacer region (11). These changes result in a palindromic dif. The third sequence, difB4 (found in strain ATCC 13102), is similar to the palindromic difB but does not have the G-to-A change in the spacer region. Previous work has shown that the sequence of dif influences excision of the GGI from the gonococcal chromosome, with strains that have a sequence more similar to consensus having a higher loss frequency (11).
Fig 5.
Alignment of difA and difB from N. gonorrhoeae and N. meningitidis. The 28-bp dif sequence is the binding site of the site-specific recombinases XerCD. difA is conserved at the XerC- and XerD-binding sites in gonococci and meningococci with and without a GGI. difB is found only in strains with a GGI. XerC- and XerD-binding sites are indicated by a line above the first sequence in the alignment. Bases that are in bold differ from the consensus dif sequence. The changes in difB of 01/241471 and 97/252675 result in a palindrome. Numbers assigned to difA and difB sequences are based on those proposed by Domínguez et al. (11).
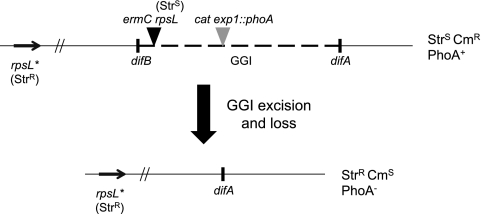
To quantify GGI loss, we utilized an assay that selects for absence of the GGI, thus allowing for detection of low-frequency events (Fig. 6) (11). The assay strain has a streptomycin sensitivity marker inserted within the GGI and a streptomycin resistance marker elsewhere on the chromosome. As streptomycin sensitivity is dominant over streptomycin resistance, a strain that has the GGI will be streptomycin sensitive, and a strain that has lost the GGI will be streptomycin resistant (11, 22). Bacteria were grown for 18 h on nonselective GCB plates, swabbed into liquid medium, and plated on GCB agar plates containing streptomycin to select for bacteria that had excised and lost the GGI.
Fig 6.
Schematic of the GGI loss assay. A strain that has both streptomycin-sensitive (Strs) and -resistant (Strr) alleles of rpsL is also marked with erythromycin resistance (Ermr) and chloramphenicol resistance (Cmr) and is positive for alkaline phosphatase activity (PhoA+). Any cell that has the GGI is Strs, Cmr, and PhoA+. If the GGI is excised from the chromosome and the cell undergoes division, the GGI will be lost from that cell, since the GGI appears to be incapable of replicating on its own. A cell that has lost the GGI is then Strr, Cms, and PhoA−.
Domínguez et al. (11) showed that the loss frequency for the gonococcal strain MS11 is 1.15 × 10−6. If the MS11 difB sequence is replaced with the consensus dif sequence (difA), the frequency of loss is 1.84 × 10−3. We found the loss frequency for meningococcal strain 01/241471 to be 3.45 × 10−8 ± 2.76 × 10−8. We could not detect loss for strains 97/252675, 98/250521, or ATCC 13102, indicating that the loss frequency is 6.25 × 10−9, the limit of detection, or less (Table 3).
Table 3.
Frequency of GGI loss is lower in N. meningitidis than in N. gonorrhoeae
| Parenta | Strain | dif type | Loss frequency |
|---|---|---|---|
| MS11 (Gc) | MS11 | AB | 1.15 × 10−6b |
| MS11 (Gc) | ND569 | AA | 1.84 × 10−3b |
| 01/241471 (Mc) | KL1035 | AB | 3.45 × 10−8 ± 2.76 × 10−8 |
| 97/252675 (Mc) | KL1036 | AB | <6.25 × 10−9 ± 1.16 × 10−8 |
| 98/250521 (Mc) | KL1034 | AB | <3.14 × 10−9 ± 1.22 × 10−9 |
| 13102 (Mc) | KL1045 | AB | <5.28 × 10−10 ± 1.57 × 10−9 |
Each parent strain was modified by transforming with a rpsL(K43R) marker and then inserting an ermC-rpsL cassette and an mTnCmPhoA cassette in the GGI and a tetracycline resistance marker in recA.
