Abstract
In the era after the introduction of the meningococcal serogroup C conjugate vaccine, from 1 January 2003 to 31 December 2010, serogroup B meningococci were the major cause of invasive meningococcal disease in the province of Québec, Canada, being responsible for 72% of all meningococcal disease cases. Of the 334 invasive serogroup B Neisseria meningitidis strains analyzed, 53.9% belonged to the ST-269 clonal complex (CC). Since it first emerged in 2003, the percentage of invasive serogroup B isolates that belonged to the ST-269 CC had increased from 35% in 2003 to 76% in 2010. Among the 180 meningococci in the ST-269 CC, 91.7% belonged to a single ST (ST-269). The most common PorA genotypes identified in the ST-269 CC were (i) VR1 19-1, VR2 15-11, VR3 36 (84%) and (ii) VR1 18-7, VR2 9, VR3 35-1 (9%). Cases of invasive disease due to the ST-269 CC were commonly found in those aged 11 to 19 years (30.5%) and 20 to 40 years (25.5%). Meningococci of the ST-269 CC were uncommon in other Canadian provinces. In contrast to the ST-269 CC, invasive serogroup B meningococci that belonged to the ST-41/44 CC were much more diverse genetically. However, one ST (ST-571), which is uncommon in the United States, accounted for 35% of all cases due to this CC. The current finding suggests that the ST-269 clone may indeed represent an emerging hypervirulent clone of meningococci.
INTRODUCTION
Invasive meningococcal disease (IMD), is caused by the strict human pathogen Neisseria meningitidis, which is divided into 12 different serogroups based on unique capsular polysaccharide structures on the surface of the bacteria that can be recognized by specific serogrouping antisera (53). Further phenotypic subdivisions are based on 2 major outer membrane proteins, PorB and PorA, which contain antigens, detectable by a panel of monoclonal antibodies, corresponding to serotypes and serosubtypes, respectively (24). Genetic analysis by multilocus sequence typing (MLST), which is based on nucleotide sequence differences in 7 housekeeping enzyme genes, allows the bacteria to be grouped into clonal groups or clonal complexes (CCs) (34).
Most meningococcal infections are not noticed due to asymptomatic transmissions occurring between healthy individuals (carriers), but for reasons not completely understood, sometimes infections may result in serious invasive disease, with mortality rates reaching 10% or more (43). IMD occurs globally as endemic or sporadic disease, and occasionally outbreaks or epidemics occur. Analysis of N. meningitidis isolates from sporadic disease cases or asymptomatic carriers shows that the organism is composed of a highly heterogeneous population of bacteria, while isolates recovered from outbreaks or epidemics usually belong to only a limited number of clones, so-called hypervirulent clones or lineages (10, 41).
In Canada, the last major meningococcal epidemic occurred in the late 1930s to early 1940s and was caused by serogroup A bacteria, with a peak incidence of close to 13 cases per 100,000 population (36). Another major event in more recent history was the identification of the serogroup C, serotype 2a, electrophoretic type (ET) 15 clone, which emerged in 1986 (2) to cause two waves of outbreaks in different provinces in Canada in 1989 to 1991 (17, 22, 28, 46, 48, 54) and again in 2000 to 2001 (40, 49, 50). This clone also spread to the United States (29) and other parts of the world (31).
As a result of the serogroup C meningococcal outbreaks, the province of Québec instituted two rounds of province-wide vaccination campaigns in the last 20 years. The first campaign was launched between December 1992 and March 1993 using plain tetravalent (A, C, Y, W135) or bivalent (A, C) polysaccharide vaccines (17). The second province-wide vaccination campaign was launched in May to June 2001 using the serogroup C glycoconjugate vaccine (Menjugate; Chiron, Emeryville, CA), which was offered to all residents age 2 months to 20 years (18, 19).
In the winter of 2004 to 2005 and 3 years after the second round of province-wide vaccination against serogroup C N. meningitidis, we identified an unique clone of serogroup B meningococci, with the antigenic formula B:17:P1.19 and multilocus sequence type (ST) 269, causing a cluster of IMD cases in a region (Chaudiere-Appalaches, region 12) near the capital city of the province (32). During a roughly 2-year period from March 2003 to June 2005, 38 cases due to this clone had been identified. In the subsequent years, serogroup B meningococci have been identified as the predominant cause of IMD in Québec.
In this study, we characterized all serogroup B N. meningitidis isolates recovered from culture-confirmed IMD cases in the province of Québec during the last 8 years, from January 2003 to December 2010. The objectives of this study were to analyze the clonal diversity of invasive serogroup B strains in the province and to provide an update on the characteristics of this emerging clone of ST-269.
MATERIALS AND METHODS
IMD cases and bacterial isolates.
Since IMD is a notifiable disease, clinicians and laboratories are required to report cases to the public health authority, and laboratories are invited to submit isolates from IMD cases to the provincial public health laboratory, Laboratorie de Santé Publique du Québec (LSPQ). Only culture-confirmed IMD cases were included in this study, and age information was recorded from the specimen requisition form. When N. meningitidis was recovered from multiple specimen types, only one isolate was included for the analysis.
As most cases reported by clinicians can be linked to laboratory isolates, we concluded that most isolates have been captured from culture-confirmed meningococcal disease cases in the province by the existing surveillance system. Only about 15% of the IMD cases were not included here, since they were diagnosed by PCR and therefore cultures were not available for further study.
