Abstract
Endometrial stromal tumors are divided into three types: benign stromal nodules, endometrial stromal sarcomas, and undifferentiated endometrial sarcomas. A variety of cytogenetic abnormalities involving chromosome 7 have been reported in endometrial stromal sarcomas, including a recurrent t(7;17)(p15;q21). We have identified two zinc finger genes, which we have termed JAZF1 and JJAZ1, at the sites of the 7p15 and 17q21 breakpoints. Analyses of tumor RNA indicate that a JAZF1/JJAZ1 fusion is present in all types of endometrial stromal tumors; however, the fusion appears to be rarer among endometrial stromal sarcomas that would be considered high-grade according to certain classification schemes. These findings suggest that the less malignant endometrial stromal tumors may evolve toward more malignant types, but that some endometrial stromal sarcomas with relatively abundant mitotic activity may compose a biologically distinct group.
Endometrial stromal tumors of the uterus constitute a spectrum of neoplasms that exhibit varying degrees of malignancy and can present a number of challenges with respect to diagnosis and classification (1). Stromal nodules lie at the benign end of this spectrum of neoplasms. These nodules consist of well-circumscribed tumors composed of uniform cells resembling those of normal endometrial stroma during the proliferative phase of the menstrual cycle. Endometrial stromal sarcomas (ESSs) are malignant neoplasms that occupy the middle of the spectrum and are histologically similar to stromal nodules except for infiltration of the myometrium and/or vascular invasion. Tumors that depart significantly in histologic appearance from normal endometrial stroma are referred to as undifferentiated endometrial sarcomas (or undifferentiated uterine sarcomas) and represent the most malignant end of this tumor spectrum.
In the older literature, all malignant endometrial stromal tumors were categorized as ESSs and were subclassified into low-grade and high-grade types (2, 3). High-grade ESSs were distinguished from low-grade ESSs by an increased frequency of mitoses (>10 per high power microscopic field) and were generally assumed to have a worse prognosis. More recently, it has been argued that the number of mitoses within ESSs is largely irrelevant to outcome, which is said to be almost exclusively a function of stage at diagnosis (4–6).
Diagnosis and classification of endometrial stromal tumors has until now been based primarily on histologic criteria. In recent years, specific genetic alterations identified in different types of human tumors have provided useful diagnostic markers, led to insights into the basic biology of both neoplastic and normal tissues, and increasingly contributed to the development of rational forms of cancer therapy (7–9). With these considerations in mind, we have characterized the DNA and genes surrounding the breakpoints of a recurrent chromosomal translocation, the t(7;17)(p15;q21), reported in several cases of low-grade ESS (10–13). We have found that recombination at these breakpoints results in fusion of two previously unknown genes, which we have termed JAZF1 and JJAZ1. Fusion of these genes appears to be common in low-grade ESSs but is not limited to these neoplasms and can be found in other types of endometrial stromal tumors as well. However, the incidence of this fusion appears to be reduced among ESSs that might be classified as high-grade.
Materials and Methods
Tissues and Cells.
Initial molecular genetic studies were performed on four cases of low-grade ESS found to have the t(7;17)(p15;q21)(see Table 1 for the full karyotypes). In case BWH-42, the tissue was obtained from a metastatic retroperitoneal mass in a 58-yr-old woman who had undergone a hysterectomy for a low-grade ESS 16 years earlier; in case BWH-665, from a metastatic vaginal mass in a 69-yr-old woman who had undergone a hysterectomy for a low-grade ESS 18 years earlier; and in both case LU-550 and case LU-965, from primary uterine tumors in 41-yr-old women. Cells cultured directly from the tumors were grown in RPMI medium 1640 supplemented with 20% FCS, Mito + Serum Extender (Becton Dickinson), and bovine pituitary extract (Becton Dickinson) under standard conditions.
Table 1.
Karyotypes of low-grade ESSs initially studied
| Case | Karyotype |
|---|---|
| BWH-42 | 46, XX, t(7;13)(p15;p13), t(7;17)(p15;q21) |
| BWH-665 | 46, XX, t(7;17)(p15;q21) |
| LU-550 | 46, XX, t(7;17)(p15;q21) |
| LU-954 | 45, XX, −7, t(7;17)(p15;q21) |
Yeast Artificial Chromosomes (YACs), Bacterial Artificial Chromosomes (BACs), and cDNAs.
YACs and BACs were purchased from Research Genetics (Huntsville, AL). Methods for growth of yeast and bacteria, as well as procedures for isolation of YAC and BAC DNA, were as previously described (14, 15). cDNAs were identified in lambda bacteriophage libraries prepared from human fetal brain (Stratagene) and human umbilical vein endothelial cells (HUVEC) (16) by hybridization of probes to plaque lifts on nylon membranes. Phage DNA was isolated from clones in the HUVEC library according to standard methods (17), and phagemid cDNA clones were rescued from the brain library according to the supplier's instructions. Plasmids containing expressed sequence tag cDNAs (ESTs) were purchased from Genome Systems (St. Louis). Plasmid DNA was purified from bacteria following standard protocols (18).