Frequency as reported by Domínguez et al. (11).
GGI acquisition by meningococci.
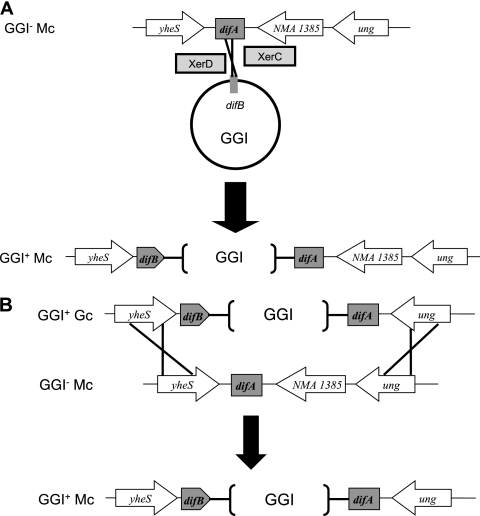
The sequence of the DNA flanking the GGI also provides insight into the mechanism of GGI acquisition by meningococci. The genomes of all 15 sequenced Mc strains currently available in the National Center for Biotechnology Information (NCBI) database show the presence of a gene, NMA1385, between difA and ung. This gene is not present in gonococcal genome sequences, and no gene is present between difA and ung in Gc (14). If GGI acquisition occurred by site-specific recombination mediated by XerCD, NMA1385 would still be present in meningococci with a GGI (Fig. 7A). However, if GGI transfer occurred by homologous recombination between gonococci and meningococci, recombination would be predicted to occur at yheS and ung, resulting in the loss of NMA1385 (Fig. 7B). We used PCR to amplify the region between parA and ung and found a fragment of increased size relative to that in gonococci. Sequencing revealed that the fragment contained NMA1385, indicating that GGI integration in Mc likely occurred via site-specific recombination.
Fig 7.
Possible mechanisms of GGI integration into the meningococcal chromosome. (A) Integration by XerCD site-specific recombination. The GGI as a mobile element contained a sequence homologous to the dif sequence. The site-specific recombinases XerC and XerD acted on the dif sequence in the GGI and dif sequence in the meningococcal chromosome, resulting in integration of the GGI. The gene NMA1385 is still present in this strain with the GGI. (B) Integration by homologous recombination. After uptake of the GGI from gonococci by a meningococcal cell, the GGI is integrated into the chromosome by homologous recombination at yheS and ung. The gene NMA1385 is no longer present.
Coculture transformation.
Gonococci with a functional T4SS show significantly higher DNA donation frequencies in coculture transformation than strains without a functional T4SS (10, 15, 32). We utilized coculture transformation to detect T4SS-dependent DNA secretion in meningococci. The two strains of meningococci that were used as parental strains, ATCC 13102 and 01/241471, were selected because they contained the most complete T4SS-encoding region of the GGI. The donor strains, KL1048 and KL1018, were marked with erythromycin resistance by inserting ermC into the pacA gene. The donors also had a kanamycin marker inserted into the pilT gene so that DNA uptake could not occur, ensuring that the donor could not act as a recipient in natural transformation. A second pair of donors, KL1050 and KL1020, had a deletion in traH, in addition to having pacA::ermC and pilT::kan. In gonococci, mutation of traH abolishes T4SS-dependent DNA secretion (14, 15). The recipients were marked with a streptomycin resistance marker.
