Abstract
The genus Bartonella includes numerous species with varied host associations, including several that infect humans. Development of a molecular diagnostic method capable of detecting the diverse repertoire of Bartonella species while maintaining genus specificity has been a challenge. We developed a novel real-time PCR assay targeting a 301-bp region of the ssrA gene of Bartonella and demonstrated specific amplification in over 30 Bartonella species, subspecies, and strains. Subsequent analysis of ssrA sequences was sufficient to discriminate Bartonella species and provided phylogenetic data consistent with that of gltA, a commonly used gene for differentiating Bartonella genotypes. Using this assay, we identified Bartonella DNA in 29% and 47% of blood specimens from elk in Wyoming and cattle in the Republic of Georgia, respectively. Sequence analysis of a subset of genotypes from elk specimens revealed a cluster most closely related to Bartonella capreoli, and genotypes from cattle were identified as Bartonella bovis, both Bartonella species commonly found in wild and domestic ruminants. Considering the widespread geographic distribution and infectivity potential to a variety of hosts, this assay may be an effective diagnostic method for identification of Bartonella infections in humans and have utility in Bartonella surveillance studies.
INTRODUCTION
The genus Bartonella includes over 30 different species and subspecies that infect a wide variety of mammalian hosts. Bartonella bacteria are transmitted by hematophagous insects such as ticks, fleas, and lice (4, 5, 8, 12, 15, 18), which can lead to widespread infection among localized populations of mammalian hosts. High prevalence of Bartonella bacteremia has been reported in populations of rodents, cats, and ruminants worldwide (3, 5, 6, 9, 13, 14, 26). Although members of the Bartonella genus infect a broad spectrum of mammalian hosts, most species exhibit restricted host specificity (25). Humans are also susceptible to Bartonella infection; to date, at least 10 Bartonella species have been implicated in human disease (2, 7, 16, 19, 21). The broad geographical distribution, wide spectrum of available reservoir hosts, and zoonotic potential of Bartonella species necessitate continued investigation of the biology and epidemiology of bacteria belonging to this genus.
Reliable detection methods are needed to facilitate ongoing epidemiological and ecological studies of Bartonella. Bacterial culture remains the preferred method for identification of Bartonella infections. However, culturing methods are laborious and time-consuming, and recovery of organisms is often low. Real-time PCR assays for detecting Bartonella have been reported previously (1, 10, 11, 24), but none of these are able to detect all species due to the significant genetic diversity within the genus. Furthermore, molecular detection methods must be applicable to a diverse array of specimen types, including insect and mammalian blood and tissue. In particular, amplification from whole blood remains a significant challenge to Bartonella detection (20).
Beyond detection, genetic targets that provide sufficient sequence diversity to allow identification to the species level are required to fully understand the distribution and host specificity of various Bartonella species and allow identification of the strains associated with human illness. The citrate synthase gene (gltA) is a common genetic target for Bartonella detection and is considered a reliable tool for distinguishing genotypes (22). One limitation of targeting this locus is its homology with sequences found in some host genomes, such as mouse, rat, and human, along with other human pathogens (11). This cross-reactivity creates a need for improved molecular diagnostics for Bartonella.
We developed a novel real-time PCR assay for detection of Bartonella spp. targeting the ssrA gene and demonstrated the ability to detect over 30 unique species, subspecies, and strains within this genus. SsrA RNA, also known as transfer-mRNA (tmRNA), is a single-copy prokaryotic-specific molecule involved in processing of incomplete peptides and resolution of stalled ribosomes during translation (17). This target has not been exploited previously for Bartonella detection. Furthermore, we demonstrate the utility of sequence-based species identification using the ssrA amplicon. Using this real-time PCR assay in combination with sequencing, we successfully amplified Bartonella DNA from ruminant blood specimens and identified the resulting genotypes.
MATERIALS AND METHODS
Bacterial strains and DNA extraction.