The age distribution of cases due to isolates belonging to the serogroup B ST-269 clonal complex (CC) was compared to the age distribution of cases due to isolates belonging to other CCs by using the χ2 test. Analyses were performed by using SAS software version 9.2 (SAS Institute, Cary, NC). A P value of <0.05 was considered statistically significant.
Identification, serogrouping, and typing of meningococci.
N. meningitidis was identified by standard biochemical tests, and serogrouping was done by the bacterial agglutination test with rabbit serogrouping antisera at the LSPQ (42). Serotyping and serosubtyping were done at the National Microbiology Laboratory (NML) using a whole-cell enzyme-linked immunosorbent assay (ELISA) (1) with a monoclonal antibody typing kit (Rijksinstituut voor Volksgezondheid en Milieu, National Institute of Public Health, Bilthoven, The Netherlands). Monoclonal antibodies to serotype antigens 17 and 19 and serosubtype antigen P1.19 were kind gifts from Wendell Zollinger of the Walter Reed Army Institute of Research.
Multilocus sequence typing (MLST) was done according to the method described by Maiden et al. (34), and isolates were assigned to STs according to the Neisseria MLST website (http://pubmlst.org/neisseria). Analysis of the MLST data from the ST-269 CC was done by eBURST (23). Sequencing of porA genes was done following the protocols of Sacchi et al. (44) and Clarke et al. (12). The PorA variable-region (VR) types were described according to the nomenclature given in the N. meningitidis PorA variable-region database (http://neisseria.org/perl/agdbnet/agdbnet.pl?file=poravr.xml) and by de Filippis et al. (13).
RESULTS
Serogroup distribution of invasive Neisseria meningitidis isolates from 2003 to 2010.
During the period of 1 January 2003 to 31 December 2010, a total of 466 culture-confirmed IMD cases were identified based on N. meningitidis isolates provided by the LSPQ to the NML, with an average of 58 cases per year. A retrospective audit of isolates at the two facilities found 95% to 100% agreement of the yearly data among the 466 case isolates identified over the 8-year period, and all invasive serogroup B isolates described in this report had been accounted for at both the LSPQ and the NML. Minor discrepancies included 2 case isolates from nonresidents of Québec, which were therefore not included in the LSPQ data set. However, these 2 isolates were received at the NML and were not excluded since it was not known at the time of our analysis if the individuals contracted their infection inside or outside Québec. Overall, 72% of the cases were due to serogroup B, while there were 75 cases (16%) caused by serogroup C, 29 cases (6%) caused by serogroup Y, 26 cases (5.6%) caused by serogroup W135, and 2 cases (0.4%) due to serogroup 29E (Table 1).
Table 1.
Distribution of Neisseria meningitidis serogroups as causes of culture-positive invasive meningococcal disease cases in Québec, Canada, 2003 to 2010
| Yr | Number (%) of cases caused by serogroup: |
Total no. of cases | ||||
|---|---|---|---|---|---|---|
| B | C | Y | W135 | Other | ||
| 2003 | 29 (58) | 13 | 7 | 1 | 0 | 50 |
| 2004 | 37 (64) | 17 | 2 | 2 | 0 | 58 |
| 2005 | 42 (66) | 13 | 2 | 6 | 1 (serogroup 29E) | 64 |
| 2006 | 46 (70) | 17 | 2 | 1 | 0 | 66 |
| 2007 | 53 (75) | 7 | 5 | 5 | 1 (serogroup 29E) | 71 |
| 2008 | 37 (70) | 5 | 5 | 6 | 0 | 53 |
| 2009 | 48 (86) | 1 | 3 | 4 | 0 | 56 |
| 2010 | 42 (88) | 2 | 3 | 1 | 0 | 48 |
| All | 334 (72) | 75 | 29 | 26 | 2 | 466 |
The number of serogroup B cases has been increasing since we first documented an outbreak in this province due to an emerging clone of ST-269 (32), from 29 cases in 2003 to a high of 53 cases in 2007. Since then, the numbers decreased somewhat to 37 cases in 2008 and remained elevated in 2009 and 2010, with 48 and 42 cases, respectively. Although the total number of culture-positive IMD cases might have peaked in 2007, with a total of 71 IMD cases and 53 cases, or 75%, that were due to serogroup B, the percentage of IMD cases due to serogroup B meningococci has continued to increase to 86 and 88% in 2009 and 2010, respectively.
A total of 29 serogroup Y IMD cases were found in the past 8 years, and the number ranged from a high of 7 cases in 2003 to a low of 2 cases in each of the years from 2004 to 2006, with an average of about 3 to 4 cases per year. For serogroup W135 cases, the number also fluctuated, from a high of 6 cases in 2005 to a low of 1 case per year in 2003, 2006, and 2010.
Clonal analysis of invasive serogroup B N. meningitidis isolates.
A total of 89 sequence types (STs) were identified among the 334 invasive serogroup B isolates, and 75 of these 89 STs were grouped into 14 CCs, with the remaining 14 STs not assigned to any known CC according to the Neisseria MLST website (Table 2). The genetic diversity of the different CCs may be examined by the number of STs involved in causing invasive disease. There was greater genetic diversity in the ST-41/44 CC strains than in the ST-269 CC strains responsible for IMD in the province of Québec in Canada.
Table 2.