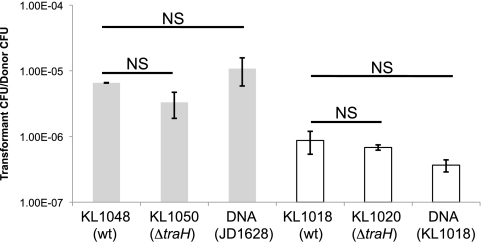
Strains were grown for 2 h in liquid culture separately and diluted, and then a donor was mixed and grown with the recipient. The recipient strains were also grown with 1 μg of purified chromosomal DNA carrying pacA::ermC to quantify the transformation frequency of the recipient with this chromosomal marker. After 2 h of coculture, bacteria were plated onto GCB agar with erythromycin (Erm), Str, or Erm and Str in order to count the number of donor, recipient, and transformant CFU (Fig. 8). Although the transformation frequency appeared to be slightly lower for the T4SS mutants, there was no significant difference in DNA donation frequency between wild-type (KL1048 or KL1018) and ΔtraH (KL1050 or KL1020) strains. Wild-type gonococcal strains show 50- to 500-fold-higher levels of DNA donation than mutants defective in type IV secretion (10, 14, 25). No such effect was seen in meningococci, indicating that these strains do not exhibit T4SS-dependent DNA secretion.
Fig 8.
N. meningitidis does not secrete DNA in a type IV secretion system-dependent manner. Differently marked strains of meningococci were grown together for 3 h and then plated with selection to identify doubly resistant transformants. Gray bars indicate that the recipient was Mc strain ATCC 13102; white bars indicate that the recipient was strain 01/241471. The frequencies shown are the geometric means from at least three independent experiments; error bars represent the standard error of the mean. NS, no significant difference by analysis of variance (ANOVA) or the Tukey-Kramer method.
Ton-independent intracellular survival.
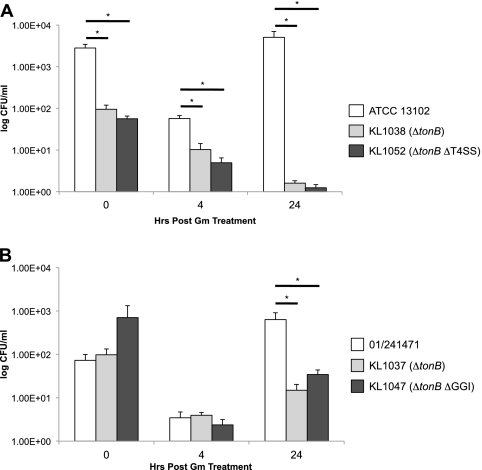
Zola et al. (42) demonstrated that gonococci that have the type IV secretion system encoded in the GGI are able to survive intracellularly under iron starvation conditions without a functional Ton complex. The Ton complex is responsible for energy transduction for a variety of outer membrane transporters, including those responsible for iron internalization (28, 42). We constructed mutations in 01/241471 and ATCC 13102 backgrounds to determine if meningococci with the GGI also exhibit Ton-independent intracellular survival. We infected ME180 cervical epithelial cells with wild-type, ΔtonB (KL1037, KL1038), ΔtonB ΔGGI (KL1047), and ΔtonB ΔT4SS (KL1052) meningococci at a multiplicity of infection (MOI) of 10 for 3 h. After the initial infection, wells were washed and treated with medium containing 25 μg/ml gentamicin (Gm) for an hour to kill any bacteria that were not inside the epithelial cells. Measurements were taken at 0, 4, and 24 h after initial Gm treatment. An hour before each of these time points, wells were treated with 25 μg/ml Gm as before. The wild-type strains 01/241471 and ATCC 13102 exhibited an initial decrease in CFU at 4 h after Gm treatment but had rebounded by 24 h posttreatment. Thus, the wild type meningococcal strains showed growth characteristics similar to those of wild-type gonococcal strains in this assay (42). Neither the ΔtonB, the ΔtonB ΔGGI, nor the ΔtonB ΔT4SS strains rebounded at 24 h (Fig. 9). There was a significant difference between the wild-type and mutant strains, but there was no significant difference between the mutant strains that carry the T4SS genes and those that do not at 24 h posttreatment. These results indicate that the T4SS in meningococci does not contribute to Ton-independent intracellular survival.
Fig 9.
T4SS+ Mc do not show Ton-independent intracellular survival. ME180 epithelial cells were infected with Mc at an MOI of 10 for 3 h followed by a 1-h treatment with 25 μg/ml gentamicin (Gm). Measurements were taken at 0, 4, and 24 h after Gm treatment. Before each additional time point, wells were treated a second time with Gm. Each experiment was performed in triplicate, with each value shown being the geometric mean from at least three independent experiments. Error bars represent the standard errors of the means. *, P < 0.05 by ANOVA and the Tukey-Kramer method.