All bacterial strains were obtained from collections at the Centers for Disease Control and Prevention in Fort Collins, CO, and Atlanta, GA. Nucleic acid was extracted from 33 Bartonella strains, including 25 defined species or subspecies using a QIAamp DNA Minikit (Qiagen, Valencia, CA). Bartonella strains included in this study are the following: B. alsatica (IBS 382), B. bacilliformis (KC584), B. birtlesii (IBS 325), B. bovis (91-4), B. capreoli (WY-Elk), B. chomelii (A828), B. clarridgeiae (Houston-2), B. doshiae (R18), B. elizabethae (F9251), B. henselae (Houston-1), B. grahamii (V2), B. japonica (Fuji 18-1T), B. koehlerae (C-29), B. melophagi (K-2C), B. phoceensis (16120), B. quintana (Fuller), B. rochalimae (BMGH), B. schoenbuchensis (R1), B. silvatica (Fuji 23-1T), B. tamiae (Th307, Th239, and Th339), B. taylorii (M16), B. tribocorum (IBS 506), B. vinsonii subsp. arupensis (OK 94-513), B. vinsonii subsp. vinsonii (Baker), B. washoensis (Sb944nv), and Bartonella isolates (Sh6397ga, Sh6396ga, Sh6537ga, Sh8784ga, Sh8200ga, and Sh8776ga). Using a MagNA Pure Compact instrument with Total Nucleic Acid Isolation Kit I (Roche Applied Science, Indianapolis, IN), nucleic acid was extracted from 61 microorganisms that are closely related genetically to Bartonella or may occupy a similar ecological niche, including the following: Afipia broomeae, Afipia clevelandensis, Afipia felis, Agrobacterium radiobacter, Agrobacterium tumefaciens, Babesia microti, Bordetella pertussis, Bordetella parapertussis, Bradyrhizobium, Brucella abortus, Brucella canis, Brucella melitensis, Brucella neotomae, Brucella ovis, Brucella suis, Campylobacter coli, Campylobacter fetus, Campylobacter jejuni, Citrobacter freundii, Enterobacter aerogenes, Enterobacter cloacae, Erwinia, Escherichia albertii (two strains), Escherichia blattae, Escherichia coli (four strains), Escherichia fergusonii, Escherichia hermannii, Escherichia vulneris, Haemophilus influenzae, Klebsiella oxytoca, Klebsiella pneumoniae, Kluyvera intermedia, Legionella pneumophila, Methylobacterium organophilum, Ochrobactrum anthropi (three strains), Ochrobactrum intermedium, Oligella urethralis (four strains), Psychrobacter phenylpyruvicus (2 strains), Raoultella planticola, Salmonella bongori, Salmonella enterica (S. enterica serovar Enteritidis, S. enterica serovar Typhi, and S. enterica serovar Typhimurium), Shigella boydii, Shigella dysenteriae, Shigella flexneri, Shigella sonnei, Toxoplasma gondii, and Vibrio cholerae. Human genomic DNA was also tested for cross-reactivity. All nucleic acid extracts were normalized to 1 ng/μl in Tris-EDTA buffer.
Real-time PCR.
Sequences of the ssrA (tmRNA) gene of five representative Bartonella species were obtained from the tmRNA Website (http://www.indiana.edu/∼tmrna/) and GenBank (accession numbers NC_005955.1, NC_005956.1, NC_010161.1, NC_012846.1, and NC_008783.1). Sequences were aligned using the Clustal W method (http://www.ebi.ac.uk/Tools/msa/clustalw2/). Primers and probes were designed using Primer Express, version 3.0, software (Applied Biosystems, Foster City, CA) with some modification for amplification of a 301-bp region of ssrA. The reaction mixture (25 μl) contained the following components: 12.5 μl of 2× PerfeCta MultiPlex qPCR SuperMix (Quanta Biosciences, Gaithersburg, MD), forward and reverse primers (ssrA-F, 5′-GCTATGGTAATAAATGGACAATGAAATAA-3′; ssrA-R, 5′-GCTTCTGTTGCCAGGTG-3′) at a final concentration of 500 nM, 6-carboxyfluorescein (FAM)-labeled probe (5′-FAM-ACCCCGCTTAAACCTGCGACG-3′-BHQ1, where BHQ is black hole quencher) at a final concentration of 100 nM, and 5 μl of extracted nucleic acid. Real-time PCR was performed on an Applied Biosystems 7500 real-time PCR instrument with the following thermocycling parameters: 1 cycle of 95°C for 2 min followed by 45 cycles of 95°C for 15 s and 60°C for 60 s, with data collection in the FAM channel. Primers and probe were tested using nuclease-free water (95 replicates of reactions) to ensure no signal in the absence of nucleic acid template.