Serogroup B IMD cases in Québec, Canada, 2003 to 2010, grouped by clonal complexes
| Clonal complex (no. of STs) | No. of serogroup B IMD cases in: |
Total no. (%) of cases | No. of cases/no. of STs | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 2003 | 2004 | 2005 | 2006 | 2007 | 2008 | 2009 | 2010 | |||
| ST-269 (11) | 10 | 17 | 21 | 27 | 28 | 17 | 28 | 32 | 180 (53.9) | 16.36 |
| ST-41/44 (31) | 8 | 11 | 7 | 8 | 12 | 12 | 12 | 8 | 78 (23.4) | 2.52 |
| ST-35 (10) | 3 | 1 | 2 | 3 | 1 | 5 | 0 | 1 | 16 (4.8) | 1.60 |
| ST-32 (5) | 0 | 0 | 3 | 0 | 4 | 1 | 3 | 0 | 11 (3.3) | 2.20 |
| ST-60 (3) | 0 | 0 | 2 | 2 | 1 | 1 | 0 | 0 | 6 (1.8) | 2.00 |
| ST-37 (2) | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 4 (1.2) | 2.00 |
| ST-212 (2) | 2 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 4 (1.2) | 2.00 |
| ST-11 (1) | 0 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 3 (0.9) | 3.00 |
| ST-213 (3) | 0 | 0 | 0 | 1 | 2 | 0 | 0 | 0 | 3 (0.9) | 1.00 |
| ST-461 (2) | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 3 (0.9) | 1.50 |
| ST-162 (1) | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 2 (0.6) | 2.00 |
| ST-254 (2) | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 2 (0.6) | 1.00 |
| ST-103 (1) | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 (0.3) | 1.00 |
| ST-1157 (1) | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 (0.3) | 1.00 |
| Unassigneda (14) | 5 | 3 | 3 | 4 | 0 | 1 | 4 | 0 | 20 (6.0) | 1.43 |
| All (89) | 29 | 37 | 42 | 46 | 53 | 37 | 48 | 42 | 334 (100.1) | 3.75 |
Not recognized as belonging to any known clonal complex according to the Neisseria MLST website.
Two CCs (ST-269 and ST-41/44) were responsible for 77% of all invasive serogroup B cases. The number of culture-positive IMD cases due to isolates within the ST-269 CC appeared to be increasing during the period of this study, from 10 cases in 2003 to 32 cases in 2010. This increase paralleled to the gradual increase in the percentage of invasive serogroup B cases due to this CC, from 35% in 2003 to 76% in 2010. In contrast, the number of cases due to isolates of the ST-41/44 CC appeared to be relatively stable throughout the study period, and this was also reflected in the percentage of cases due to this CC. The number as well as the percentage of invasive serogroup B cases due to all other clonal groups also appeared to be decreasing from 2003 to 2010.
Among the 78 individual serogroup B IMD cases caused by isolates of the ST-41/44 CC, 27 cases, or 35%, were caused by ST-571 isolates. The rest (51 cases) were caused by isolates belonging to 30 other different STs. Among these 30 other STs, 20 STs occurred only once or caused only single case, 4 STs occurred twice or caused 2 cases per ST, 2 STs occurred thrice, 3 STs occurred four times each, and ST-944 caused 5 cases.
There were 16 cases caused by isolates in the ST-35 CC, and 10 different STs were represented among the 16 case isolates, with ST-35 and ST-570 each causing 3 cases, ST-2574 and ST-3626 each causing 2 cases, and 6 other STs each causing only 1 case. Of the 11 cases caused by the ST-32 CC, 4 were due to ST-32, 3 were due to ST-2726, 2 were due to ST-33, and one case each was due to ST-2858 and ST-6544. The 6 case isolates that belonged to the ST-60 CC were highly related, with 4 isolates being ST-60 and the remaining 2 isolates being single-locus variants (SLVs) of ST-60, namely, ST-466 and ST-6546.
There were 3 cases due to the ST-11 or ET-37 CC. All 3 isolates were typed as B:2a:P1.7,1 and were of the ET-15 variant of the ST-11 or ET-37 CC.
Characteristics of the ST-269 clone in Québec, Canada.
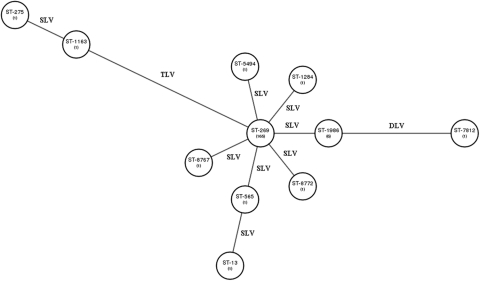
The distribution of isolates among the different STs in the ST-269 CC and their genetic relationship as revealed by the allelic profile of the multilocus sequence typing scheme are described in Fig. 1. Of the 180 isolates that belonged to the ST-269 CC, 91.7% (165 isolates) belonged to ST-269. Eleven isolates belonged to 6 STs that were SLVs of ST-269. Also related to ST-269 were 2 different STs (ST-13, which is an SLV of ST-565, and ST-7812, which is a double-locus variant of ST-1986), and each of these caused only one case. Finally 2 STs (ST-275 and ST-1163) with 1 isolate each and determined to be related, since 6 of their 7 housekeeping gene alleles were identical to each other, were somewhat more distant from the founding ST, ST-269. For example, both ST-275 and ST-1163 had three housekeeping gene alleles different from those of ST-269 and five housekeeping gene alleles different from those of ST-7812.
Fig 1.
eBURST analysis to show the relationship of different sequence types (STs) within the ST-269 clonal complex based on multilocus sequence typing data. Each ST is represented by a circle and is related to other STs by the number of allelic differences: single-locus variant (SLV), double-locus variant (DLV), and triple-locus variant (TLV). The numbers shown in parentheses represent the number of isolates found for each ST.