Association with and invasion of ME180 cervical epithelial cells.
To determine if the GGI affects the ability of meningococci to associate with or invade epithelial cells, we infected ME180 cervical epithelial cells with the wild type and an isogenic T4SS− mutant in two different backgrounds. ME180 cells were infected at an MOI of 100 for 3.5 h. There was no significant difference between either 01/241471 or ATCC 13102 and its respective T4SS− mutant for both association index and invasion index, indicating that the GGI and T4SS do not affect meningococcal adhesion or invasion of host cells under these conditions (see Fig. S1 in the supplemental material).
DISCUSSION
The presence of the gonococcal genetic island in some strains of N. meningitidis leads us to ask several intriguing questions. How did meningococci acquire the GGI? Do meningococci secrete T4SS substrates? Can GGI+ meningococci obtain iron via the T4SS? Does the presence of the GGI affect interactions between meningococci and host cells? In the process of answering these questions, we carefully mapped the GGI in seven N. meningitidis strains and found five distinct GGI types.
One way that N. meningitidis could have acquired the GGI is through natural transformation and homologous recombination with chromosomal DNA from N. gonorrhoeae. However, sequencing of the regions flanking the GGI in N. meningitidis strains suggests that this was not the method of GGI acquisition. N. meningitidis strains carry a gene, NMA1385, between difA and ung, but this gene is not found in N. gonorrhoeae. If the GGI were introduced into N. meningitidis strains by homologous recombination, then this gene would have been lost in GGI+ strains (Fig. 7). DNA sequencing revealed that this gene is present in all GGI+ N. meningitidis strains examined, suggesting that the GGI was not acquired by homologous recombination. Furthermore, sequencing of the dif sites suggests the GGI was acquired by site-specific recombination. As in gonococci, the GGI is flanked by a direct repeat, difA and difB, with difB carrying a few sequence changes relative to difA, but these sequence changes differ between N. gonorrhoeae and N. meningitidis, suggesting that the GGI was separately acquired by these two species or that the GGI was present in an ancestor of the pathogenic Neisseria species and has undergone different mutational events as these species evolved. The sequence changes in difB are found in the XerD-binding region (11, 14). Despite these mismatches to the consensus dif sequence, we were able to measure GGI loss from N. meningitidis. This result suggests that, as in gonococci, the dif sites can be acted on by XerCD to excise the GGI from the chromosome by site-specific recombination.
The GGI in meningococci varies significantly from strain to strain (Fig. 1). Five island types were identified, and these show great differences in size and gene content. Four of the five island types have insertion sequences present, and all of the island types have at least one deletion compared to the N. gonorrhoeae GGI sequences. Mc GGI type 1 is approximately 46 kb, compared with 57 kb in the gonococcal strain MS11. We did not find any insertion sequences in this island; however, there is an 11-kb deletion. The type 2 Mc GGI in ATCC 13102 has three separate insertions of IS1301 and a deletion of 8 kb. This island is approximately 51 kb. The type 3 island is about 56 kb. It has IS30 inserted in traK and has a deletion of the first 1.2 kb of the GGI, including part of traD. The island in α275, type 4, is 64 kb and contains numerous insertions, including an insertion of IS30 and umuC. UmuC is a subunit of DNA polymerase V which acts in SOS-activated DNA replication and mutagenesis (29). The type 4 GGI also has a deletion in the first 1.2 kb of the island, including yaa and the 3′ end of traD. The smallest island, type 5, is approximately 24 kb. There are two deletions, one spanning most of the type IV secretion genes that is 22 kb and another extending from yecB through parB that is 11 kb. IS1301 replaces the deleted type IV secretion genes in those strains. These data show that some GGIs are very similar to those of N. gonorrhoeae and may produce a type IV secretion system (Fig. 1 and 2). However, many of them have acquired multiple insertions and deletions and do not have the necessary components for a type IV secretion system. Through recombination and deletion, the genes are being mutated and lost. However, those that have all the type IV secretion genes appear not to have any mutations, suggesting that the type IV secretion system may function in these strains.