The limit of detection was independently determined and verified for four species (B. quintana, B. henselae, B. bovis, and B. elizabethae) by testing 10 replicates each of 10-fold serial dilutions of genomic DNA ranging from 1 ng/μl to 0.1 fg/μl. The limit of detection was identified as the lowest dilution at which amplification was observed in at least 50% of replicates.
Specificity was assessed by performing the assay using 15 ng of nucleic acid from 61 different microorganisms representing 24 genera and 48 species.
Sequencing.
Amplicons for sequencing were generated by conventional PCR with forward and reverse primers at 400 nM each using a Bio-Rad Dyad thermal cycler (Bio-Rad, Hercules, CA) and the following thermocycling conditions: 95° for 2 min, followed by 30 cycles of 95° for 15 s, 60° for 60 s, and 72° for 30 s, with a final step at 72° for 3 min. Amplicons were visualized by electrophoresis in a 1% agarose gel followed by staining with 0.05% methylene blue solution and purification using a Geneclean Turbo kit (MP Biomedicals, Solon, OH). Sequencing reactions were performed in both directions using a BigDye Terminator, version 3.1, kit (Applied Biosystems) according to the manufacturer's instructions with the same primers for the real-time PCR assay at a final concentration of 165 nM. Sequencing was performed on an Applied Biosystems 3130xl genetic analyzer.
Phylogenetic analysis.
A 253-bp region of each amplified sequence (excluding forward and reverse primers) was used for alignment and phylogenetic comparison of Bartonella species and genotypes using a Lasergene, version 8, software suite (DNAStar, Madison, WI). All ssrA sequences were aligned using the Clustal V method. Phylogenetic trees were constructed using the neighbor-joining method and bootstrapping analysis with 1,000 replicates.
Testing of animal blood.
Blood specimens collected from elk (Cervus elaphus) in Wyoming (n = 55) and cattle (Bos primigenius) in the country of Georgia (n = 89) between 2008 and 2009 were tested for Bartonella by bacterial culture using previously described methods (3). The culture results from this cohort of elk have been reported previously (3). All specimens were extracted using a DNeasy Blood and Tissue kit (Qiagen) or MagNA Pure Compact with Total Nucleic Acid Isolation Kit I (Roche Applied Science). Nucleic acid extract (5 or 10 μl) was used in each real-time PCR.
Nucleotide sequence accession numbers.
Thirty-four unique ssrA sequences obtained from Bartonella strains and isolates were submitted to GenBank (http://www.ncbi.nlm.nih.gov/) and assigned the following accession numbers: JN029776 (B. alsatica IBS 382), JN029794 (B. bacilliformis KC584), JN029775 (B. birtlesii IBS325), JN029767 (B. bovis 91-4), JN029798 (B. capreoli WY-Elk), JN029773 (B. chomelii A828), JN029768 (B. doshiae R18), JN029774 (B. elizabethae F9251), JN029785 (B. henselae Houston-1), JN029795 (B. grahamii V2), JN029784 (B. japonica Fuji 18-1T), JN029769 (B. koehlerae C-29), JN029771 (B. melophagi K-2C), JN029770 (B. phoceensis 16120), JN029766 (B. quintana Fuller), JN029797 (B. rochalimae BMGH), JN029772 (B. schoenbuchensis R1), JN029782 (B. silvatica Fuji 23-1T), JN029778 (B. tamiae Th307), JN029779 (B. tamiae Th239), JN029780 (B. tamiae Th339), JN029781 (B. taylorii M16), JN029796 (B. tribocorum IBS 506), JN029783 (B. vinsonii subsp. arupensis OK 94-513), JN029777 (B. vinsonii subsp. vinsonii Baker), JN029786 (B. washoensis Sb944nv), JN029787 (Bartonella sp. Sh6397ga), JN029791 (Bartonella sp. strain Sh8200ga), JN029793 (Bartonella sp. strain Sh8776ga), JN029788 (Bartonella sp. strain Sh6396ga), JN029790 (Bartonella sp. strain Sh8784ga), JN029792 (Bartonella sp. strain Sh9282ga), JN029789 (Bartonella sp. strain Sh6537ga), JN982716 (B. clarridgeiae Houston-2). The ssrA sequence amplified from elk blood was assigned accession number JN982717, and the sequence identified in cattle blood was identical to that of B. bovis (JN029767).