Fluorescence in Situ, Southern Blot, and Northern Blot Hybridization.
DNA probes for fluorescence in situ hybridization (FISH) were labeled by nick translation with biotin-tagged nucleoside triphosphates by using the BioPrime labeling system (Life Technologies). Hybridization was performed on slides of metaphase chromosomes prepared according to methods of Fletcher and colleagues (15, 19). Procedures for Southern blot and Northern blot hybridization have been described (15).
Inverse PCR (I-PCR) and Reverse Transcription (RT)-PCR.
I-PCR was performed following protocols previously described (15). For RT-PCR, total RNA was transcribed into cDNA with Superscript II (Life Technologies), using either 7AntisenseOuter or 17AntisenseOuter as a primer (see below). The resulting cDNA was subjected to two rounds of PCR using first the “Outer” and then the “Inner” set of primers. The JAZF1 primers were 7SenseOuter 5′-CCACAGCAGTGGAAGCCTTA-3′, 7AntisenseOuter 5′-GCTTCTCTTCCCCTCCATTCAT-3′, 7SenseInner 5′-ATCACCCCCTCCTCTTCATT-3′, and 7AntisenseInner 5′-GGACTCATCGCTGTCCGACT-3′. The JJAZ1 primers were 17SenseOuter 5′-GTTACTCGGCCTCCTCCTCCTC-3′, 17AntisenseOuter 5′-GGTTCAAATTCATTACTGGAAACTGC-3′, 17SenseInner 5′-GAGCTTTTCCTCCAGGCCTTTG-3′, and 17AntisenseInner 5′-CCGGGTTTTGTTTGATTGAGG-3′. Specific primers for glyceraldehyde 3-phosphate dehydrogenase (5′-CACATCGCTCAGACACCATG-3′ and 5′-GCCATGGAATTTGCCATGGG-3′) were used to assess the quality of the input RNA.
For RT-PCR of RNA from formalin-fixed, paraffin-embedded tissue, 10 serial 10-μm tissue sections were deparaffinized with three washes in 10 ml of xylene, washed twice with 100% EtOH, and then digested for 16 h at 60°C in 2 ml 1× digestion solution (20 mM Tris⋅HCl, pH 8.0/20 mM Na2EDTA/2% SDS/2.5 mg/ml proteinase K). Eight milliliters of Trizol was added, and the RNA was isolated according to the supplier's instructions. First strand synthesis and nested PCR were performed with the following primers: FusionOutF 5′-CACGCCACAGCAGTGGAAGC-3′, FusionOutR 5′-TTTGTTCTGGAGTTTCGATGAGACA-3′, FusionInnerF 5′-CCCACCCATCACCCCCTCCT-3′, and FusionInnerR 5′-GGTGCTATGAGATTCCGAGTTCGAAGA-3′. A 10-μm tissue section adjacent to those used for extraction of RNA was examined histologically.
Determination of Nucleotide Sequence and Computer-Assisted Analysis of Derived Sequence.
Sequence data were obtained by using the Amplicycle Sequencing Kit (Perkin–Elmer) and analyzed by using the National Center for Biotechnology Information (NCBI) blast server and the gcg software package (Genetics Computer Group, Madison, WI). Iterated sequence was suppressed in analyses for genes by use of the repeatmasker2 software program available at the MRC Mammalian Genetics Unit WWW server (http://www.mgu.har.mrc.ac.uk/). Peptides were analyzed by using software available at the Expert Protein Analysis System (ExPASy) WWW server (http://www.expasy.ch/).
Results
Identification of a YAC Spanning the Chromosome 7 Breakpoint.
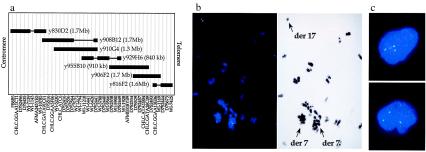
FISH was performed on metaphase preparations from the tumor of case BWH-42, using as probes YACs from the Whitehead Centre d'Étude du Polymorphisme Humain (CEPH) megaYAC contig 7.3 in the region of the HOXA gene cluster (Fig. 1a). FISH with YAC y830D2 produced hybridization signals limited to both derivative 7 (der7) chromosomes in this tumor, whereas the analysis with YAC y816F12 demonstrated hybridization to the der13 and der17 chromosomes. FISH with YACs y910G4 and y929H6 demonstrated hybridization signals on the der17 and the der7 of the t(7;13). These results indicated that the chromosome 7 breakpoints of both the t(7;17) and the t(7;13) of BWH-42 map within the 7.3 contig between YACs y830D2 and y816F12 but that the breakpoints are separated by more than 1 megabase. Furthermore, the chromosome 7 breakpoint in the t(7;17) lies between DNA contained within y830D2 and y910G4.
Figure 1.