We measured DNA transfer in coculture for the GGI+ N. meningitidis strains that appeared to have intact type IV secretion systems. We detected transfer in coculture, but DNA donation occurred at the same frequency from traH strains as from the wild-type parental strains. These data suggest that the meningococcal strains do not donate DNA via type IV secretion. These data led us to wonder whether the oriT was present in these strains. DNA sequencing of the yaf-ltgX intergenic region revealed that the oriT is conserved. However, the sequence also identified a 2-bp deletion between the putative −35 and −10 hexamers in the promoter for the operon containing the genes encoding the relaxase TraI and putative coupling protein TraD (Fig. 4) (31). This deletion may significantly diminish transcription of these genes. The promoter for the other transcript from this region had no sequence changes, suggesting that meningococci may make the structural subunits of the type IV apparatus.
Previous studies showed that meningococci and gonococci require a functional Ton complex to survive intracellularly under iron-limiting conditions (13, 21). Recently, it was demonstrated that gonococci producing the type IV secretion system exhibit Ton-independent intracellular survival (42). The mechanism of survival is unknown, but these data suggest that the T4SS is active during intracellular growth. We examined meningococci for Ton-independent intracellular survival and found that T4SS presence did not confer Ton-independent survival, indicating that the T4SS likely functions differently in meningococci than in gonococci. We also tested those same strains and isogenic T4SS− mutants for differences in association with and invasion of host cells. We found that there were no significant differences between the parental and T4SS− strains. Thus, under the conditions tested, no differences in meningococcal infection were conferred by the T4SS.
The GGI is more common in meningococci than previously thought. We identified multiple island types, and it is likely that more types exist. Some GGIs had insertions of genes, such as umuC, and undiscovered islands probably have insertions of other interesting genes, some of which may contribute to meningococcal fitness. Approximately one-sixth of Gc strains lack the atlA gene, which is required for type IV secretion of DNA, and carry in its place eppA and a variant allele of traG, just as we have found here for Mc strains (10, 20, 28). Those eppA+ Gc strains do not secrete DNA (9), and we have found that the GGI+ Mc strains similarly do not show increased DNA donation for transformation (Fig. 8). The existence of these multiple Gc and Mc strains with this different T4SS gene arrangement suggests that this putative T4SS functions differently from the MS11 T4SS and has a function in the pathogenic Neisseria that is still to be determined.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH grants AI047958 and AI072605 to J.P.D.
We thank the Active Bacterial Core Surveillance (ABCs) Team and the Emerging Infection Programs (EIP) Network of the CDC, the Wisconsin State Laboratory of Hygiene, Meningococcal Reference Laboratory of the Manchester Public Health Laboratory Service, L. H. Harrison, and D. A. Caugant for providing meningococcal isolates used in this study. We thank Nadia Domínguez for pNMD37, pNMD42, and ND518.