RESULTS
Real-time PCR for detection of Bartonella ssrA.
Amplification of the target sequence occurred with all Bartonella species (n = 24) and unclassified Bartonella strains (n = 7) tested (data not shown). Amplification curves demonstrated a sigmoidal shape and had crossing threshold (CT) values between 15 and 21 with 5 ng of DNA per reaction. No amplification was observed in no-template control (NTC) reactions (n = 95) or with DNA from other microorganisms (n = 61; listed above in “Bacterial strains and DNA extraction”) or with human genomic DNA (data not shown). The limit of detection was independently determined for four species (B. quintana, B. henselae, B. bovis, and B. elizabethae) and found to be ≤5 fg of nucleic acid per reaction (data not shown).
Bartonella phylogeny based on ssrA genotypes.
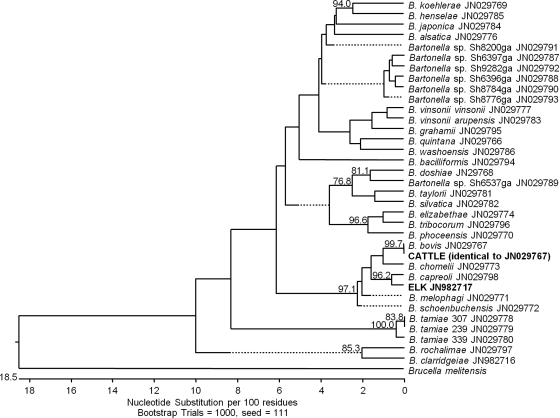
Phylogenetic analysis of ssrA sequences from each Bartonella strain or isolate showed that this region was sufficient to discriminate all Bartonella species and that separation of clades based on ssrA sequences was consistent with phylogeny based on gltA, which is considered a reliable tool for distinguishing closely related Bartonella genotypes (22). First, the ssrA sequences from ruminant-associated Bartonella, including B. chomelii, B. capreoli, B. bovis, B. melophagi, and B. schoenbuchensis, formed an independent clade; sequence identity between these species was ≥94%. Further, both subspecies of B. vinsonii (B. vinsonii subsp. vinsonii and B. vinsonii subsp. arupensis) included in this study formed a separate grouping in the tree with 98% identity, as did three strains of the recently identified pathogenic Bartonella species B. tamiae (≥97.2% identity) (21). Among all ssrA sequences, the lowest percent identity (75.3 to 84.1%) was observed for strains of B. tamiae relative to other Bartonella species, thus supporting the separation of B. tamiae as a novel species (21). The division of two additional clades which are similarly separated by gltA comparison, one consisting of B. elizabethae and B. tribocorum and the other including B. henselae and B. koehlerae, was also supported by the phylogenetic analysis of ssrA. Overall, the separation of major Bartonella clades based on ssrA sequences was consistent with phylogeny based on gltA (20, 23).
Detection and identification of Bartonella in animal blood.
