Abstract
New Delhi metallo-β-lactamase 1 (NDM-1), which is associated with resistance to carbapenem, was first reported in 2008. A sensitive and rapid molecular assay to detect the plasmid blaNDM-1 in clinical isolates is needed to control its spread. We describe a loop-mediated isothermal amplification (LAMP) assay for the rapid detection of blaNDM-1 from pure culture and sputum, urine, and fecal samples. Eight sets of primers were designed to recognize six or eight distinct sequences on target blaNDM-1, and one set was selected as the most appropriate set of primers for its rapid detection. The specificity and sensitivity of the primers in the LAMP reactions for blaNDM-1 detection were determined. The sensitivity of the LAMP assay for blaNDM-1 detection in sputum, urine, and fecal samples was also tested. Two methods, namely, monitoring of turbidity and addition of calcein to the reaction tube, were used to determine negative and positive results. The results showed that target DNA was amplified and visualized by the two detection methods within 70 min at an isothermal temperature of 65°C. The sensitivity of LAMP, with a detection limit of 10.70 pg/μl DNA, was 100-fold greater than that of PCR. Thirteen infection bacterial strains without blaNDM-1 were selected for testing of specificity, and the results of the amplification were negative, which showed that the primers had good levels of specificity. The LAMP method reported here is demonstrated to be a potentially valuable means for the detection of blaNDM-1 and rapid clinical diagnosis, being fast, simple, and low in cost.
INTRODUCTION
Pathogens carrying the plasmid blaNDM-1 are known for their high rates of resistance to carbapenems and were first reported in August 2010. Kumarasamy et al. identified 44 isolates carrying New Delhi metallo-β-lactamase 1 (NDM-1) in Chennai, India; 26 in Haryana, India; 37 in the United Kingdom; and 73 in other sites in India and Pakistan (14). NDM-1 was mostly found among Escherichia coli and Klebsiella pneumoniae strains (6, 18) which were highly resistant to all antibiotics except tigecycline and colistin. NDM-1 was reported worldwide within 6 months (1, 25, 30; European Antimicrobial Resistance Surveillance Network database [http://ecdc.europa.eu/en/activities/surveillance/EARS-Net/Pages/Database.aspx]). Bacteria with resistance to carbapenems conferred by blaNDM-1 are potentially a major global health problem. The WHO has urged countries to implement infection control measures in hospitals to limit the spread of multidrug-resistant strains and to reinforce national policies on the prudent use of antibiotics (32). Thus, a sensitive and rapid method for detection of blaNDM-1 is needed to prevent its spread. We isolated Acinetobacter baumannii XM with blaNDM-1 for the first time in 2010 from an advanced lung cancer patient and used this strain in the present study.
Recently, real-time PCR assays for the rapid detection of blaNDM-1 have been reported (13). While these assays for rapid, sensitive, and specific detection appear to be promising, PCR requires specialized high-cost instruments and consumables. In addition, Taq DNA polymerase in PCR assays can be inactivated by inhibitors present in crude biological samples (5). Thus, another rapid, simple, and cost-effective assay is needed to complement current PCR methods. The recently developed loop-mediated isothermal amplification (LAMP) method requires only a temperature-controlled water bath. It is based on autocycling strand displacement DNA synthesis in the presence of Bst DNA polymerase under isothermal conditions within 1 h (19, 29). The LAMP method was evaluated and optimized for blaNDM-1 detection. Because four or six specific primers that recognize six or eight different sequences on the DNA target are used, LAMP amplifies DNA with a high specificity; and LAMP technology has been widely used in clinical diagnosis (6, 9, 21); qualitative and quantitative detection of epidemic bacteria (7, 20, 28, 34), viruses (10, 27, 31), and parasites (3, 12); as well as in fetal sex identification (8), among other applications. In the study described in this communication, we first designed eight sets of primers and optimized the LAMP assay for detection of blaNDM-1. Second, the specificity and sensitivity of the primers in the LAMP reactions for blaNDM-1 detection were determined. Finally, the sensitive LAMP assay for blaNDM-1 detection in sputum, urine, and fecal samples was tested.
MATERIALS AND METHODS
Bacterial strains and preparation of templates.