The antigenic characteristics and PorA genotypes of the ST-269 isolates are summarized in Table 3. One hundred sixty-five of the 180 ST-269 CC isolates belonged to ST-269, and 160 were serotype 17, of which 140 expressed the serosubtype antigen P1.19. Sixteen isolates were B:17:P1.9, two were B:17:P1.6, and two were B:17:P1.-.
Table 3.
Antigenic formulas and PorA genotypes of Québec serogroup B isolates belonging to the ST-269 clonal complex
| ST(s) (n) | Antigenic formula (n) | PorA genotype(s) (VR1/VR2/VR3) |
|---|---|---|
| ST-269 (165) | B:17:P1.19 (140) | 19-1/15-11/36 |
| B:17:P1.9 (16) | 18-7/9/35-1 | |
| B:17:P1.6 (2) | 18-1/3/38 | |
| B:17:P1.- (2) | 7-2/30-3/38, 18-1/30-30/38 | |
| B:NT:P1.19 (2) | 19-1/13-1/35-1, 19-1/15-11/36 | |
| B:4:P1.- (1) | 19-1/15-11/36 | |
| B:14:P1.15 (1) | 19/15/36 | |
| B:NT:P1.6 (1) | 18-1/3/38 | |
| Other (15) | ||
| ST-1986 | B:17:P1.19 (6) | 19-1/15-11/36 |
| ST-1284, ST-5495, ST-8767, ST-8772 | B:17:P1.19 (4) | 19-1/15-11/36 |
| ST-275 | B:17:P1.9 (2) | 22/9/35-1 |
| ST-565 | B:NT:P1.13 (1) | 19/13/35-1 |
| ST-13 | B:NT:P1.16 (1) | 31/16/37-1 |
| ST-7812 | B:19:P1.7,1 (1) | 71-/1/35-1 |
The most common PorA genotypes were (i) VR1 19-1, VR2 15-11, VR3 36 (152 [84.4%] for the ST-269 CC and 142 isolates [86%] for ST-269 alone), (ii) VR1 18-7, VR2 9, VR3 35-1 (16 isolates of ST-269), and (iii) VR1 18-1, VR2 3, VR3 38 (3 isolates).
Characteristics of culture-confirmed IMD cases due to isolates belonging to serogroup B ST-269 CC.
In 115 cases (64%), the bacteria were isolated from cerebrospinal fluid (CSF) cultures of patients, including one case with the isolate obtained from both blood and CSF cultures. In contrast, 64 cases (36%) had positive blood cultures, and in one case the isolate was obtained from the synovial fluid. Although almost twice as many patients had positive CSF cultures than had positive blood cultures, the clinical presentations of the cases were not known, as this information is not routinely captured by our laboratory surveillance activities.
More than 50% of the cases occurred in those aged 11 to 40 years, with 55 cases (30.5%) among those aged 11 to 19 years and 46 cases (25.5%) in those aged 20 to 40 years. There were 30 cases (16.7%) in those aged 1 to 5 years, and 19 cases (10.6%) occurred in those aged 41 to 60 years. In contrast, there were only 9 cases (5%) in infants less than 12 months old, 9 cases (5%) in those aged 6 to 10 years, and only 12 cases (6.7%) in those over the age of 60 years. The age distribution of cases due to isolates belonging to serogroup B ST-269 CC was significantly different from those of cases due to isolates belonging to other CCs (P < 0.0001 by chi-square test) (Table 4). There were almost twice as many cases in those aged 11 to 40 years due to the ST-269 CC (56%) compared to other CCs (30%). In contrast, there were almost 5 times fewer children <1year of age among cases due to the ST-269 CC (5%) compared to other CCs.
Table 4.
Comparison of the age distributions of invasive serogroup B meningococcal disease cases according to the clonal nature of the Neisseria meningitidis strains
| Age group (yr) | No. (%) of: |
|
|---|---|---|
| ST-269 CC cases | Other serogroup B cases | |
| <1 | 9 (5) | 36 (23.4) |
| 1-5 | 30 (16.7) | 37 (24.0) |
| 6-10 | 9 (5) | 6 (3.9) |
| 11-19 | 55 (30.5) | 20 (13.0) |
| 20-40 | 46 (25.5) | 26 (16.9) |
| 41-60 | 19 (10.6) | 15 (9.7) |
| >60 | 12 (6.7) | 14 (9.1) |
| All | 180 | 154 |
DISCUSSION
To the best of our knowledge, this study represents the largest single report of serogroup B IMD cases due to highly related isolates belonging to the ST-269 CC. In the past, epidemic spread of serogroup B strains involved clones or CCs such as ET-5 or ST-32 (e.g., in Norway in the mid-1970s with subsequent intercontinental spread [6] and more recently in the state of Oregon in the United States [20]), ST-41/44 (e.g., in New Zealand beginning in 1991 [35]), and ST-8 or cluster A4 (e.g., in The Netherlands in the 1960s [9] and in many European countries and North America in the1970s [7, 8]). In the province of Québec, with a population of approximately 7.5 million in 2005 and 7.9 million in 2010 according to Statistics Canada, we have documented a total of 180 serogroup B IMD cases due to the ST-269 CC over the last 8 years. Ninety-two percent of the 180 cases were due to one single ST (ST-269), and 140 (85%) of the ST-269 isolates presented with the serotype and serosubtype antigens 17:P1.19 and PorA genotype VR1 19-1, VR2 15-11, VR3 36. This coupled with the fact that 30% of the cases were in those aged 11 to 19 years and another 26% in those aged 20 to 40 years all pointed toward a hyperendemic or epidemic spread of a single clone causing IMD in Québec (38). Thus, the present findings suggest that this clone may indeed represent an emerging hypervirulent clone of meningococci. A detailed epidemiological analysis of this clone as well as other invasive serogroup B clones in Québec will be described separately in the near future.