Next, we utilized this assay to screen elk and cattle blood specimens for the presence of Bartonella and compared results to bacterial culture (Table 1). Bartonella DNA was detected in 16 of 55 (29.1%) and 42 of 89 (47.2%) specimens from elk and cattle, respectively. The appropriate amplicon size was confirmed for positive samples (data not shown). Using traditional culturing methods, Bartonella was recovered from only 4 of 55 (7.3%) elk and 34 of 89 (38.2%) cattle specimens (Table 1).
Table 1.
Detection of Bartonella in ruminant blood by real-time PCR and culture
| Detection method | Elk specimen results (n = 55) |
Cattle specimen results (n = 89) |
||
|---|---|---|---|---|
| No. (%) of Bartonella-positive specimens | Species identified by ssrA sequence | No. (%) of Bartonella-positive specimens | Species identified by ssrA sequence | |
| Real-time PCR (ssrA) | 16 (29.1) | B. capreoli | 42 (47.2) | B. bovis |
| Culture | 4 (7.3)a,b | 34 (38.2)b | ||
Culture results previously reported (3).
All culture-positive specimens were also positive by real-time PCR.
Since comparison of ssrA genotypes from Bartonella reference strains showed that this sequence provides sufficient information to discriminate Bartonella genotypes, we performed sequencing analysis of a subset of ssrA sequences amplified from elk (n = 3) and cattle (n = 5) specimens in order to identify the Bartonella species present. Analysis of ssrA sequences from elk blood revealed one genotype which clustered most closely with B. capreoli (Fig. 1), a Bartonella species found in wild and domestic ruminants (3). These results are consistent with previous identification of B. capreoli isolated from these samples using sequencing analysis of gltA (3). Similarly, a single ssrA genotype present in cattle blood was found to be identical to that of B. bovis (99.7% similarity) (Fig. 1). This result corroborates previous identification of B. bovis from these cattle specimens by analysis of gltA (data not shown).
Fig 1.
Phylogenetic relationships between ssrA sequences of all Bartonella species, subspecies, and isolates tested. GenBank accession numbers are shown for each genotype. Both ssrA genotypes obtained from ruminant blood were closely related to Bartonella species found in wild and domestic ruminants. The genotype identified from elk blood (JN982717) clustered closely with B. capreoli (96.2% similarity), and the single genotype identified in cattle blood was identical to B. bovis (99.7%). Only bootstrap replicates of >70% are noted.
DISCUSSION
Significant genetic diversity within the Bartonella genus presents a challenge for developing a molecular assay for detection of all species. The real-time PCR assay described here is able to detect all Bartonella species tested and demonstrated utility for screening primary specimens from animal hosts. Moreover, we established that this assay can also be used in combination with sequencing analysis to distinguish Bartonella species. Collectively, both of these accomplishments provide a significant advancement for studying Bartonella.
Unlike some previously reported PCR assays for Bartonella detection that display cross-reactivity with similar sequences present in host genomes (11), the genetic target of the current assay is a 301-bp region of the ssrA gene, which has not been identified in eukaryotic organisms to date, thereby reducing the potential for false-positive amplification in mammalian specimens. In addition, we performed extensive assessment of the specificity of this assay using a large collection of genetically or ecologically relevant organisms to ensure specific amplification of Bartonella species.
Identification of Bartonella by culture requires up to 4 weeks, whereas nucleic acid extraction and real-time PCR require only a few hours. Our results suggest that the ssrA real-time PCR assay is more sensitive than culture or other PCR assays and, therefore, represents an important advancement in Bartonella detection. Amplification of Bartonella DNA from whole-blood samples has been a challenge in some Bartonella studies. Using the new assay, we detected 100% of culture-positive ruminant blood specimens. Multiple factors, including extraction method, improved real-time PCR reagents, and oligonucleotide design, have likely contributed to the improvement in Bartonella detection from whole blood reported here. This assay is ideally suited for use as a screening tool to identify Bartonella-positive samples, which can then be subjected to the more laborious culture procedure in order to obtain an isolate.