Analysis of YACs in the vicinity of the 7p15 breakpoint. (a) Map of YAC clones at the 7p15 site. STSs are shown at the top without reference to the genetic distances between them. Solid, boxed areas indicate the presence of the STS within the DNA of the YAC; horizontal lines indicate the absence of the STS, presumably because of a deletion in the YAC insert. (b) FISH analysis of tumor BWH-42 with YAC probes derived from chromosome 7. Chromosomes and cells were hybridized with biotin-labeled DNA, and hybridization was detected with avidin-conjugated FITC. (Left) FISH was performed with YAC y908B12 and the chromosomal DNA counterstained with 4′,6-diamidino-2-phenylindole (DAPI). The hybridization signal is split between two derivative 7 chromosomes–that of the t(7;17) to the left and of the t(7;13) to the right–and one derivative 17 chromosome, as identified in the contrast-enhanced image of the DAPI stain on the right. (c) FISH performed with the y908B12 probe on interphase nuclei isolated from paraffin-embedded tumor in case BWH-665.
YAC y908B12, which contains DNA between clones y830D2 and y910G4, was then used as a probe and showed hybridization to both der7 chromosomes and the der17 chromosome of BWH-42, thereby demonstrating that y908B12 spans the chromosome 7 breakpoint of the t(7;17) (Fig. 1b). Additionally, analysis with YAC y908B12 of interphase nuclei from a second case of low-grade ESS, BWH-665, produced three hybridization signals (Fig. 1c).
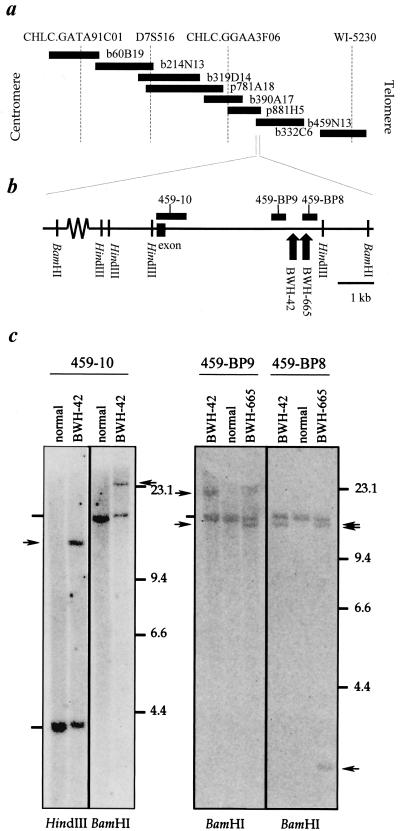
Construction of a BAC Contig Across the Chromosome 7 Breakpoint.
An arrayed BAC library was screened by PCR using primers for the four sequence-tagged sites (STSs) CHLC.GATA91C01, D7S516, CHLC.GGAA3F06, and WI-5230, which lie at the centromeric end of the y908B12 YAC insert. This screen identified BACs b60B19, b319D14, b390A17, and b332C6. To determine overlaps and find additional BACs in the region, the sequences at the ends of the inserts of the BACs were determined. A search of the GenBank database indicated that DNA within a PAC clone, p881H5, included the telomeric sequence within b390A17. DNA of this PAC clone had been sequenced by the Genome Sequencing Center (GSC) at Washington University (http://genome.wustl.edu/est/esthmpg.html), and further examination of chromosome 7 sequence data on the GSC web site using the end sequences of the BAC inserts resulted in construction of the contig shown in Fig. 2a. FISH analyses on cases BWH-42 and BWH-665 using pooled probes from BACs b60B19 through b459N13 in this contig demonstrated an identical hybridization pattern to that seen with YAC y908B12 (data not shown). These results indicated that the BAC contig spans the chromosome 7 breakpoint in the t(7;17) of case BWH-42.
Figure 2.
Localization of the 7p15 breakpoint to the centromeric end of BAC b459N13. (a) Map of the BAC and PAC clones in the centromeric region of YAC y908B12. STSs are shown at the top of the map at relative positions based on DNA sequence available for the region. (b) Position of Southern blot hybridization probes within DNA near the centromeric end of BAC b459N13. The approximate positions of chromosome 7p15 breakpoints in cases BWH-42 and -665, as determined in the Southern blots in c, are illustrated with upward arrows near the telomeric end of the region shown. (c) Southern blot analyses of DNA derived from the tumors in cases BWH-42 and -665. Normal DNA was extracted from fibroblasts of the patient in case BWH-42. The probes used for each blot are indicated above the autoradiograms; the restriction enzymes used are shown below. Arrowheads indicate the positions of rearranged restriction fragments; dashes the positions of the germline restriction fragments.
Identification of a Gene in the Chromosome 7p15 BAC/PAC Contig and Detection of the Breakpoint by Southern Blot Analysis.
The EST database was queried for matches to the genomic sequence available for this region. Probes for Southern blot analysis of tumor DNA were then constructed by PCR amplification of repeat-free segments of genomic DNA adjacent to or included in the expressed regions (Fig. 2b). Such a probe derived from the region of overlap between BAC b459N13 and PAC p881H5 (probe 459-10) detected non-germline bands in both BamHI and HindIII digests of tumor DNA from case BWH-42 but not in similar digests of DNA from normal fibroblasts (Fig. 2c). The probe 459-10 also detected a non-germline band in a BamHI digest of tumor DNA from case BWH-665.