A total of 14 bacterial strains were used in this study, and their sources are listed in Table 1. Isolate A. baumannii XM was from an advanced lung cancer patient in Fujian Province and was responsible for China's first NDM-1 infection reported by the Academy of Military Medical Sciences. blaNDM-1 was validated by PCR-based sequencing, and the sequence of the blaNDM-1 gene showed 100% identity with the sequences of previously reported genes. blaNDM-1 was located on the chromosome by analysis of the pulsed-field gel electrophoresis (PFGE) profile and Southern blot hybridization (data not shown). These bacteria were cultured at 37°C in brain heart infusion (BHI) broth according to a standard protocol. The Chelex method was used to extract the bacterial whole genomic DNA (including plasmid; the same method was used for the assay described below) as follows: the bacterial pellet, in 200 μl phosphate-buffered saline (PBS), was mixed with an equal volume of Chelex DNA extraction buffer (25 mM NaOH, 10 mM Tris-HCl, 1% Triton X-100, 1% NP-40, 0.1 mM EDTA, 2% Chelex-100). The mixture was incubated at 100°C for 10 min and then immediately placed on ice. Finally, the mixture was centrifuged at 14,000 × g for 2 min. The supernatant was used as the template in both the LAMP assay and PCRs (17). In order to estimate the sensitivity and specificity of the LAMP assay under real conditions, pure genomic DNA was extracted from A. baumannii XM using a Wizard genomic DNA purification kit from Promega. The genomic DNA was then prepared by serial 10-fold dilutions to give concentrations ranging from 1,070 ng/μl to 0.107 pg/μl.
Table 1.
Bacterial strains used in this study
| Species | Source |
|---|---|
| Acinetobacter baumannii XM (with blaNDM-1) | Clinical isolate |
| A. baumannii H949 | Our microorganism center |
| A. baumannii F398 | Our microorganism center |
| A. baumannii B260 | Our microorganism center |
| A. baumannii H18 | Our microorganism center |
| Shigella sonnei 2531 | Our microorganism center |
| S. flexneri 4536 | Our microorganism center |
| Salmonella enteritidis 50326-1 | Our microorganism center |
| Vibrio carchariae 5732 | Our microorganism center |
| V. parahaemolyticus 5474 | Our microorganism center |
| S. enterica serotype Paratyphi 86423 | Our microorganism center |
| Enteroinvasive E. coli 44825 | Our microorganism center |
| Enterotoxigenic E. coli 44824 | Our microorganism center |
| Enteropathogenic E. coli 2348 | Our microorganism center |
Preparation of pure culture.
A 200-μl sample from an overnight culture of bacteria was subjected to centrifugation at 8,000 rpm for 10 min, and the supernatant was discarded. The bacterial cells were resuspended in 200 μl PBS, and then the Chelex method (discussed above) was used to extract the bacterial genomic DNA.
Preparation of sputum samples, urine samples, and stool samples.
Sputum samples, urine samples, and stool samples were collected from healthy donors. For sputum and urine samples, a 200-μl sample was extracted directly with the Chelex method. For stool samples, 100 mg of the fecal sample was suspended in 0.9 ml distilled water by vigorous shaking for 5 min. After 7 min, precipitation took place and 200 μl of supernatant was mixed with an equal volume of Chelex DNA extraction buffer to prepare the templates. Then, the pure genomic DNA extracted from A. baumannii XM was placed into the different sample templates and concentrations of pure genomic DNA were made up as follows: 107.0 ng/μl, 10.70 ng/μl, 1.070 ng/μl, 107.0 pg/μl, 10.70 pg/μl, 1.070 pg/μl, 0.107 pg/μl, and 0.010 pg/μl.
Primer design.
To design blaNDM-1-specific LAMP primers, the sequence of blaNDM-1 with accession number FN396876 was downloaded from the NCBI GenBank database (35). The sequence was further analyzed by Primer Explorer (version 4) software (http:/primerexplorer.jp/lamp), and the outer forward primer (F3), outer backward primer (B3), forward inner primer (FIP), and backward inner primer (BIP) (19) were designed. An additional two loop primers (loop F and loop B) were designed to accelerate the amplification reaction. To obtain the most appropriate primers, we designed eight sets of primers. The FIP and BIP primers recognize both sense and antisense strands and were linked by a four-thymidine spacer (TTTT). To compare the sensitivity and specificity of PCR, normal PCR was performed with the primers named NDM1-F and NDM1-R. The primers were synthesized commercially (Beijing AUGCT DNA-SYN Biotechnology Co., Ltd., Beijing, China).