Due to the improved sensitivity compared to culture, our data indicate a much higher prevalence of Bartonella bacteremia among elk species than previously reported (3). These results are more consistent with previous reports of high prevalence of Bartonella bacteremia among ruminant populations in other parts of the world (5, 6, 13). This assay may serve as an effective tool for investigating the dynamics of bacteremia in ruminants and other animals and for assessing the veterinary and medical importance of Bartonella infection in ruminants and potential zoonotic threat to humans.
Our analysis of ssrA genotypes showed that diversity within this region is sufficient to distinguish Bartonella species. Phylogenetic analysis revealed grouping of some ssrA genotypes obtained from Bartonella type strains according to host specificity. For example, Bartonella species associated with ruminants formed a single clade in the phylogenetic tree (Fig. 1). Also, separation of the two other clades of Bartonella including B. elizabethae with B. tribocorum and B. henselae with B. koehlerae was also supported by ssrA sequence analysis. In addition, three strains of B. tamiae, a recently identified pathogenic Bartonella species, formed an outlying group based on ssrA genotypes. These results are consistent with phylogenetic analysis of concatenated sequences from six genetic targets, including 16S rRNA, gltA, rpoB, ftsZ, groEL, and the 16S-23S rRNA intergenic spacer (ITS) previously reported by Kosoy and colleagues (21). According to La Scola et al., of the commonly used genetic targets for Bartonella identification, only gltA and rpoB sequences provide sufficient discriminatory power and interspecies diversity to allow discrimination of Bartonella species (22). Our data suggest that inclusion of ssrA sequences in phylogenetic analysis based on multiple genetic targets may provide additional supportive evidence for accurate identification of Bartonella isolates.
Considering the widespread geographic distribution and ability of Bartonella to infect a variety of animal species, this novel assay may be useful for determining the prevalence of Bartonella in large-scale surveillance studies. Furthermore, this assay may serve as an effective diagnostic method for identification of Bartonella infections in humans. Even within a given host organism, the types of specimens collected may vary based on different clinical manifestations; for instance, human clinical specimens may include whole blood, heart valve, or other tissue types. Therefore, PCR-based methods for Bartonella detection must be applicable to a variety of sample types. Additional studies are needed to assess the performance of this assay using human clinical specimens and other sample types.
ACKNOWLEDGMENTS
This work was supported in part by an appointment to the Emerging Infectious Diseases Fellowship Program administered by the Association of Public Health Laboratories and funded by the Centers for Disease Control and Prevention (CDC). Funding was also provided by the Global Disease Detection program at the CDC.
We thank Jessica Waller, Rebecca Napoliello, and Daniel Burken for assisting with real-time PCR assays.
Footnotes
Published ahead of print 29 February 2012
REFERENCES
- 1. Angelakis E, et al. 2009. Molecular detection of Bartonella species in rodents from the Lao PDR. Clin. Microbiol. Infect. 15(Suppl 2):95–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Avidor B, et al. 2004. Bartonella koehlerae, a new cat-associated agent of culture-negative human endocarditis. J. Clin. Microbiol. 42:3462–3468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bai Y, Cross PC, Malania L, Kosoy M. 2011. Isolation of Bartonella capreoli from elk. Vet. Microbiol. 148:329–332 [DOI] [PubMed] [Google Scholar]