The breakpoint region was further narrowed by hybridizing Southern blots of tumor DNA with two probes, 459-BP8 and 459-BP9, amplified from sequence lying 3-kb telomeric to 459-10 and separated from each other by 798bp. Probe 459-BP9 detected bands identical to those seen with probe 459-10 in the two cases. In both HindIII and BamHI digests of DNA from BWH-42, probe 459-BP8 detected non-germline bands distinct from those in case BWH-665 whereas, in case BWH-665, probe 459-BP8 detected the non-germline band seen with probe 459-10, as well as a new non-germline band. Taken together, these results indicated that the chromosome 7 breakpoint in case BWH-42 lies in DNA between that contained in probes 459-BP8 and 459-BP9 and that the breakpoint in case BWH-665 lies within DNA sequence contained in probe 459-BP8.
Detection of a Tumor-Specific Transcript with Hybridization Probes for DNA Near the Chromosome 7 Breakpoint.
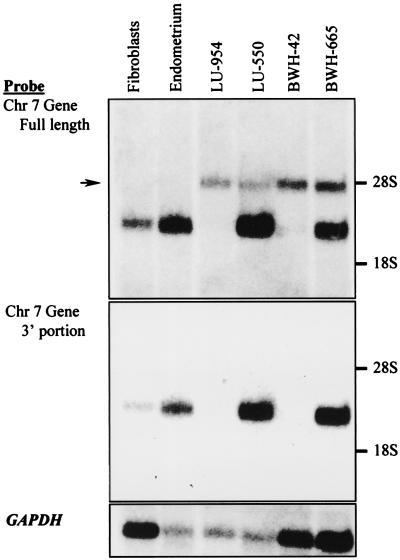
The chromosome 7 breakpoint in cases BWH-42 and BWH-665 mapped to the region of putative intronic sequence predicted by the alignment of the EST AA431106 with the sequences of b459N13 and p881H5. A series of overlapping ESTs presumably derived from a single gene were found in the database, and a hybridization probe consisting of the three most 3′ exons of the gene was constructed. This probe was hybridized to Northern blots containing polyadenylated RNA purified from normal human fibroblasts, normal human endometrium, and four cases of low-grade ESSs known to contain a t(7;17) (Fig. 3). A band corresponding to a transcript of ≈3.2 kb appeared in analyses of RNA from both normal tissues and two tumors, and a second band corresponding to a length of about 4.5 kb appeared in analyses of RNA only from the four tumors.
Figure 3.
Northern blot analyses of poly(A)+ RNA from ESSs and normal control tissues. The source of normal fibroblasts was the same as that in Fig. 2, whereas normal endometrium was obtained from an unrelated patient. (a) Analysis performed with a probe for the 5′ end of EST AA431106 (nucleotides 74 to 1561 of JAZF1 in Fig. 5a). The arrow indicates the position of a band corresponding to a ≈4.5-kb transcript seen only in tumor samples. (b) Analysis performed with a probe for the 3′ end of EST AA431106 (nucleotides 1120 to 2883 of JAZF1 in Fig. 5a). (c) Analysis with a probe for the GAPDH gene used as a control for the quality and amount of RNA on the blot.
To determine which part of the normal chromosome 7 gene hybridized to the 4.5-kb transcript found in tumors, probes were designed to represent the portions of the gene on the centromeric and telomeric sides of the chromosome 7 breakpoint. Northern blot analysis with a centromeric probe yielded a pattern of bands identical to that seen with the original probe, and analysis with a telomeric probe yielded only the 3.2-kb band (data not shown). These findings indicated that the chromosome 7 gene is expressed in normal endometrium and that disruption of the penultimate intron of the gene in at least some tumors with the t(7;17) results in the production of an abnormal transcript that contains only the centromeric portion of the gene.
Identification of a BAC Clone Spanning the Breakpoint on Chromosome 17.
Two sets of nested PCR primers were constructed to be oriented in a tail-to-tail configuration and complementary to sequence 450 bp apart on each side of the telomeric HindIII site in Fig. 2b. Template for I-PCR with these primers was prepared by HindIII digestion and subsequent self-ligation of genomic DNA from tumor tissue in case BWH-42 and from normal fibroblasts of the same patient. I-PCR amplification of these templates produced a predicted, unrearranged 3.5-kb product from both tumor and normal DNA and a predicted, rearranged 1.6-kb product only from the tumor DNA (data not shown).
The 1.6-kb I-PCR product was ligated into the pCR2.1Topo vector, and the sequences at the ends of the insert of the resulting clone were determined. One end sequence corresponded to chromosome 7 sequence up to a HindIII site and then immediately diverged into non-chromosome 7 sequence. When this new sequence was used to search the GenBank database, it was found that a BAC, b307A16, from chromosome 17 contained matching sequence. These results indicated that the breakpoint at chromosome 17q21 in case BWH-42 was contained in DNA of b307A16.
