Abstract
Early targeted antimicrobial therapy helps decrease costs and prevents the spread of antimicrobial resistance, including in Escherichia coli, the most frequent Gram-negative bacterium that causes sepsis. Therefore, rapid susceptibility testing represents the major prerequisite for knowledge-based successful antimicrobial treatment. To accelerate testing for antibiotic susceptibility, we have developed a new mass spectrometry-based assay for antibiotic susceptibility testing (MAAST). For proof of principle, we present an ampicillin susceptibility test for E. coli with a turnaround time of 90 min upon growth detection.
TEXT
Immediate empirically based broad-spectrum antibiotic therapy is a cornerstone in treatment of sepsis (8, 9). However, the early de-escalation of broad-spectrum antibiotic therapy based on knowledge of microbial identification and susceptibility not only reduces therapeutic costs (1, 2, 12) but also reduces the spread of antimicrobial resistance (3, 5, 10).
Unfortunately, the turnaround time (TAT) of classical antibiotic susceptibility testing (AST) based on microbial growth inhibition is 7.5 to 14 h for direct AST from blood culture (BC) fluid or 31.5 to 36 h for indirect AST from subcultured bacteria (4). We have developed a mass spectrometry (MS)-based assay for antibiotic susceptibility testing (MAAST) that allows us to provide meaningful health care information on AST results in less than 90 min after primary microbial growth is detected in a BC that has turned positive.
MAAST is based on the detection, identification, and quantification of antibiotics and their corresponding metabolization/inactivation products as generated by β-lactamases. The feasibility of this test principle for the detection of ampicillin susceptibility in Escherichia coli is based on the hydrolysis of ampicillin by β-lactamases into ampicillin-penicilloic acid. The hydrolysis of ampicillin results in a mass shift of ampicillin from m/z 350 Da to m/z 368 Da, which allows easy detection of all compounds by MS (Fig. 1A). To increase the specificity of the assay, compounds were quantified after identification by collision-induced dissociation (CID) into compound-specific dissociation products.
Fig 1.
(A) MS full-scan analysis showing the detection of ampicillin (m/z 350 Da) and its hydrolysis products ampicillin-penicilloic acid (m/z 368 Da) and penilloic acid (m/z 324 Da). Ampicillin and ampicillin-penicilloic acid were chosen for collision fragmentation into the specific products m/z 350→160 Da and m/z 368→324 Da. (B) Separations and quantifications were performed by LC-MS using an Agilent series 1100 LC system with an Zorbax Eclipse XDB-C18 column and a nonisocratic mobile phase that consisted of a continuous gradient of ammonium formiate and methanol at a temperature of 60°C and with an HCT Ultra mass spectrometer as detector. Specific masses with the specific CID products were eluted by 7.5 and 12 min. (C) MAAST linearity for the detection of ampicillin ranges from 0.1 to 100 μg/liter (coefficient of variation, <20%). (D) Kinetic analysis of bacterial ampicillin hydrolysis shows the decreasing amount of ampicillin and inverse gain of ampicillin-penicilloic acid. Ampicillin is completely degraded after 120 min. The drug/metabolite ratio allows a secure differentiation of ampicillin susceptibility and resistance after 30 min of incubation.
As known from drug monitoring, multiple compounds can be separated by high-pressure liquid chromatography (HPLC) and quantified by calculation of the peak area of the separated compounds. The combination of HPLC and mass spectrometry (LC-MS) allows simultaneous detection, identification, and quantification of ampicillin and metabolites (Fig. 1B). HPLC separation was performed using an Agilent series 1100 LC system (Agilent Technologies, Germany) with a Zorbax Eclipse XDB-C18 column (Agilent Technologies) and a nonisocratic mobile phase consisted of a continuous gradient of 4 mM ammonium formiate and 90% methanol at 60°C. For MS detection, we used an HCT Ultra mass spectrometer (Bruker Daltonics) with an electrospray ionization (ESI) interface. Nitrogen served as a nebulizer and drying gas. Argon was used for CID. Mass spectrometric data were continuously collected during the complete HPLC run.