LAMP reaction.
The LAMP reactions were carried out in 25-μl reaction mixtures (DNA amplification kit; Eiken Chemical Co., Ltd., Tochigi, Japan) containing the following reagents (final concentrations): 20 mM Tris-HCl (pH 8.8), 10 mM KCl, 10 mM (NH4)2SO4, 0.1% Tween 20, 0.8 M betaine, 8 mM MgSO4, 1.4 mM each deoxynucleoside triphosphate, and 8 U Bst DNA polymerase. The amount of primer needed for one reaction was 40 pmol for FIP and BIP, 20 pmol for LF and LB, and 5 pmol for F3 and B3. Finally, an appropriate amount of template genomic DNA was added to the reaction tube. The reaction was carried out in the reaction tube (Reaction Tube; Eiken Chemical Co., Ltd., Tochigi, Japan) at 65°C for 50 min and inactivated at 80°C for 5 min in dry bath incubators.
Detection of LAMP products.
Two different methods were used to detect LAMP products. For direct visual inspection, 1 μl of calcein (fluorescent detection reagent; Eiken Chemical Co., Ltd., Tochigi, Japan) was added to 25 μl of LAMP products before the LAMP reaction. For a positive reaction, the color changed from orange to green, while a negative reaction failed to turn green and remained orange. The color change could be observed by the naked eye under natural light or with the aid of UV light at 365 nm. For monitoring of turbidity (15), real-time amplification by the LAMP assay was monitored through spectrophotometric analysis by recording the optical density at 400 nm every 6 s with the help of a Loopamp real-time turbidimeter (LA-230; Eiken Chemical Co., Ltd., Tochigi, Japan).
PCR detection.
PCRs were carried out with 25-μl reaction mixtures containing 12.5 μl PCR master mix reagents (Tiangen Biotech Co., Ltd., Beijing, China), 1 μmol/liter NDM1-F and NDM1-R primers, and the same amount of DNA template used in the LAMP reaction. The reaction was initially carried out at 94°C for 2 min, followed by 35 cycles at 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s. The final extension step was carried out at 72°C for 10 min. The PCR-amplified products were analyzed by 2% agarose gel (Amresco) electrophoresis and stained with ethidium bromide. Images were documented by a Bio-Rad Gel Doc EQ imaging system.
RESULTS
The most appropriate primers for rapid detection of blaNDM-1.
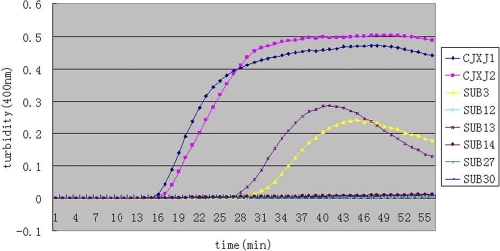
We designed eight sets of primers for detection of blaNDM-1. Under the same reaction conditions, we observed that four turbidity curves occurred after reaction for 15 min, that four of eight sets amplified the target sequence, and that the primers in the CJXJ1 set, which could amplify the target gene in the shortest time, were the fastest and the most optimal reaction primers (Fig. 1). Our data show that the use of loop primers shortened the amplification time by approximately one-third, which agreed with reported data (16). The primers in the CJXJ1 primer set (Table 2) were chosen as the final primers for blaNDM-1 detection by LAMP.
Fig 1.
Eight sets of primers amplified the target gene under the same conditions. Turbidity was monitored by a Loopamp real-time turbidimeter at 400 nm every 6 s. Assays with the CJXJ1 and CJXJ2 primer sets were performed with Loop primers; assays with the other sets were performed without Loop primers.
Table 2.