Identification of a Gene in the Chromosome 17 Breakpoint Region.
Analysis of DNA within b307A16 revealed that about 3.8 kb of a 4.44-kb
EST, KIAA0160, map within genomic DNA beginning less than 20 kb away
from the chromosome 17 breakpoint; however, the first 647 bp of this
EST is not present anywhere in b307A16 or the overlapping BAC b542B22.
This observation suggested that a deletion had occurred in the genomic
DNA contained in b307A16 and that the predicted gene extends across the
chromosome 17 breakpoint. If this gene were disrupted by the chromosome
17 breakpoint, the orientation of the gene would be consistent with a
fusion of this gene with the gene disrupted by the breakpoint in
chromosome 7. To test this possibility, RNA derived from tumor BWH-42
and from control fibroblasts was analyzed by RT-PCR using primers
complementary to sense sequence at the 5′ end of both the 7 and 17
genes and anti-sense sequence located at the 3′ end of both genes, with
the primers paired in all four possible ways. In these experiments, the
primer pairs for the intact chromosome 17 gene amplified products from
the RNA of both the normal tissue and the tumor, but the recombinant 5′
chromosome 7–3′ chromosome 17 pair amplified a product of about 440 bp
only from the tumor RNA (Fig. 4). The
reciprocal recombinant primer pair failed to amplify a product from
either of the RNA samples. The 5′ chromosome 7–3′ chromosome 17 primer
pair used in an RT-PCR assay of RNA derived from the three other tumors
analyzed previously in Northern blots yielded similar findings. These
results demonstrated that the chromosome 17 gene was disrupted by the
translocation and that a fusion transcript containing the 5′ end of the
chromosome 7 gene and the 3′ end of the chromosome 17 gene had been
created. Sequence analysis of the RT-PCR products confirmed that RNA
sequence transcribed from chromosome 7 had been joined to RNA sequence
transcribed from chromosome 17 at the sites indicated in Fig.
5
 .
.
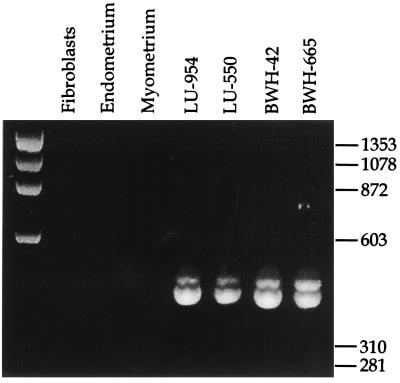
Figure 4.
RT-PCR analysis of the RNA sequence transcribed across the site of recombination in the t(7;17)(p15;q21). Ethidium bromide-stained agarose gel of RT-PCR products amplified from total RNA of normal tissues and the four low-grade ESSs known to carry the translocation. The source of fibroblasts and normal endometrium was the same as that in Fig. 3; the normal myometrium was obtained from the same specimen as the endometrium.
Figure 5.
Nucleotide and amino acid sequences of the chromosome 7 gene (JAZF1) and chromosome 17 gene (JJAZ1) joined in the t(7;17)(p15;q21). (a) The conceptually translated sequence of the largest ORF is shown below the sequence of the sense strand of the JAZF1 cDNA. The shaded boxes indicate the amino acids that code for C2H2 zinc finger domains. The breakpoint in the four cases of low-grade ESS analyzed by Northern blot and RT-PCR divided the coding sequence at the position of the vertical line (after nucleotide 435). (b) The sequence of JJAZ1 displayed as in a. The double underline indicates the amino acids that code a possible bipartite nuclear localization signal. The breakpoint in the four cases of low-grade ESS divided the coding sequence at the position of the vertical line (before nucleotide 469).
Structure of the Genes Identified at 7p15 and 17q21.
Fig. 5a presents the sequence of the chromosome 7 gene, as deduced from the GenBank and EST databases plus an additional 176 bp at the 5′ end acquired through screening of two cDNA libraries derived from human brain and human umbilical vein endothelial cells (HUVECs). The entire sequence of the chromosome 17q21 gene is contained in the EST KIAA0160 (Fig. 5b). The true 5′ ends of these transcripts remain unknown because the ORFs continue to the 5′ ends of the sequences assembled so far.
Conceptual translation of the ORF of the chromosome 7 gene revealed an N-proximal zinc finger domain as well as a nearly tandem pair of zinc finger domains close to the C terminus. Similar analysis of the chromosome 17 gene sequence revealed a zinc finger domain in the C-terminal third of the coding sequence. All four of these zinc finger domains are of the Cys2His2 class. Other than a possible bipartite nuclear localization sequence in the chromosome 17 gene, these domains represented the only identifiable motifs in the coding sequences and the only significant similarity to other known genes. In light of these structural features, we refer to the chromosome 7 gene by the acronym JAZF1, for Juxtaposed with Another Zinc Finger gene, and the chromosome 17 gene by JJAZ1, for Joined to JAZF1.