Using this setting, the assay range for the quantification was found to be 4 orders of magnitude, with a coefficient of variation of <20% (Fig. 1C), allowing the detection of high but also very low β-lactamase activity.
A time course analysis of bacterial ampicillin inactivation by E. coli revealed complete hydrolysis of ampicillin within 2 h (Fig. 1D). All measurements were normalized and expressed as metabolite/ampicillin peak ratios which improve the inter- and intra-assay levels of precision of MAAST. Using this peak ratio for classification, the diagnostic accuracy of MAAST was 100% for incubation periods beyond 30 min, even if ampicillin hydrolysis is only partially completed at this time. From subcultured bacteria, the overall assay time was determined to be <60 min.
The robustness and efficiency of this assay system were validated using 60 E. coli isolates from clinical samples previously characterized using a Vitek 2 system. Twenty strains were characterized as ampicillin susceptible (≤8 μg/ml; CLSI), and 40 strains had been typed as resistant (≥32 μg/ml; CLSI). Twenty of 40 isolates had been phenotypically characterized to harbor an extended-spectrum β-lactamase (ESBL) (Vitek 2 card AST-118; bioMérieux). We included ESBL E. coli because the catalytic activity of ESBLs might differ from those of other β-lactamases. Specifically, their catalytic center is mutated to also bind broad-spectrum cephalosporins (11).
Defined bacterial inocula were suspended in 10 μg/ml ampicillin in deionized water. After 30 min of incubation at 37°C, bacteria were sedimented by centrifugation at 10,000 × g for 3 min. Ampicillin and its hydrolysis product were quantified within the bacterium-free supernatant.
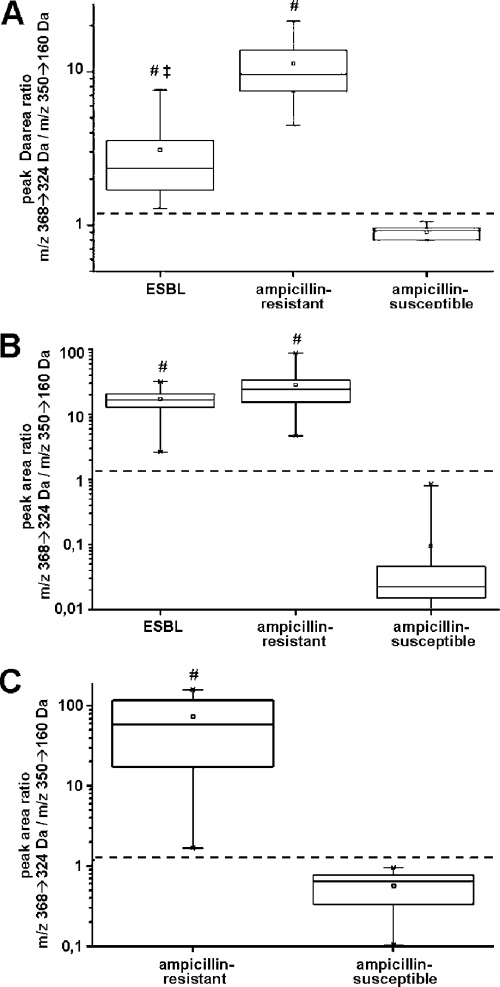
Initially, we tested MAAST with single colonies of each E. coli strain. As shown in Fig. 2A, ampicillin-susceptible E. coli strains show an ampicillin/metabolite ratio of around 1, thereby completely separating them from the ratios seen in the resistant strains. Specifically, a signal/noise ratio of 3 was observed from the ESBL strains, and a ratio of 10 was seen in non-ESBL ampicillin-resistant E. coli strains. Interestingly, the ESBL subset displayed a significantly lower activity than the bacteria carrying non-ESBL β-lactamases, possibly due to lower ampicillin conversion activities in ESBL (Fig. 2A). Accordingly, the increase in bacterial cell mass in the MAAST (i.e., 3 colonies/strain) improved the distinction of ESBL strains from ampicillin-sensitive bacteria. Specifically, MAAST separated ampicillin-susceptible strains from all resistant strains, with differences in peak ratios of 2 to 3 orders of magnitude, while it reduced the peak ratio difference between ESBL and non-ESBL β-lactamase strains (Fig. 2B). This observation has now prompted future studies using a broad-spectrum cephalosporin as the substrate.