Footnotes
Published ahead of print 24 February 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Bille E, et al. 2005. A chromosomally integrated bacteriophage in invasive meningococci. J. Exp. Med. 201:1905–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Blakely G, Colloms S, May G, Burke M, Sherratt D. 1991. Escherichia coli XerC recombinase is required for chromosomal segregation at cell division. New Biol. 3:789–798 [PubMed] [Google Scholar]
- 3. Blakely G, et al. 1993. Two related recombinases are required for site-specific recombination at dif and cer in E. coli K-12. Cell 75:351–361 [DOI] [PubMed] [Google Scholar]
- 4. Boschiroli ML, et al. 2002. The Brucella suis virB operon is induced intracellularly in macrophages. Proc. Natl. Acad. Sci. U. S. A. 99:1544–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Caugant DA, Hoiby EA, Rosenqvist E, Froholm LO, Selander RK. 1992. Transmission of Neisseria meningitidis among asymptomatic military recruits and antibody analysis. Epidemiol. Infect. 109:241–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Christie PJ. 1997. Agrobacterium tumefaciens T-complex transport apparatus: a paradigm for a new family of multifunctional transporters in eubacteria. J. Bacteriol. 179:3085–3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dillard JP. 2011. Genetic manipulation of Neisseria gonorrhoeae. Curr. Protoc. Microbiol. 23:4A.1.1–4A.1.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dillard JP, Hackett KT. 2005. Mutations affecting peptidoglycan acetylation in Neisseria gonorrhoeae and Neisseria meningitidis. Infect. Immun. 73:5697–5705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dillard JP, Salgado-Pabón W, Turner N, Kohler PL. 2008. DNA processing and secretion by the gonococcal type IV secretion system, abstr. O02. Abstr. 16th Int. Pathog. Neisseria Conf., Rotterdam, The Netherlands [Google Scholar]
- 10. Dillard JP, Seifert HS. 2001. A variable genetic island specific for Neisseria gonorrhoeae is involved in providing DNA for natural transformation and is found more often in disseminated infection isolates. Mol. Microbiol. 41:263–278 [DOI] [PubMed] [Google Scholar]
- 11. Domínguez NM, Hackett KT, Dillard JP. 2011. XerCD-mediated site-specific recombination leads to loss of the 57-kilobase gonococcal genetic island. J. Bacteriol. 193:377–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gerbase AC, Rowley JT, Mertens TE. 1998. Global epidemiology of sexually transmitted diseases. Lancet 351(Suppl. 3):2–4 [DOI] [PubMed] [Google Scholar]
- 13. Hagen TA, Cornelissen CN. 2006. Neisseria gonorrhoeae requires expression of TonB and the putative transporter TdfF to replicate within cervical epithelial cells. Mol. Microbiol. 62:1144–1157 [DOI] [PubMed] [Google Scholar]
- 14. Hamilton HL, Domínguez NM, Schwartz KJ, Hackett KT, Dillard JP. 2005. Neisseria gonorrhoeae secretes chromosomal DNA via a novel type IV secretion system. Mol. Microbiol. 55:1704–1721 [DOI] [PubMed] [Google Scholar]
- 15. Hamilton HL, Schwartz KJ, Dillard JP. 2001. Insertion-duplication mutagenesis of Neisseria: use in characterization of DNA transfer genes in the gonococcal genetic island. J. Bacteriol. 183:4718–4726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Immel SK, Hamilton HL, Dillard JP. 2003. Mutagenesis of novel genes in the type IV secretion system region of the gonococcal genetic island, abstr. D-142, p. 227 Abstr. 103rd Gen. Meet. Amer. Soc. Microbiol. American Society for Microbiology, Washington, DC. [Google Scholar]
- 17. Jain S, Kahnt J, van der Does C. 2011. Processing and maturation of the pilin of the type IV secretion system encoded within the gonococcal genetic island. J. Biol. Chem. 286:43601–43610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kellogg DS, Jr, Peacock WL, Jr, Deacon WE, Brown L, Pirkle CL. 1963. Neisseria gonorrhoeae. I. Virulence genetically linked to clonal variation. J. Bacteriol. 85:1274–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Klee SR, et al. 2000. Molecular and biological analysis of eight genetic islands that distinguish Neisseria meningitidis from the closely related pathogen Neisseria gonorrhoeae. Infect. Immun. 68:2082–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kohler PL, Hamilton HL, Cloud-Hansen K, Dillard JP. 2007. AtlA functions as a peptidoglycan lytic transglycosylase in the Neisseria gonorrhoeae type IV secretion system. J. Bacteriol. 