Analysis of Endometrial Stromal Tumors for the JAZF1/JJAZ1 Fusion Transcript.
RT-PCR was performed on RNA extracted from paraffin-embedded, formalin-fixed tissues of five archival cases of low-grade ESS, for which cytogenetic analysis had not been done, of three stromal nodules, and of seven high-grade ESSs. RT-PCR performed with sense primers for both JAZF1 and JJAZ1 and nested antisense JJAZ1 primers demonstrated amplifiable template from each case, as judged by the generation of JJAZ1 product. Each of the three stromal nodules was also positive for the fusion transcript. Among the ESSs, all low-grade cases were positive in this assay, but only three of seven high-grade cases were positive (Table 2). Two of the high-grade tumors would be considered undifferentiated endometrial sarcomas in the more recently proposed classification system, and, of these, one was positive for the fusion and the other not.
Table 2.
Summary of RT-PCR results for JAZF1/JJAZ1 fusion RNA in archival tissue specimens
| Tumor type | Total cases | Positive cases |
|---|---|---|
| Stromal nodule | 3 | 3 |
| Low-grade ESS | 5 | 5 |
| High-grade ESS* | 7 | 3 |
| Normal endometrium | 10 | 0 |
Includes two cases classifiable as undifferentiated endometrial sarcoma, of which one was positive.
Discussion
Chromosomal translocations generally affect the malignant phenotype of cells that contain them through various kinds of alterations in genes at or near the breakpoints in DNA of one or both of the participating chromosomes (20). The t(7;17)(p15;q21) appears to be an example of a chromosomal translocation resulting in a gene fusion, in which the 5′ end of one gene, termed here JAZF1, on chromosome 7 is joined to the 3′ end of a second gene, termed JJAZ1, on chromosome 17. In this respect, the t(7;17)(p15;q21) resembles the majority of chromosomal translocations so far analyzed in soft tissue sarcomas, most of which contain fusions of EWS or a related gene to a second gene encoding a DNA-binding protein (21, 22). Whether detection of the JAZF1/JJAZ1 fusion always represents the presence of the t(7;17)(p15;q21), or may be found in cells without a translocation identifiable by cytogenetic analysis, remains to be determined.
The cDNAs for JAZF1 and JJAZ1 appear to specify the synthesis of proteins composed of 243 and 739 aa, respectively. Determination of the amino acid sequences encoded by the genes is complicated by the fact that large ORFs are continuous with the 5′ ends of the cDNA sequences assembled for the two genes; however, several features of the cDNA sequences for both genes support the conclusion that the first methionine codons represent the actual beginning of the coding sequences. For instance, the total length of the cDNA sequences for the two genes coincides with the estimated size of mRNA in bands identified by Northern blot hybridization. Furthermore, in RNase protection assays, probes generated from genomic DNA overlapping sequence at the 5′ end of the JAZF1 cDNA protected a fragment of RNA from liver and lung cells corresponding to a size consistent with the cDNA representing the full length of the transcript for this gene (comparable experiments for JJAZ1 were not possible because genomic sequence at the 5′ end of this gene has not yet been identified). Also, the nucleotides surrounding the putative initiation codon in both genes (CACCATGA in JAZF1 and CGCGATGG in JJAZ1) conform well to the consensus rules proposed for vertebrate translational start sites (23). Finally, the mouse ortholog of JAZF1 (contained in the overlapping murine ESTs AI595264, AA061309, and AI428135) shows striking homology to the human cDNA over the entire presumptive ORF but diverges from the human cDNA 5′ of the position of the codon provisionally assigned as the first methionine. Similarly, the JJAZ1 Drosophila ortholog (encoding a conceptually translated protein AAF49094) in genomic fly DNA contains termination codons in all three reading frames shortly upstream of the position corresponding to the putative first methionine in the human gene.
The specific functions of JAZF1 and JJAZ1 and the reason for their involvement in a gene fusion associated with neoplasia are not directly apparent from the sequences of the cDNAs. The only recognizable regions within the two cDNAs resembling sequences within known genes in humans and other species encode zinc finger motifs, as often found in DNA binding proteins (24, 25). The two C-proximal zinc finger domains encoded by JAZF1 are particularly similar in structure and relative spacing to two zinc fingers in the yeast protein Sfp1p, although the homology between these two proteins is limited to these two paired zinc fingers. This homology, albeit slight, might seem noteworthy in a cancer-related gene because Sfplp negatively regulates the G2/M transition in the cell cycle of Saccharomyces cerevesiae by serving as a transcriptional activator of the gene PDS1 (26, 27), the product of which inhibits Esp1p (28, 29), a protein that dissociates the cohesin complexes responsible for holding together sister chromatids during G2 (30). However, in transfected cultured cell lines, JAZF1 protein appears to have no significant effect on transcription of reporter genes driven by the promoter of PTTG1 (31, 32), the human ortholog of PDS1 (data not shown).