Fig 2.
Differentiation of ampicillin-sensitive and -resistant E. coli strains by MAAST. Sixty E. coli strains were obtained from clinical isolates and previously identified and characterized by a Vitek 2 system, cryo-conserved, and subcultured on Luria-Bertani agar plates for 24 h at 37°C prior to MAAST. (A) The bacterial mass corresponding to a single colony was used for analysis. Using this low inoculum, ESBL and ampicillin-resistant non-ESBL strains showed a significantly higher drug/metabolite ratio than ampicillin-susceptible strains without overlap. Additional, ESBL strains showed a significantly lower drug/metabolite ratio than ampicillin-resistant non-ESBL strains. (B) Using the bacterial mass corresponding to 3 colonies, ESBL and ampicillin-resistant non-ESBL strains showed a significantly higher drug/metabolite ratio than ampicillin-susceptible strains without overlap. (C) Bacteria in positive BC flasks of 24 clinical specimens were biotyped as E. coli using MALDI-TOF MS and used for analysis. MAAST results were retrospectively compared to results of classical AST with a Vitek 2 system. As seen in for subcultures, direct MAAST from BC showed an accuracy of 100% for the detection of β-lactamase activity. #, P < 0.005 versus ampicillin sensitive; ‡, P < 0.05 versus β-lactamase positive (Student's t test).
MAAST was further evaluated using another 24 E. coli strains directly obtained from positive blood culture (BC) bottles without prior processing. For the recovery of bacteria from BC, 5-ml suspensions were taken from BC flasks and centrifuged at 3,000 × g for 10 min in serum Monovette tubes containing separation gel (Sarstedt, Germany). Bacteria now located on top of the gel layer were suspended in 1 ml 0.9% NaCl solution, washed twice, and counted in a UF1000i urine flow cytometer (Sysmex, Hamburg) to adjust to a density of approximately 1,000 bacteria/μl. A 180-μl bacterial suspension was supplemented with 10 μg/ml ampicillin, incubated for 30 min at 37°C, and sedimented by centrifugation at 10,000 × g for 3 min. Ampicillin and its hydrolysis product were quantified within the supernatant. Using this setting, MAAST was sufficient to differentiate ampicillin-susceptible from -resistant strains with 100% accuracy (Fig. 2C). The total assay time, including the extraction and preparation of bacteria, was well below 90 min.
Taken together, we have demonstrated in this proof-of-principle study that MAAST is capable of safely determining resistance of E. coli to ampicillin within 90 min of the first detection of microbial growth in BC. Compared to conventional growth inhibition tests, MAAST dramatically accelerates the antibiotic susceptibility diagnosis and thereby may allow an earlier de-escalation of antibiotic therapy. In light of the well-known importance of nosocomial infections, rapid AST has been shown to save time, reduce antibiotic use and, consequently, therapeutic costs (7), and aid in the prevention of spreading antibiotic resistance (3, 5, 10).
As the principle of MAAST is based on the degradation of antibiotics—in our model case ampicillin—the assay is currently limited to resistance mechanisms caused by substrate metabolization. Since β-lactams are frequently used to treat hospital patients, MAAST could be helpful in many cases, especially in susceptibility testing of Gram-negative bacteria.