Sequence of primers used for specific amplification of blaNDM-1
| Primer | Type | Sequence (5′–3′) |
|---|---|---|
| CJXJ1F3 | Forward outer | GCATAAGTCGCAATCCCCG |
| CJXJ1B3 | Backward outer | GGTTTGATCGTCAGGGATGG |
| CJXJ1FIP | Forward inner | CTGGCGGTGGTGACTCACGTTTTGCATGCAGCGCGTCCA |
| CJXJ1BIP | Backward inner | CGCGACCGGCAGGTTGATCTTTTGGTCGATACCGCCTGGAC |
| CJXJ1LF1 | Loop forward | GCATCAGGACAAGATGGGC |
| CJXJ1LB1 | Loop backward | TCCAGTTGAGGATCTGGGC |
| NDM1-F | PCR forward | CAGCACACTTCCTATCTC |
| NDM1-R | PCR backward | CCGCAACCATCCCCTCTT |
Temperature of blaNDM-1 LAMP reaction.
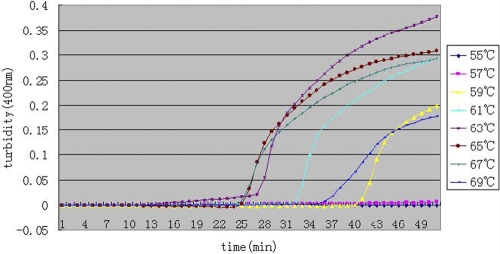
To optimize the reaction conditions of the primers for blaNDM-1 in the LAMP reaction, we observed different temperatures from 55°C to 69°C at 2°C intervals. As shown in Fig. 2, 63°C to 67°C is the most suitable reaction temperature range. Finally, we chose 65°C as the reaction temperature.
Fig 2.
Different temperatures of the LAMP reaction for detection of NDM-1. Turbidity was monitored by a Loopamp real-time turbidimeter at 400 nm every 6 s.
Specificity of NDM-1 LAMP.
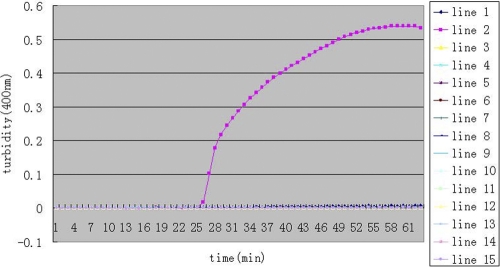
To test the specificity of LAMP for blaNDM-1, we used A. baumannii XM with blaNDM-1 as the positive strain and distilled water as the negative control. Thirteen infectious bacterial strains without blaNDM-1 were selected. As depicted in Fig. 3, we observed that the increased turbidity curve appeared only when A. baumannii XM with blaNDM-1 was used as the template and not with the negative control (double-distilled water) and other bacterial species, including A. baumannii H949, A. baumannii F398, A. baumannii B260, A. baumannii H18, S. sonnei 2531, S. flexneri 4536, S. enterica serotype Enteritidis 50326-1, S. enterica serotype Paratyphi 86423, enteroinvasive E. coli 44825, enterotoxigenic E. coli 44824, enteropathogenic E. coli 2348, V. carchariae 5732, and V. parahaemolyticus 5474. These results suggest that these primers could be used to detect blaNDM-1.
Fig 3.
Specificity of the LAMP reaction for detection of blaNDM-1. Turbidity was monitored by a Loopamp real-time turbidimeter at 400 nm every 6 s. Amplification was performed at 65°C for 65 min. Lines: 1, negative control (double-distilled water); 2, A. baumannii XM; 3, A. baumannii H949; 4, A. baumannii F398; 5, A. baumannii B260; 6, A. baumannii H18; 7, S. sonnei 2531; 8, S. flexneri 4536; 9, S. enterica serotype Enteritidis 50326-1; 10, V. carchariae 5732; 11, S. enterica serotype Paratyphi 86423; 12, enteroinvasive E. coli 44825; 13, enterotoxigenic E. coli 44824; 14, enteropathogenic E. coli 2348; and 15, V. parahaemolyticus 5474.
Sensitivity of NDM-1 LAMP.