Another finding possibly relevant to the role of JAZF1 in oncogenesis is the lack of JAZF1 RNA in tumors BWH-42 and LU-954. The reason for the absence of this RNA is different in the two cases: tumor LU-954 is monosomic for chromosome 7, whereas tumor BWH-42 contains unrearranged DNA for JAZF1 on the allele not broken by the t(7;17), as demonstrated by Southern blot analysis. Failure to detect RNA in the latter case may therefore be due to transcriptional silencing of the normal copy of JAZF1, as has been noted in the unrearranged alleles of MYC and BCL2 in some tumors bearing chromosomal translocations with breakpoints in or near these genes (33, 34). Loss of expression for normal versions of JAZF1 in multiple tumors suggests a possible role of this gene as a tumor suppressor, similar to the situation observed for TEL, which is involved in chromosomal translocations and has tumor suppressor activity in some acute lymphoblastic leukemias (35–37).
The distribution of the JAZF1/JJAZ1 fusion among uterine stromal tumors carries a number of implications for the biology of these neoplasms. For example, the finding of the fusion in stromal nodules raises the possibility that malignant endometrial stromal tumors can develop from stromal proliferations that are initially benign. Additionally, of the seven high-grade ESSs studied, only three showed evidence of the fusion, suggesting that some high-grade ESSs may arise by a different pathogenetic mechanism than low-grade ESSs, despite the current tendency to consider all of these tumors as clinically homogeneous. Alternatively, this discrepancy between high and low grade tumors could be due to the relatively small sample of cases studied or to the fact that we analyzed tumors for only one form of the fusion and that functionally equivalent genetic recombinations between JAZF1 and JJAZ1 at intragenic sites that would not have been detected by us may be present in some tumors. These explanations notwithstanding, if, after study of more high-grade ESSs, the fusion is confirmed to be present only within a subset of such tumors, it will be important to correlate possible differences in clinical behavior with the presence or absence of this genetic lesion.
Acknowledgments
We thank Megan Smith for valuable technical assistance. This work was supported by the National Foundation for Cancer Research and National Institutes of Health-National Cancer Institute Grant CA-85995.
Abbreviations
- ESS
endometrial stromal sarcoma
- YAC
yeast artificial chromosome
- BAC
bacterial artificial chromosome
- EST
expressed sequence tag
- FISH
fluorescence in situ hybridization
- RT-PCR
reverse transcription–PCR
- STS
sequence-tagged site
- I-PCR
inverse PCR
References
- 1.Zaloudek C, Norris H J. In: Blaustein's Pathology of the Female Genital Tract. Kurman R J, editor. New York: Springer; 1994. pp. 487–528. [Google Scholar]
- 2.Norris H J, Taylor H B. Cancer. 1966;19:755–766. doi: 10.1002/1097-0142(196606)19:6<755::aid-cncr2820190604>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 3.Tavassoli F A, Norris H J. Histopathology. 1981;5:1–10. doi: 10.1111/j.1365-2559.1981.tb01761.x. [DOI] [PubMed] [Google Scholar]
- 4.Evans H L. Cancer. 1982;50:2170–2182. doi: 10.1002/1097-0142(19821115)50:10<2170::aid-cncr2820501033>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 5.Chang K L, Crabtree G S, Lim-Tan S K, Kempson R L, Hendrickson M R. Am J Surg Pathol. 1990;14:415–438. doi: 10.1097/00000478-199005000-00002. [DOI] [PubMed] [Google Scholar]
- 6.Hendrickson M R, Longacre T A, Kempson R L. In: Diagnostic Surgical Pathology. Sternberg S S, editor. Philadelphia: Lippincott Williams & Wilkins; 1999. pp. 2203–2305. [Google Scholar]
- 7.Pegram M, Hsu S, Lewis G, Pietras R, Beryt M, Sliwkowski M, Coombs D, Baly D, Kabbinavar F, Slamon D. Oncogene. 1999;18:2241–2251. doi: 10.1038/sj.onc.1202526. [DOI] [PubMed] [Google Scholar]
- 8.Heise C, Sampson-Johannes A, Williams A, McCormick F, Von Hoff D D, Kirn D H. Nat Med. 1997;3:639–645. doi: 10.1038/nm0697-639. [DOI] [PubMed] [Google Scholar]