Over the past 8 years as the ST-269 clone expanded and caused IMD cases in the province, a PorA antigenic variant (B:17:P1.9) with a PorA genotype of VR1 18-7, VR2 9, VR3 35-1) appeared in 2007 to 2010 and accounted for 16 cases, or 9% of all ST-269 CC cases. This may suggest a recombination of the porA genes of the parent ST-269 clone and another meningococcus bearing the PorA antigen of P1.9. It is of interest to note that PorA antigen P1.9 is a common antigen among serogroup B meningococci in Québec (data not shown). Based on multilocus sequence analysis, only 2 singleton isolates or 2 STs (ST-275 and ST-1163) were found among the 180 ST-269 CC isolates, which were distantly related to the parent clone of ST-269 (Fig. 1). Although these 2 isolates were also typed as B:17:P1.9, both had PorA genotypes of VR1 22, VR2 9, VR3 35-1. Also, both STs did not seem to have expanded and were not detected in subsequent years.
The stability of the ST-269 clone in Québec during the past 8 years can be evidenced by a single predominant ST that represented 92% of all the isolates found in this CC, suggesting limited diversification or mutations in their housekeeping enzyme genes. Other single- or double-locus variants of the ST-269 clone also were restricted mostly to causing single cases, other than ST-1986, which caused a total of 6 cases over the years. Also, most (83%) of the ST-269 CC isolates detected in this province had the antigens B:17:P1.19 with a PorA genotype of VR1 19-1, VR2 15-11, VR3 36. Stability of the PorA antigen and genotype during a meningococcal disease epidemic in New Zealand over a period of 13 years has also been reported (16). However, upon intercontinental spread, diversifications do appear to occur, as has been reported for the ET-5 (5) and ET-15 (30) clones. This is most likely the result of genetic recombinations occurring between the epidemic clone and endemic meningococci unique to the locality.
According to the Neisseria meningitidis MLST website (http://neisseria.org/nm/typing/mlstdb/), ST-269 was first identified in the United Kingdom in 1975 in a serogroup B invasive disease isolate with serotype 4 and a PorA genotype of P1.18-7,9. Invasive isolates identified in 1985 with serotype antigen 15 and a PorA genotype of P1.19-1, 15-11 were also described on the N. meningitidis MLST website. However, the ST-269 clone causing invasive disease in Québec, Canada, is quite different from the ST-269 CC that is also prevalent among recent IMD isolates in England and Wales (33). For example, of the 168 IMD isolates that belonged to the ST-269 CC recovered from 2000 to 2008 in England and Wales, 38 distinct STs were found, and 4 major STs were identified: ST-269 represented 38% of the isolates, ST-275 represented 14%, ST-1161 represented 10%, and ST-283 represented 3%. Also, over the years the number of ST-269 isolates had decreased while the number of ST-275 isolates had increased. The genetic diversity of the ST-269 clonal complex found in England and Wales was also reflected in the PorA genotypes found among their isolates, with 30 different PorA subtypes identified. Nevertheless the predominant PorA subtype found there was also P1.15-1, 19-11. While the ST-275 cluster has been increasing in England and Wales, there were only 2 such isolates detected in the province of Québec and both occurred in 2006; none was detected in the last 4 years. However, it is interesting to note that the ST-275 isolate and its SLV of ST-1163 found in Québec also had the PorA genotype of VR1 22, VR2 9, similar to that found in England and Wales (33).
Besides the cases that we had documented in Québec, Canada, 2 other smaller clusters or outbreaks of IMD cases due to the ST-269 CC have been described in the literature. The first report was from Belgium and involved 39 cases from 2006 to 2009 (3). The isolates there were typed as B:NT:P1.14 with a PorA genotype of VR1 22, VR2 14. The second report described a community outbreak in southwest France from December 2008 to September 2009, with 11 cases in a 20-km-diameter area with a population of 375,000 (14). The isolates involved in this outbreak were typed as B:NT:P1.-, ST-269 with a PorA genotype of VR1 19-1, VR2 15-11.
Invasive serogroup B isolates that belong to the ST-269 CC are not very common in other Canadian provinces, with only about 50 such isolates identified in the past 8 years. Also, no particular ST appears to predominate, and as many as 14 distinct STs have been identified from Canadian provinces other than Québec (unpublished data from the NML).
In the United States, among the 520 serogroup B isolates analyzed from the period from 1 January 2000 to 31 December 2005, only 20 isolates, or 3.8%, belonged to the ST-269 CC. Heterogeneity of the ST-269 CC was reflected by 15 distinct STs being represented among the 20 isolates analyzed, with no single predominant ST found (27). In Asia, ST-269 was either rare or not described at all in the 3 studies reported from that region, namely, from China (55), Japan (47), and Taiwan (11).
In contrast to the ST-269 CC with very limited heterogeneity, invasive serogroup B isolates in Québec that belonged to the ST-41/44 CC were much more diverse genetically. Nevertheless, one ST (ST-571) appeared to be more common, being responsible for 35% of all the ST-41/44 IMD cases. Sixteen ST-571 cases (59%) occurred in the 3-year period from 2006 to 2008, while only 2 cases occurred in both 2003 and 2004. In 2009 and 2010, 3 ST-571 cases were identified each year. However, isolates of ST-571 were not common in the United States. In a recent study there, among the 130 invasive serogroup B isolates that belonged to the ST-41/44 CC recovered between 2000 and 2005, ST-571 strains were either absent or caused no more than one case (27). Similarly, ST-571 was either absent or infrequent among the ST-41/44 CC isolates analyzed in a study involving isolates from the Czech Republic, Greece, and Norway (56).