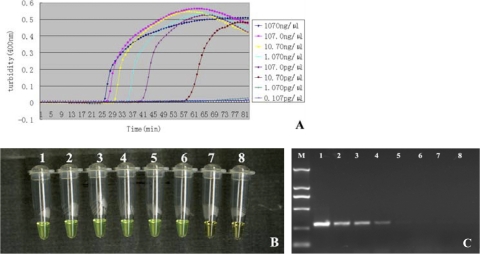
To determine the sensitivity of primers in LAMP detection of blaNDM-1, pure genomic DNA was extracted from A. baumannii XM using a Wizard genomic DNA purification kit, and the genomic DNA was subject to serial 10-fold dilutions to give concentrations ranging from 1,070 ng/μl to 0.107 pg/μl. As shown in Fig. 4A, the detection limit of the LAMP assay for blaNDM-1 was 10.70 pg/μl. Moreover, we monitored the results of the blaNDM-1 analysis by LAMP by a direct visual method. Before the LAMP reaction, we added 1 μl of fluorescent detection reagent to 25 μl of the LAMP reaction mixture. When the reaction was finished, all positive reactions turned green, while the negative ones remained orange (Fig. 4B). Therefore, we concluded that these two detection methods had the same sensitivity. For comparison purposes, PCR using the NDM1-F and NDM1-R primers with the same amount of A. baumannii with blaNDM-1 was also carried out. We observed that the detection limit for PCR was 1.07 ng/μl (Fig. 4C).
Fig 4.
Comparison of sensitivity between the LAMP reaction and PCR for detection of the blaNDM-1 gene. The pure genomic DNA extracted from A. baumannii XM was diluted in a serial 10-fold dilution. Both LAMP reactions (A and B) and PCRs (C) were carried out in duplicate for each dilution point. Tubes and lanes: 1, 1,070 ng/μl; 2, 107.0 ng/μl; 3, 10.70 ng/μl; 4, 1.070 ng/μl; 5, 107.0 pg/μl; 6, 10.70 pg/μl; 7, 1.070 pg/μl; 8, 0.107 pg/μl. (A) Turbidity was monitored by a Loopamp real-time turbidimeter at 400 nm every 6 s; (B) 1 μl of fluorescent detection reagent was added to 25 μl of LAMP reaction mixture before the LAMP reaction; (C) the PCR products were analyzed by 2% agarose gel electrophoresis and stained with ethidium bromide.
Evaluation of blaNDM-1 LAMP with sputum, urine, and fecal samples.
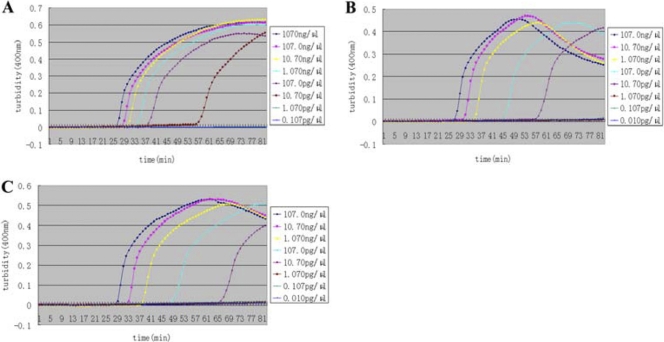
The blaNDM-1 analysis with the LAMP assay was then further evaluated with sputum, urine, and fecal samples. The preparation of three samples was described previously. We can see from Fig. 5A to C that, compared with the pure samples (Fig. 4A), the sensitivity for the simulated samples was unchanged (10.70 pg/μl). We conclude that LAMP is less affected by extraneous components in samples than is PCR.
Fig 5.
Detection of the blaNDM-1 gene in simulated sputum samples (A), simulated urine samples (B), and simulated fecal samples (C) by a Loopamp real-time turbidimeter at 400 nm every 6 s. The concentration of pure genomic DNA extracted from A. baumannii XM in each simulated sputum sample is shown.
Clinical sample detection.
