Abstract
Carcinogenic human papillomavirus (HPV) infections are necessary causes of most anogenital cancers. Viral load has been proposed as a marker for progression to cancer precursors but has been confirmed only for HPV16. Challenges in studying viral load are related to the lack of validated assays for a large number of genotypes. We compared viral load measured by Linear Array (LA) HPV genotyping with the gold standard, quantitative PCR (Q-PCR). LA genotyping and Q-PCR were performed in 143 cytology specimens from women referred to colposcopy. LA signal strength was measured by densitometry. Correlation coefficients and receiver operating characteristic (ROC) analyses were used to evaluate analytical and clinical performance. We observed a moderate to strong correlation between the two quantitative viral load measurements, ranging from an R value of 0.61 for HPV31 to an R value of 0.86 for HPV52. We also observed agreement between visual LA signal strength evaluation and Q-PCR. Both quantifications agreed on the disease stages with highest viral load, which varied by type (cervical intraepithelial neoplasia grade 2 [CIN2] for HPV52, CIN3 for HPV16 and HPV33, and cancer for HPV18 and HPV31). The area under the curve (AUC) for HPV16 Q-PCR at the CIN3 cutoff was 0.72 (P = 0.004), and the AUC for HPV18 LA at the CIN2 cutoff was 0.78 (P = 0.04). Quantification of LA signals correlates with the current gold standard for viral load, Q-PCR. Analyses of viral load need to address multiple infections and type attribution to evaluate whether viral load has clinical value beyond the established HPV16 finding. Our findings support conducting comprehensive studies of viral load and cervical cancer precursors using quantitative LA genotyping data.
INTRODUCTION
Infections with carcinogenic human papillomaviruses (HPV) are the necessary cause of cervical cancer. Most HPV infections clear after a few months while a few persist and may progress to cervical precancer and, eventually, invasive cancer. Two functional states of HPV infections have been recognized based on the viral expression and replication patterns and their role in cervical carcinogenesis (2): productive infections are characterized by production and release of large amounts of viral particles, while expression patterns in transforming infections shift to expression of viral oncogenes, resulting in cellular transformation and low virus particle production.
HPV viral load is a product of the number of cells infected and number of viruses per infected cell and is therefore influenced by two main factors: (i) the extent of an HPV infection on the cervical surface and (ii) the level of viral production in the area of infection. Viral load has been suggested to be a potential biomarker for cervical intraepithelial neoplasia grade 2 (CIN2) or greater, but currently there is no consistent evidence that a one-time measurement of viral load is a useful marker of prevalent disease or disease progression (9). HPV16 is the only genotype for which there is some indication that viral load may predict viral persistence and progression to precancer (1, 4, 18).
One limitation in defining a role for HPV viral load in previous analyses has been the lack of standardized assays for a broad range of HPV genotypes. Quantitative real-time PCR methods are considered to be the gold standard for HPV load assessment, but these have not been developed and validated for the wide spectrum of carcinogenic HPV types often encountered in cervical samples. Although signal intensities from Hybrid Capture 2 (HC2) or various endpoint PCR-based assays have been proposed and partly used as surrogates for viral load, these approaches have limitations (6, 13). HC2 only gives aggregate signal strength for a pool of 13 carcinogenic types, and commercial genotyping assays, such as the Roche Linear Array (LA) or Innogenetics InnoLiPA (line probe assay), do not formally report quantitative results. Furthermore, some of these surrogates have not been properly validated against quantitative PCR (Q-PCR), the gold standard of HPV viral load measurement. A validated method to measure viral load based on a widely used genotyping assay such as the LA would allow the evaluation of the biological and clinical importance of viral load for many HPV genotypes in large epidemiological studies.
We previously described an automated densitometry method to quantify signal intensity using the LA HPV genotyping test (7). We demonstrated that automated evaluation can improve manual visual inspection of LA strips. Here, we compared the visual and automated quantification of LA genotyping signals to a validated viral load assay for HPVs 16, 18, 31, 33, and 52 using the same DNA isolated from liquid-based cytology samples.
MATERIALS AND METHODS
Study population.