Two candidate serogroup B meningococcal vaccines are in advanced stages of development. The 4CMenB investigational vaccine contains four components: factor H binding protein (fHBP), neisserial heparin binding antigen (NHBA), neisserial adhesin A (NadA), and the PorA antigen P1.4 (25). The other investigational MenB vaccine is a bivalent fHBP (39). Therefore, our next goal will be to examine the genetic and antigenic diversity of the multiple investigational vaccine antigens among our collection of invasive serogroup B isolates. A small pilot study involving DNA sequencing of the antigen genes in a dozen ST-269 isolates recovered from 2003 to 2010 has indicated the absence of NadA and P1.4 and the presence of fHBP allele 15 (or Novartis variant 1 subvariant 15) and NHBA subvariant 21 (data not shown). This is similar to the type of 4CMenB vaccine antigens found among invasive serogroup B ST-269 isolates collected from England and Wales (33).
Since the Québec ST-269 CC isolates did not express the PorA P1.4 or the NadA antigen present in the 4CMenB investigation vaccine, the effectiveness of this candidate vaccine against the ST-269 CC isolates will depend solely on (i) how well the vaccine components fHBP and NHBA will match with the corresponding antigens found in the clinical isolates and (ii) how well these antigens are expressed in vivo by the clinical isolates. Since a significant proportion of the ST-269 CC cases in Québec were due to one homogenous clone of defined PorA antigens, with 94% of the strains possessing the PorA VR2 type 19-11 or 9 (Table 3), it may be feasible to develop a bivalent PorA vaccine that will be ST-269 specific for use in the province of Québec. An advantage of a PorA-specific vaccine is its proven track record in effectiveness against a homogeneous population of meningococci expressing homologous PorA antigens (4, 15, 37, 45). However, vaccines with narrow specificities that target only one or a limited number of serogroups or serotypes, such as the capsular polysaccharide vaccines, in meningococci or pneumococci have a high propensity to lead to the development of vaccine escape mutants due to the phenomenon of capsule switching (52) or capsule replacement (51) and even phase variation in antigen expression (26). Unfortunately, in N. meningitidis, a single species-specific virulence factor that would allow development of an effective vaccine, such as the diphtheria or tetanus toxoid, has not been identified. In search of a comprehensive vaccine against serogroup B meningococci, some investigators have proposed that multiple components may enhance the effectiveness of the vaccine coverage and at the same time minimize the possibility of engendering vaccine escape mutants (21). However, this theory has not been proven in a real-world situation, and therefore further studies involving the epidemiology and laboratory characterization of strains, as well as the postmarketing of any such vaccines, are now required.
ACKNOWLEDGMENTS
We thank the staff of the National Microbiology Laboratory's DNA Core Facility for providing primers and help with DNA sequencing. The members of LSPQ thank Québec's hospital laboratories for their participation in the province's IMD laboratory surveillance program. R.S.W.T. thanks Laura Chihara, Carleton College, for helpful discussion.
This study made use of the Neisseria Multi Locus Sequence Typing website (http://pubmlst.org/neisseria/), developed by Keith Jolly and Man-Suen Chan and sited at the University of Oxford. The development of this site has been funded by the Wellcome Trust and European Union. Some of our MLST data have been submitted to this website.
Footnotes
Published ahead of print 15 February 2012
REFERENCES
- 1. Abdillahi H, Poolman JT. 1987. Whole cell ELISA for typing Neisseria meningitidis with monoclonal antibodies. FEMS Microbiol. Lett. 48:367–371 [PubMed] [Google Scholar]
- 2. Ashton FE, et al. 1991. Emergence of a virulent clone of Neisseria meningitidis serotype 2a that is associated with meningococcal group C disease in Canada. J. Clin. Microbiol. 29:2489–2493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bertrand, et al. 2011. Detection of a geographical and endemic cluster of hyper-invasive meningococcal strains. Microbes Infect. 13:684–690 [DOI] [PubMed] [Google Scholar]
- 4. Bjune G, et al. 1991. Effect of outer membrane vesicle vaccine against serogroup B meningococcal disease in Norway. Lancet 338:1093–1096 [DOI] [PubMed] [Google Scholar]
- 5. Bygraves JA, et al. 1999. Population genetic and evolutionary approaches to analysis of Neisseria meningitidis isolates belonging to the ET-5 complex. J. Bacteriol. 181:5551–5556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Caugant, et al. 1986. Intercontinental spread of a genetically distinctive complex of clones of Neisseria meningitidis causing epidemic disease. Proc. Natl. Acad. Sci. U. S. A. 83:4927–4931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Caugant DA, et al. 1987. Genetic structure of Neisseria meningitidis populations in relation to serogroup, serotype, and outer membrane protein patterns. J. Bacteriol. 169:2781–2792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Caugant DA, et al. 1987. Genetic relationships and clonal populations of serotype 2 strains of Neisseria meningitidis. Infect. Immun. 55:1503–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Caugant DA, Bol P, Hoiby EA, Zanen HC, Froholm LO. 1990. Clones of serogroup B Neisseria meningitidis causing systemic disease in the Netherlands, 1958 through 1986. J. Infect. Dis. 162:867–874 [DOI] [PubMed] [Google Scholar]