While this work was in progress, a different approach had been reported employing matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) MS analysis to detect carbapenem resistance from subcultured isolates of Enterobacteriaceae, including E. coli, Klebsiella pneumoniae, and Pseudomonas aeruginosa (6). However, for reasons of sensitivity, different carbapenemases like KPC-2, NDM-1, different IMPs, VIM1, and VIM2 required complete substrate hydrolysis to safely detect metabolites. This is due to the incapability of MALDI-TOF MS to quantify substrates and the unfavorable low molecular masses of these molecules. By comparison, MAAST employs LC and tandem MS (MS/MS) to detect minute β-lactamase activities and allows quantification of both the mother substance and metabolite. In conclusion, MAAST represents a powerful, fast, and versatile tool to rapidly detect antibiotic resistance in human bacterial pathogens.
ACKNOWLEDGMENTS
We gratefully acknowledge the technical assistance of Christina Haese, Hüsseyin Yörük, and Korinna Mosbach.
Footnotes
Published ahead of print 8 February 2012
REFERENCES
- 1. Beekmann SE, Diekema DJ, Chapin KC, Doern GV. 2003. Effects of rapid detection of bloodstream infections on length of hospitalization and hospital charges. J. Clin. Microbiol. 41:3119–3125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Coleman RW, Rodondi LC, Kaubisch S, Granzella NB, O'Hanley PD. 1991. Cost-effectiveness of prospective and continuous parenteral antibiotic control: experience at the Palo Alto Veterans Affairs Medical Center from 1987 to 1989. Am. J. Med. 90:439–444 [PubMed] [Google Scholar]
- 3. Fraser A, et al. 2006. Benefit of appropriate empirical antibiotic treatment: thirty-day mortality and duration of hospital stay. Am. J. Med. 119:970–976 [DOI] [PubMed] [Google Scholar]
- 4. Gherardi G, et al. 2011. Comparative evaluation of the Vitek-2 Compact and Phoenix systems for rapid identification and antibiotic susceptibility testing directly from blood cultures of Gram-negative and Gram-positive isolates. Diagn. Microbiol. Infect. Dis. 72:20–31 [DOI] [PubMed] [Google Scholar]
- 5. Goldmann DA, et al. 1996. Strategies to prevent and control the emergence and spread of antimicrobial-resistant microorganisms in hospitals. A challenge to hospital leadership. JAMA 275:234–240 [PubMed] [Google Scholar]
- 6. Hooff GP, et al. 2011. Characterization of beta-lactamase enzyme activity in bacterial lysates using MALDI-mass spectrometry. J. Proteome Res. 11:79–84 [DOI] [PubMed] [Google Scholar]
- 7. Kerremans JJ, et al. 2008. Rapid identification and antimicrobial susceptibility testing reduce antibiotic use and accelerate pathogen-directed antibiotic use. J. Antimicrob. Chemother. 61:428–435 [DOI] [PubMed] [Google Scholar]
- 8. Leibovici L, et al. 1998. The benefit of appropriate empirical antibiotic treatment in patients with bloodstream infection. J. Intern. Med. 244:379–386 [DOI] [PubMed] [Google Scholar]
- 9. Lodise TP, McKinnon PS, Swiderski L, Rybak MJ. 2003. Outcomes analysis of delayed antibiotic treatment for hospital-acquired Staphylococcus aureus bacteremia. Clin. Infect. Dis. 36:1418–1423 [DOI] [PubMed] [Google Scholar]
- 10. McGowan JE., Jr 1994. Do intensive hospital antibiotic control programs prevent the spread of antibiotic resistance? Infect. Control Hosp. Epidemiol. 15:478–483 [DOI] [PubMed] [Google Scholar]
- 11. Pfaller MA, Segreti J. 2006. Overview of the epidemiological profile and laboratory detection of extended-spectrum beta-lactamases. Clin. Infect. Dis. 42(Suppl 4):S153–S163 [DOI] [PubMed] [Google Scholar]
- 12. Tumbarello M, et al. 2010. Costs of bloodstream infections caused by Escherichia coli and influence of extended-spectrum-beta-lactamase production and inadequate initial antibiotic therapy. Antimicrob. Agents Chemother. 54:4085–4091 [DOI] [PMC free article] [PubMed] [Google Scholar]