- 9.le Coutre P, Mologni L, Cleris L, Marchesi E, Buchdunger E, Giardini R, Formelli F, Gambacorti-Passerini C. J Natl Cancer Inst. 1999;91:163–168. doi: 10.1093/jnci/91.2.163. [DOI] [PubMed] [Google Scholar]
- 10.Sreekantaiah C, Li F P, Weidner N, Sandberg A A. Cancer Genet Cytogenet. 1991;55:163–166. doi: 10.1016/0165-4608(91)90073-4. [DOI] [PubMed] [Google Scholar]
- 11.Dal Cin P, Aly M S, De Wever I, Moerman P, van den Berghe H. Cancer Genet Cytogenet. 1992;63:43–46. doi: 10.1016/0165-4608(92)90062-d. [DOI] [PubMed] [Google Scholar]
- 12.Pauwels P, Dal Cin P, Van de Moosdijk C N, Vrints L, Sciot R, van den Berghe H. Histopathology. 1996;29:84–87. [PubMed] [Google Scholar]
- 13.Hennig Y, Caselitz J, Bartnitzke S, Bullerdiek J. Cancer Genet Cytogenet. 1997;98:84–86. doi: 10.1016/s0165-4608(96)00393-7. [DOI] [PubMed] [Google Scholar]
- 14.Philippsen P, Stotz A, Scherf C. Methods Enzymol. 1991;194:169–182. doi: 10.1016/0076-6879(91)94014-4. [DOI] [PubMed] [Google Scholar]
- 15.Morgan J A, Yin Y, Borowsky A D, Kuo F, Nourmand N, Koontz J I, Reynolds C, Soreng L, Griffin C A, Graeme-Cook F, et al. Cancer Res. 1999;59:6205–6213. [PubMed] [Google Scholar]
- 16.Ginsburg D, Handin R I, Bonthron D T, Donlon T A, Bruns G A, Latt S A, Orkin S H. Science. 1985;228:1401–1406. doi: 10.1126/science.3874428. [DOI] [PubMed] [Google Scholar]
- 17.Lech K, Reddy K J, Sherman L A. In: Current Protocols in Molecular Biology. Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Vol. 1. New York: Wiley; 1990. pp. 1.13.11–11.13.10. [Google Scholar]
- 18.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 19.Fletcher J. In: Current Protocols in Human Genetics. Dracopoli N C, Haines J L, Korf B R, Moir D T, Morton C C, Seidman C E, Seidman J G, Smith D R, editors. Vol. 1. New York: John Wiley & Sons; 1994. pp. 10.13.11–10.13.10. [Google Scholar]
- 20.Rabbitts T H. Nature (London) 1994;372:143–149. doi: 10.1038/372143a0. [DOI] [PubMed] [Google Scholar]
- 21.Ladanyi M. Diagn Mol Pathol. 1995;4:162–173. doi: 10.1097/00019606-199509000-00003. [DOI] [PubMed] [Google Scholar]
- 22.Dei Tos A P, Dal Cin P. Virchows Arch. 1997;431:83–94. doi: 10.1007/s004280050073. [DOI] [PubMed] [Google Scholar]
- 23.Kozak M. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berg J M, Shi Y. Science. 1996;271:1081–1085. doi: 10.1126/science.271.5252.1081. [DOI] [PubMed] [Google Scholar]
- 25.Clarke N D, Berg J M. Science. 1998;282:2018–2022. doi: 10.1126/science.282.5396.2018. [DOI] [PubMed] [Google Scholar]
- 26.Xu Z, Norris D. Genetics. 1998;150:1419–1428. doi: 10.1093/genetics/150.4.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nasmyth K, Peters J-M, Uhlmann F. Science. 2000;288:1379–1384. doi: 10.1126/science.288.5470.1379. [DOI] [PubMed] [Google Scholar]
- 28.Yamamoto A, Guacci V, Koshland D. J Cell Biol. 1996;133:99–110. doi: 10.1083/jcb.133.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamamoto A, Guacci V, Koshland D. J Cell Biol. 1996;133:85–97. doi: 10.1083/jcb.133.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ciosk R, Zachariae W, Michaelis C, Shevchenko A, Mann M, Nasmyth K. Cell. 1998;93:1067–1076. doi: 10.1016/s0092-8674(00)81211-8. [DOI] [PubMed] [Google Scholar]
- 31.Zou H, McGarry T J, Bernal T, Kirschner M W. Science. 1999;285:418–421. doi: 10.1126/science.285.5426.418. [DOI] [PubMed] [Google Scholar]
- 32.Kakar S S. Gene. 1999;240:317–324. doi: 10.1016/s0378-1119(99)00446-1. [DOI] [PubMed] [Google Scholar]
- 33.Bentley D L, Groudine M. Mol Cell Biol. 1986;6:3481–3489. doi: 10.1128/mcb.6.10.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cleary M L, Smith S D, Sklar J. Cell. 1986;47:19–28. doi: 10.1016/0092-8674(86)90362-4. [DOI] [PubMed] [Google Scholar]
- 35.Golub T R, Barker G F, Lovett M, Gilliland D G. Cell. 1994;77:307–316. doi: 10.1016/0092-8674(94)90322-0. [DOI] [PubMed] [Google Scholar]
- 36.Golub T R, Barker G F, Stegmaier K, Gilliland D G. Curr Top Microbiol Immunol. 1996;211:279–288. doi: 10.1007/978-3-642-85232-9_28. [DOI] [PubMed] [Google Scholar]
- 37.Takeuchi S, Seriu T, Bartram C R, Golub T R, Reiter A, Miyoshi I, Gilliland D G, Koeffler H P. Leukemia. 1997;11:1220–1223. doi: 10.1038/sj.leu.2400743. [DOI] [PubMed] [Google Scholar]