- 10. Caugant DA. 1998. Population genetics and molecular epidemiology of Neisseria meningitidis. APMIS 106:505–525 [PubMed] [Google Scholar]
- 11. Chiou CS, et al. 2006. Molecular epidemiology and emergence of worldwide epidemic clones of Neisseria meningitidis in Taiwan. BMC Infect. Dis. 6:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Clarke SC, Diggle MA, Molling P, Unemo M, Olcén P. 2003. Analysis of PorA variable region 3 min meningococci: implications for vaccine policy? Vaccine 21:2468–2473 [DOI] [PubMed] [Google Scholar]
- 13. de Filippis I, Gopalan V, Huyen Y. 2011. A web-based resource for the determination of PorA VR3 alleles of Neisseria meningitidis. Infect. Gene Evol. 11:248–249 [DOI] [PubMed] [Google Scholar]
- 14. Dellsie E, et al. 2010. Community outbreak of group B meningococcal disease in southwest France—December 2008 to September 2009. Euro Surveill. 15:pii=19665. http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId-19665 [PubMed] [Google Scholar]
- 15. de Moraes JC, et al. 1992. Protective efficacy of a serogroup B meningococcal vaccine in Sao Paulo, Brazil. Lancet 340:1074–1078 [DOI] [PubMed] [Google Scholar]
- 16. Devoy AF, Dyet KH, Martin DR. 2005. Stability of PorA during a meningococcal disease epidemic. J. Clin. Microbiol. 43:832–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. De Wals P, et al. 1996. Impact of a mass immunization campaign against serogroup C meningococcus in the province of Quebec, Canada. Bull. World Health Organ. 74:407–411 [PMC free article] [PubMed] [Google Scholar]
- 18. De Wals P, Deceuninck G, Boulianne N, De Serres G. 2004. Effectiveness of a mass immunization campaign using serogroup C meningococcal conjugate vaccine. JAMA 292:2491–2494 [DOI] [PubMed] [Google Scholar]
- 19. De Wals P, Deceuninck G, Lefebvre B, Boulianne N, De Serres G. 2011. Effectiveness of serogroup C meningococcal conjugate vaccine. A 7-year follow-up in Québec, Canada. Pediatr. Infect. Dis. J. 30:566–569 [DOI] [PubMed] [Google Scholar]
- 20. Diermayer M, et al. 1999. Epidemic serogroup B meningococcal disease in Oregon. JAMA 281:1493–1497 [DOI] [PubMed] [Google Scholar]
- 21. Donnelly J, et al. 2010. Qualitative and quantitative assessment of meningococcal antigens to evaluate the potential strain coverage of protein-based vaccines. Proc. Natl. Acad. Sci. U. S. A. 107:19490–19495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Farley JD, Osei W. 1992. Invasive meningococcal disease, British Columbia December 1991-March 1992. Can. J. Public Health 83:138–140 [PubMed] [Google Scholar]
- 23. Francisco AP, Bugalho M, Ramirez M, Carrico IA. 2009. Global optimal eBURST analysis of multilocus typing data using a graphic matroid approach. BMC Bioinformatics 10:152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Frasch CE, Zollinger WD, Poolman JT. 1985. Serotyping antigens of Neisseria meningitidis and a proposed scheme for designation of serotypes. Rev. Infect. Dis. 7:504–510 [DOI] [PubMed] [Google Scholar]
- 25. Giuliani MM, et al. 2006. A universal vaccine for serogroup B meningococcus. Proc. Natl. Acad. Sci. U. S. A. 103:10834–10839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hammerschmidt S, et al. 1996. Capsule phase variation in Neisseria meningitidis serogroup B by slipped-strand mis-pairing in the polysialyltransferase gene (siaD): correlation with bacterial invasion and the outbreak of meningococcal disease. Mol. Microbiol. 20:1211–1220 [DOI] [PubMed] [Google Scholar]
- 27. Harrison LH, et al. 2010. Population structure and capsular switching of invasive Neisseria meningitidis isolates in the pre-meningococcal conjugate vaccine era—United States, 2000–2005. J. Infect. Dis. 201:1208–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Heimann GA, et al. 1989. Meningococcal disease in Ontario during the winter of 1988-89. Can. Dis. Wkly. Rep. 15:59–62 [PubMed] [Google Scholar]
- 29. Jackson LA, Schuchat A, Reeves MW, Wenger JD. 1995. Serogroup C meningococcal outbreaks in the United States, an emerging threat. JAMA 273:383–389 [PubMed] [Google Scholar]