A total of 336 clinical samples and clinical swabs of the environment in intensive care units were collected for LAMP-based surveillance of blaNDM-1 from 7 hospitals. We detected the other three strains with blaNDM-1, which were A. lwoffii SJH, Stenotrophomonas maltophilia JKYJ-01, and Enterococcus faecalis PIJ. These results were validated by PCR-based sequencing, and the sequences of the blaNDM-1 genes showed 100% identity with those of previously reported genes. These three NDM-1-producing strains were resistant to carbapenems, cephalosporins, and the β-lactam inhibitor combinations tested and susceptible to colistin. Transfer of blaNDM-1 from the three isolates was done with E. coli J53, the clinical strains of S. flexneri 4536 and S. enterica serotype Enteritidis as recipients. However, no transconjugant was observed. The genetic locations of the blaNDM-1 in the three strains were determined by analysis of PFGE profiles and Southern blot hybridization. The data clearly showed that blaNDM-1 is on the chromosome in A. lwoffii SJH, S. maltophilia JKYJ-01, and E. faecalis PIJ (data not shown). Further experiments are in progress to investigation the molecular nature of these strains. These findings prove the validity of the method that we have established. We are now working on larger-scale screening using the LAMP method in the hope of better understanding NDM-1 prevalence.
DISCUSSION
In the past several years, numerous multiple-drug-resistant bacteria have been identified: methicillin-resistant Staphylococcus aureus, the so-called ESKAPE organisms (an acronym for Enterococcus faecium, S. aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species), and others. They are notorious because of their resistance to multiple antimicrobial agents rather than enhanced virulence or pathogenicity. More recently, blaNDM-1, which is a new mobile class B enzyme or metallo-β-lactamase, has emerged and has added to the antibiotic resistance problem. Bacteria with resistance to carbapenem conferred by New Delhi metallo-β-lactamase 1 (NDM-1) are now becoming a major global health problem. The majority of cases have been reported in India (14) and Pakistan (23). Cases have also been reported in Australia (25), Greece (30), Canada (22), Singapore (2), and the United States (1) and, according to recent reports, in China (36), Japan (4), Kenya (26), Oman (24), and China's Taiwan region (33). Development of a sensitive and reliable test for NDM-1 is therefore a priority for early diagnosis and control.
To meet this challenge, we evaluated and optimized a novel LAMP assay for blaNDM-1 detection that was able to specifically detect blaNDM-1 in pathogens that carry blaNDM-1 within 90 min, including DNA extraction. In evaluating the sensitivity of the LAMP assay for blaNDM-1 detection, we observed that the LAMP assay was 100-fold more sensitive than the PCR assay. In addition, the PCR is carried out under temperature-cycling conditions, which is time-consuming, and the reaction also depends on the high precision of the PCR instruments. Compared to PCR, the LAMP reaction was carried out in a constant-temperature environment, and it does not require temperature cycling, so a temperature-controlled water bath or other device that can heat stably is sufficient. Moreover, LAMP reaction primers specifically recognize target sequences of four or six of the six or eight independent target sequence regions, whereas PCR primers recognize target sequences of two independent regions. The specificity and sensitivity are thereby greatly enhanced, and the probability of false-positive results is decreased. Kaneko et al. found that the LAMP reaction is not susceptible to the influence of different components in clinical samples, and so purification of DNA from the sample is not necessary (11). On the other hand, the sensitivity of the PCR can be greatly reduced in the presence of exogenous DNA and inhibitors. Therefore, the LAMP method is more suitable than PCR for rapid detection of blaNDM-1 in clinical samples. Although the amplification principle of the LAMP method is complex, it is a simple procedure whose rapidity, high sensitivity, and specificity make it suitable for blaNDM-1 detection, especially for routine diagnostic and infection control purposes.
Although there are many advantages of LAMP assay (19) (LAMP amplifies DNA with high efficiency under isothermal conditions, LAMP is highly specific for the target sequence, and LAMP is simple and easy to perform), it shows a high rate of false-positive results in its current performance. This is because the amplification efficiency of the LAMP assay is extremely high and the LAMP assay is capable of synthesizing 20 μg of specific DNA in a 25-μl reaction mixture within 60 min (15). Strict spatial separation of reagent preparation and performance of the test is very necessary to avoid contamination. At present, we added low-melting-point paraffin wax to the reaction tubes, after adding reaction solution, to prevent the spread of amplification products. Now it seems that this approach works well for avoidance of contamination.
In conclusion, a specific, sensitive, rapid, and cost-effective LAMP assay for blaNDM-1 detection in pathogens was established. The LAMP assay will be very useful for rapid detection of blaNDM-1 in primary health care units.