- 4. Billeter SA, et al. 2009. Infection and replication of Bartonella species within a tick cell line. Exp. Appl. Acarol. 49:193–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Breitschwerdt EB, Kordick DL. 2000. Bartonella infection in animals: carriership, reservoir potential, pathogenicity, and zoonotic potential for human infection. Clin. Microbiol. Rev. 13:428–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chang CC, et al. 2000. Bartonella spp. isolated from wild and domestic ruminants in North America. Emerg. Infect. Dis. 6:306–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chomel BB, Boulouis HJ, Breitschwerdt EB. 2004. Cat scratch disease and other zoonotic Bartonella infections. J. Am. Vet. Med. Assoc. 224:1270–1279 [DOI] [PubMed] [Google Scholar]
- 8. Chomel BB, et al. 1996. Experimental transmission of Bartonella henselae by the cat flea. J. Clin. Microbiol. 34:1952–1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chomel BB, Kasten RW, Henn JB, Molia S. 2006. Bartonella infection in domestic cats and wild felids. Ann. N. Y. Acad. Sci. 1078:410–415 [DOI] [PubMed] [Google Scholar]
- 10. Ciervo A, Ciceroni L. 2004. Rapid detection and differentiation of Bartonella spp. by a single-run real-time PCR. Mol. Cell Probes 18:307–312 [DOI] [PubMed] [Google Scholar]
- 11. Colborn JM, et al. 2010. Improved detection of Bartonella DNA in mammalian hosts and arthropod vectors by real-time PCR using the NADH dehydrogenase gamma subunit (nuoG). J. Clin. Microbiol. 48:4630–4633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cotte V, et al. 2008. Transmission of Bartonella henselae by Ixodes ricinus. Emerg. Infect. Dis. 14:1074–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dehio C, et al. 2001. Bartonella schoenbuchii sp. nov., isolated from the blood of wild roe deer. Int. J. Syst. Evol. Microbiol. 51:1557–1565 [DOI] [PubMed] [Google Scholar]
- 14. Heller R, et al. 1997. Prevalence of Bartonella henselae and Bartonella clarridgeiae in stray cats. J. Clin. Microbiol. 35:1327–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Higgins JA, Radulovic S, Jaworski DC, Azad AF. 1996. Acquisition of the cat scratch disease agent Bartonella henselae by cat fleas (Siphonaptera: Pulicidae). J. Med. Entomol. 33:490–495 [DOI] [PubMed] [Google Scholar]
- 16. Jacomo V, Kelly PJ, Raoult D. 2002. Natural history of Bartonella infections (an exception to Koch's postulate). Clin. Diagn. Lab. Immunol. 9:8–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Karzai AW, Roche ED, Sauer RT. 2000. The SsrA-SmpB system for protein tagging, directed degradation and ribosome rescue. Nat. Struct. Biol. 7:449–455 [DOI] [PubMed] [Google Scholar]
- 18. Kim CM, et al. 2005. Detection of Bartonella species from ticks, mites and small mammals in Korea. J. Vet. Sci. 6:327–334 [PubMed] [Google Scholar]
- 19. Koehler JE. 1996. Bartonella infections. Adv. Pediatr. Infect. Dis. 11:1–27 [PubMed] [Google Scholar]
- 20. Kosoy M, et al. 2010. Identification of Bartonella infections in febrile human patients from Thailand and their potential animal reservoirs. Am. J. Trop. Med. Hyg. 82:1140–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kosoy M, et al. 2008. Bartonella tamiae sp. nov., a newly recognized pathogen isolated from three human patients from Thailand. J. Clin. Microbiol. 46:772–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. La Scola B, Zeaiter Z, Khamis A, Raoult D. 2003. Gene-sequence-based criteria for species definition in bacteriology: the Bartonella paradigm. Trends Microbiol. 11:318–321 [DOI] [PubMed] [Google Scholar]
- 23. Maillard R, et al. 2004. Bartonella chomelii sp. nov., isolated from French domestic cattle (Bos taurus). Int. J. Syst. Evol. Microbiol. 54:215–220 [DOI] [PubMed] [Google Scholar]
- 24. Morick D, et al. 2009. Detection of Bartonella spp. in wild rodents in Israel using HRM real-time PCR. Vet. Microbiol. 139:293–297 [DOI] [PubMed] [Google Scholar]
- 25. Vayssier-Taussat M, Le Rhun D, Bonnet S, Cotte V. 2009. Insights in Bartonella host specificity. Ann. N. Y. Acad. Sci. 1166:127–132 [DOI] [PubMed] [Google Scholar]
- 26. Ying B, Kosoy MY, Maupin GO, Tsuchiya KR, Gage KL. 2002. Genetic and ecologic characteristics of Bartonella communities in rodents in southern China. Am. J. Trop. Med. Hyg. 66:622–627 [DOI] [PubMed] [Google Scholar]