189:5421–5428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Larson JA, Higashi DL, Stojiljkovic I, So M. 2002. Replication of Neisseria meningitidis within epithelial cells requires TonB-dependent acquisition of host cell iron. Infect. Immun. 70:1461–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lederberg J. 1951. Streptomycin resistance: a genetically recessive mutation. J. Bacteriol. 61:549–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Marra A, Blander SJ, Horwitz MA, Shuman HA. 1992. Identification of a Legionella pneumophila locus required for intracellular multiplication in human macrophages. Proc. Natl. Acad. Sci. U. S. A. 89:9607–9611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Morse SA, Bartenstein L. 1974. Factors affecting autolysis of Neisseria gonorrhoeae. Proc. Soc. Exp. Biol. Med. 145:1418–1421 [DOI] [PubMed] [Google Scholar]
- 25. Pachulec E. 2010. Ph.D. thesis. University of Groningen, Groningen, The Netherlands [Google Scholar]
- 26. Petering H, et al. 1996. Genes associated with meningococcal capsule complex are also found in Neisseria gonorrhoeae. J. Bacteriol. 178:3342–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8:785–786 [DOI] [PubMed] [Google Scholar]
- 28. Ramsey ME, Woodhams KL, Dillard JP. 2011. The gonococcal genetic island and type IV secretion in the pathogenic Neisseria. Front. Microbiol. 2:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Reuven NB, Arad G, Maor-Shoshani A, Livneh Z. 1999. The mutagenesis protein UmuC is a DNA polymerase activated by UmuD′, RecA, and SSB and is specialized for translesion replication. J. Biol. Chem. 274:31763–31766 [DOI] [PubMed] [Google Scholar]
- 30. Rosenstein NE, Perkins BA, Stephens DS, Popovic T, Hughes JM. 2001. Meningococcal disease. N. Engl. J. Med. 344:1378–1388 [DOI] [PubMed] [Google Scholar]
- 31. Salgado-Pabón W, et al. 2010. Increased expression of the type IV secretion system in piliated Neisseria gonorrhoeae variants. J. Bacteriol. 192:1912–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Salgado-Pabón W, Jain S, Turner N, van der Does C, Dillard JP. 2007. A novel relaxase homologue is involved in chromosomal DNA processing for type IV secretion in Neisseria gonorrhoeae. Mol. Microbiol. 66:930–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 34. Schulein R, Dehio C. 2002. The VirB/VirD4 type IV secretion system of Bartonella is essential for establishing intraerythrocytic infection. Mol. Microbiol. 46:1053–1067 [DOI] [PubMed] [Google Scholar]
- 35. Seifert HS. 1997. Insertionally inactivated and inducible recA alleles for use in Neisseria. Gene 188:215–220 [DOI] [PubMed] [Google Scholar]
- 36. Seifert HS, Chen EY, So M, Heffron F. 1986. Shuttle mutagenesis: a method of transposon mutagenesis for Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 83:735–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Snyder LAS, Jarvis SA, Saunders NJ. 2005. Complete and variant forms of the ‘gonococcal genetic island' in Neisseria meningitidis. Microbiology 151:4005–4013 [DOI] [PubMed] [Google Scholar]
- 38. Sparling PF. 2000. Biology of Neisseria gonorrhoeae. In Homes KK, Sparling PF, Lemon SM, Stamm WE, Piot P, Wasserheit JN. (ed), Sexually transmitted diseases, 2nd ed McGraw Hill, New York, NY [Google Scholar]
- 39. Swanson J, Kraus SJ, Gotschlich EC. 1971. Studies on gonococcus infection. I. Pili and zones of adhesion: their relation to gonococcal growth patterns. J. Exp. Med. 134:886–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vayssier-Taussat M, et al. 2010. The Trw type IV secretion system of Bartonella mediates host-specific adhesion to erythrocytes. PLoS Pathog. 6:e1000946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wright CJ, Jerse AE, Cohen MS, Cannon JG, Seifert HS. 1994. Nonrepresentative PCR amplification of variable gene sequences in clinical specimens containing dilute, complex mixtures of microorganisms. J. Clin. Microbiol. 32:464–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zola TA, Strange HR, Domínguez NM, Dillard JP, Cornelissen CN. 2010. Type IV secretion machinery promotes Ton-independent intracellular survival of Neisseria gonorrhoeae within cervical epithelial cells. Infect. Immun. 78:2429–2437 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.