ACKNOWLEDGMENTS
We are indebted to the 174th Hospital of PLA for kindly providing Acinetobacter baumannii XM and helpful information. We are grateful to Christian Riedel for advice on the preparation of the manuscript and Fengjing Li for technical assistance and helpful discussions.
We have no conflicts of interest to declare.
This work was supported by grants from the National Natural Science Foundation of China (no. 81071321 and no. 81071399) and a grant from the Mega-Projects of Science and Technology Research of China (grant no. 2011ZX10004-001) to Jing Yuan.
Footnotes
Published ahead of print 22 February 2012
REFERENCES
- 1. Centers for Disease Control and Prevention 2010. Detection of Enterobacteriaceae isolates carrying metallo-beta-lactamase United States, 2010. MMWR Morb. Mortal. Wkly. Rep. 59:750. [PubMed] [Google Scholar]
- 2. Chan HL, Poon LM, Chan SG, Teo JW. 2011. The perils of medical tourism: NDM-1-positive Escherichia coli causing febrile neutropenia in a medical tourist. Singapore Med. J. 52:299–302 [PubMed] [Google Scholar]
- 3. Chen R, et al. 2011. Loop-mediated isothermal amplification: rapid detection of Angiostrongylus cantonensis infection in Pomacea canaliculata. Parasit. Vectors 4:204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chihara S, et al. 2011. First case of New Delhi metallo-beta-lactamase 1 producing Escherichia coli infection in Japan. Clin. Infect. Dis. 52:153–154 [DOI] [PubMed] [Google Scholar]
- 5. de Franchis R, Cross NC, Foulkes NS, Cox TM. 1988. A potent inhibitor of Taq polymerase copurifies with human genomic DNA. Nucleic Acids Res. 16:10355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Endo S, et al. 2004. Detection of gp43 of Paracoccidioides brasiliensis by the loop-mediated isothermal amplification (LAMP) method. FEMS Microbiol. Lett. 234:93–97 [DOI] [PubMed] [Google Scholar]
- 7. Hara-Kudo Y, et al. 2005. Loop-mediated isothermal amplification for the rapid detection of Salmonella. FEMS Microbiol. Lett. 253:155–161 [DOI] [PubMed] [Google Scholar]
- 8. Hirayama H, et al. 2006. Rapid sexing of water buffalo (Bubalus bubalis) embryos using loop-mediated isothermal amplification. Theriogenology 66:1249–1256 [DOI] [PubMed] [Google Scholar]
- 9. Ihira M, et al. 2004. Rapid diagnosis of human herpesvirus 6 infection by a novel DNA amplification method, loop-mediated isothermal amplification. J. Clin. Microbiol. 42:140–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Imai M, et al. 2006. Development of H5-RT-LAMP (loop-mediated isothermal amplification) system for rapid diagnosis of H5 avian influenza virus infection. Vaccine 24:6679–6682 [DOI] [PubMed] [Google Scholar]
- 11. Kaneko H, Kawana T, Fukushima E, Suzutani T. 2007. Tolerance of loop-mediated isothermal amplification to a culture medium and biological substances. J. Biochem. Biophys. Methods 70:499–501 [DOI] [PubMed] [Google Scholar]
- 12. Kong QM, et al. 2012. Loop-mediated isothermal amplification (LAMP): early detection of Toxoplasma gondii infection in mice. Parasit. Vectors 5:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Krüttgen A, Razavi S, Imöhl M, Ritter K. 2011. Real-time PCR assay and a synthetic positive control for the rapid and sensitive detection of the emerging resistance gene New Delhi metallo-β-lactamase-1 (blaNDM-1). Med. Microbiol. Immunol. 200:137–141 [DOI] [PubMed] [Google Scholar]
- 14. Kumarasamy KK, et al. 2010. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological and epidemiological study. Lancet Infect. Dis. 10:597–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mori Y, et al. 2001. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem. Biophys. Res. Commun. 289:150–154 [DOI] [PubMed] [Google Scholar]
- 16. Nagamine K, et al. 2002. Accelerated reaction by loop-mediated isothermal amplification using loop primers. J. Mol. Cell. Probes 16:223. [DOI] [PubMed] [Google Scholar]
- 17. Nagamine K, Watanabe K, Ohtsuka K, Hase T, Notomi T. 2001. Loop-mediated isothermal amplification reaction using a nondenatured template. Clin. Chem. 47:1742–1743 [PubMed] [Google Scholar]