- 30. Jelfs J, Munro R, Wedege E, Caugant DA. 2000. Sequence variation in the porA gene of a clone of Neisseria meningitidis during epidemic spread. Clin. Diagn. Lab. Immunol. 7:390–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jelfs J, Munro R, Ashton FE, Caugant DA. 2000. Genetic characterization of a new variant within the ET-37 complex of Neisseria meningitidis associated with outbreaks in various parts of the world. Epidemiol. Infect. 125:285–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Law DKS, et al. 2006. Invasive meningococcal disease in Quebec, Canada, due to an emerging clone of ST-269 serogroup B meningococci with serotype antigen 17 and serosubtype antigen P1.19 (B:17:P1.19). J. Clin. Microbiol. 44:2743–2749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lucidarme J, et al. 2009. Characterization of fHbp, nhba (gna2132), nadA, porA, sequence type (ST), and genomic presence of IS1301 in group B meningococcal ST-269 clonal complex isolates from England and Wales. J. Clin. Microbiol. 47:3577–3585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Maiden MCJ, et al. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. U. S. A. 95:3140–3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Martin DR, Walker SJ, Baker MG, Lennon DR. 1998. New Zealand epidemic of meningococcal disease identified by a strain with phenotype B:4:P1.4. J. Infect. Dis. 177:497–500 [DOI] [PubMed] [Google Scholar]
- 36. National Advisory Committee on Immunisation 2007. Statement on conjugate meningococcal vaccine for serogroups A, C, Y and W135. Can. Commun. Dis. Rep. 33(ACS-3):1–24 [PubMed] [Google Scholar]
- 37. Oster P, et al. 2005. MeNZBTM: a safe and highly immunogenic tailor-made vaccine against the New Zealand Neisseria meningitidis serogroup B disease epidemic strain. Vaccine 23:2191–2196 [DOI] [PubMed] [Google Scholar]
- 38. Peltola H, Kataja JM, Makela PH. 1982. Shift in the age-distribution of meningococcal disease as predictor of an epidemic. Lancet ii:595–597 [DOI] [PubMed] [Google Scholar]
- 39. Pillai S, et al. 2005. Outer membrane protein (OMP) based vaccine for Neisseria meningitidis serogroup B. Vaccine 23:2206–2209 [DOI] [PubMed] [Google Scholar]
- 40. Pollard AJ, Tam TWS, National Advisory Committee on Immunization 2001. An Advisory Committee statement: statement on recommended use of meningococcal vaccines. Can. Commun. Dis. Rep. 27:2–36 [PubMed] [Google Scholar]
- 41. Reeves MW, Perkins BA, Diermayer M, Wenger J. 1995. Epidemic-associated Neisseria meningitidis detected by multilocus enzyme electrophoresis. Emerg. Infect. Dis. 1:53–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ringuette L, Lorange M, Ryan A, Ashton F. 1995. Meningococcal infections in the province of Quebec, Canada, during the period 1991 to 1992. J. Clin. Microbiol. 33:53–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rosenstein NE, Perkins BA, Stephens DS, Popovic T, Hughes JM. 2001. Meningococcal disease. N. Engl. J. Med. 344:1378–1388 [DOI] [PubMed] [Google Scholar]
- 44. Sacchi CT, et al. 1998. Proposed standardization of Neisseria meningitidis PorA variable-region typing nomenclature. Clin. Diagn. Lab. Immunol. 5:845–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sierra GVG, et al. 1991. Vaccine against group B Neisseria meningitidis: protection trial and mass vaccination results in Cuba. NIPH Ann. 14:195–210 [PubMed] [Google Scholar]
- 46. Sweet L. 1992. The Prince Edward Island meningococcal immunization program. January—February 1992. Can. J. Public Health 83:129–130 [PubMed] [Google Scholar]
- 47. Takahashi H, et al. 2004. Characterization of Neisseria meningitidis isolates collected from 1974 to 2003 in Japan by multilocus sequence typing. J. Med. Microbiol. 53:657–662 [DOI] [PubMed] [Google Scholar]
- 48. Tolomeo O, Buffett C, Richardson E. 1998. Management of a cluster of cases of invasive group C Neisseria meningitidis infections in the Hamilton-Wentworth region. Can. Commun. Dis. Rep. 24:122–126 [PubMed] [Google Scholar]
- 49. Tsang RSW, et al. 2004. Phenotypic and genetic characterization of a unique variant of serogroup C ET-15 meningococci (with the antigenic formula C:2a:P1.7,1) causing invasive meningococcal disease in Quebec, Canada. J. Clin. Microbiol. 42:1460–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tsang RSW, Law DKS, Henderson AM, Cameron ML, Stoltz J. 2006. Increase in serogroup C meningococcal disease in Canada is associated with antigenic changes in the protein antigens of the ET-15 clone of Neisseria meningitidis. J. Infect. Dis. 194:1791–1792 [DOI] [PubMed] [Google Scholar]
- 51. Tsang RSW. 2007. Capsule switching and capsule replacement in vaccine preventable bacterial diseases. Lancet Infect. Dis. 7:569–570 [DOI] [PubMed] [Google Scholar]
- 52. Tyler SD, Tsang R. 2004. Genetic analysis of Canadian isolates of C:2a:P1.2,5 and B:2a:P1.2,5 Neisseria meningitidis strains belonging to the hypervirulent clone of ET-15. Can. J. Microbiol. 50:433–443 [DOI] [PubMed] [Google Scholar]
- 53. Vedros NA. 1987. Development of meningococcal serogroups. p 33–37 In Vedros NA. (ed), Evolution of meningococcal disease. CRC Press, Boca Raton, FL [Google Scholar]
- 54. Whalen CM, Hockin JC, Ryan A, Ashton F. 1995. The changing epidemiology of invasive meningococcal disease in Canada, 1985 through 1992. JAMA 273:390–394 [PubMed] [Google Scholar]
- 55. Yang L, et al. 2008. Genotypic characterization of Neisseria meningitidis serogroup B strains circulating in China. J. Infect. 56:211–218 [DOI] [PubMed] [Google Scholar]
- 56. Yazdankhah SP, et al. 2004. Distribution of serogroups and genotypes among disease-associated and carried isolates of Neisseria meningitidis from the Czech Republic, Greece, and Norway. J. Clin. Microbiol. 42:5146–5153 [DOI] [PMC free article] [PubMed] [Google Scholar]