- 18. Nordmann P, Cuzon G, Naas T. 2009. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect. Dis. 9:228–236 [DOI] [PubMed] [Google Scholar]
- 19. Notomi T, et al. 2000. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28:E63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ohtsuka K, et al. 2005. Detection of Salmonella enterica in naturally contaminated liquid eggs by loop-mediated isothermal amplification, and characterization of Salmonella isolates. Appl. Environ. Microbiol. 71:6730–6735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Okafuji T, et al. 2005. Rapid diagnostic method for detection of mumps virus genome by loop-mediated isothermal amplification. J. Clin. Microbiol. 43:1625–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Peirano G, et al. 2011. The characteristics of NDM-producing Klebsiella pneumoniae from Canada. Diagn. Microbiol. Infect. Dis. 71:106–109 [DOI] [PubMed] [Google Scholar]
- 23. Perry JD, et al. 2011. Prevalence of faecal carriage of Enterobacteriaceae with NDM-1 carbapenemase at military hospitals in Pakistan, and evaluation of two chromogenic media. J. Antimicrob. Chemother. 66:2288–2294 [DOI] [PubMed] [Google Scholar]
- 24. Poirel L, et al. 2011. NDM-1-producing Klebsiella pneumoniae isolated in the Sultanate of Oman. J. Antimicrob. Chemother. 66:304–306 [DOI] [PubMed] [Google Scholar]
- 25. Poirel L, Lagrutta E, Taylor P, Pham J, Nordmann P. 2010. Emergence of metallo-β-lactamase NDM-1-producing multi drug resistant Escherichia coli in Australia. Antimicrob. Agents Chemother. 54:4914–4916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Poirel L, Revathi G, Bernabeu S, Nordmann P. 2011. Detection of NDM-1-producing Klebsiella pneumoniae in Kenya. Antimicrob. Agents Chemother. 55:934–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Poon LL, et al. 2004. Rapid detection of the severe acute respiratory syndrome (SARS) corona virus by a loop-mediated isothermal amplification assay. Clin. Chem. 50:1050–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Song T, et al. 2005. Sensitive and rapid detection of Shigella and enteroinvasive Escherichia coli by a loop-mediated isothermal amplification method. FEMS Microbiol. Lett. 243:259–263 [DOI] [PubMed] [Google Scholar]
- 29. Ushikubo H. 2004. Principle of LAMP method—a simple and rapid gene amplification method. Uirusu 54:107–112 (In Japanese.) [DOI] [PubMed] [Google Scholar]
- 30. Vatopoulos A. 2008. High rates of metallo-beta-lactamase-producing Klebsiella pneumoniae in Greece: a review of the current evidence. Euro Surveill. 13(4):pii=8023 [PubMed] [Google Scholar]
- 31. Wang Y, et al. 2011. Rapid detection of newly isolated Tembusu-related Flavivirus by reverse-transcription loop-mediated isothermal amplification assay. Virol. J. 8:553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. World Health Organization WHO urges measures to combat antimicrobial resistance. World Health Organization, Geneva, Switzerland: http://www.who.int/mediacentre/news/releases/2010/amr_20100820/en/index.html [Google Scholar]
- 33. Wu HS, et al. 2010. First identification of a patient colonized with Klebsiella pneumoniae carrying blaNDM-1 in Taiwan. J. Chin. Med. Assoc. 73:596–598 [DOI] [PubMed] [Google Scholar]
- 34. Yeh HY, et al. 2005. Evaluation of a loop-mediated isothermal amplification method for rapid detection of channel fish Ictalurus punctatus important bacterial pathogen Edwardsiella ictaluri. J. Microbiol. Methods 63:36–44 [DOI] [PubMed] [Google Scholar]
- 35. Yong D, et al. 2009. Characterization of a new metallo-β-lactamase gene, NDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53:5046–5054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhou Z, et al. 2012. Identification of New Delhi metallo-β-lactamase gene (NDM-1) from a clinical isolate of Acinetobacter junii in China. Can. J. Microbiol. 58:112–115 [DOI] [PubMed] [Google Scholar]