This analysis was based on DNA extracted from cervical cells collected in the Study to Understand Cervical Cancer Early Endpoints and Determinants (SUCCEED) conducted at the University of Oklahoma Health Sciences Center (OUHSC). The study details are reported elsewhere (17). In brief, SUCCEED enrolled women referred for colposcopy following an abnormal Pap result. Women under 18 years of age, women who had prior treatment with chemotherapy or radiation for any cancer, and women pregnant at the time of their visit were excluded from the study. Participants provided written informed consent prior to enrollment into the study. Study procedures were approved by the OUHSC and National Cancer Institute (NCI) Institutional Review Boards. For the purposes of this study, we randomly selected 104 women from the SUCCEED study population who had linear array signal strength measurements previously assessed and sufficient DNA for additional viral load measurements. The selection was restricted to women who tested positive (hybridization signals equal to or above the lowest visual cutoff, extremely weak signal) for at least one of five HPV types (HPV16, HPV18, HPV31, HPV33, or HPV52) by the Linear Array HPV Genotyping Test (Roche Molecular Diagnostics, Branchburg, NJ). The same DNA used in the LA assay was used to assess viral load in singleplex Q-PCR assays. Individual samples were tested using only those Q-PCR assays corresponding to the HPV type(s) detected by LA (i.e., if a specimen was positive for HPV16 or HPV31 by LA, the sample would be tested using Q-PCR assays designed to detect HPV16 or HPV31). In addition, for each of the five types, we randomly selected 5 to 10% (based on the number of positive specimens tested) LA-negative samples to evaluate the negative agreement, including four specimens negative for HPV16 and HPV52 and two specimens each negative for HPVs 18, 31, and 33. All of these 14 specimens were confirmed negative for the respective HPV types by Q-PCR. In total, 143 quantitative PCR assays were run using the 104 specimens, 55 for HPV16, 28 for HPV18, 19 for HPV31, 20 for HPV33, and 21 for HPV52. Table 1 shows the cross-tabulation of cytology and histology in the 104 women sampled for this analysis, demonstrating the full range of cytology and histology results.
Table 1.
Histology and cytology results among 104 women included in the analysis
| Cytologya | Histology |
||||
|---|---|---|---|---|---|
| <CIN2 | CIN2 | CIN3 | Cancer | Total | |
| No. of NILM | 15 | 1 | 3 | 1 | 20 |
| Row % | 75.0 | 5.0 | 15.0 | 5.0 | 19.2 |
| Col % | 35.7 | 3.2 | 16.7 | 7.7 | |
| No. of ASCUS | 3 | 2 | 0 | 0 | 5 |
| Row % | 60.0 | 40.0 | 0.0 | 0.0 | 4.8 |
| Col % | 7.1 | 6.5 | 0.0 | 0.0 | |
| No. of LSIL | 11 | 4 | 1 | 0 | 16 |
| Row % | 68.8 | 25.0 | 6.3 | 0.0 | 15.4 |
| Col % | 26.2 | 12.9 | 5.6 | 0.0 | |
| No. of HSIL | 13 | 24 | 14 | 12 | 63 |
| Row % | 20.6 | 38.1 | 22.2 | 19.1 | 60.6 |
| Col % | 31.0 | 77.4 | 77.8 | 92.3 | |
| Total | 42 | 31 | 18 | 13 | 104 |
| Row % | 40.4 | 29.8 | 17.3 | 12.5 | 100.0 |
Row %, percentage of row frequency; Col %, percentage of column frequency. For example, for <CIN2, there were 15 patients with NILM, representing 75% of the total number of NILM cases and 35.7% of the total number of instances of <CIN2.
Specimen collection and linear array genotyping.
Prior to colposcopic examination, cervical cell samples were collected and rinsed directly into PreservCyt solution (Hologic, Boxborough, MA) as previously described (16). The cytology specimen was used for ThinPrep (Hologic, Boxborough, MA) cytology and for HPV genotyping using the LA HPV Genotyping Test, which detects 37 HPV genotypes (HPV types 6, 11, 16, 18, 26, 31, 33, 35, 39, 40, 42, 45, 51, 52, 53, 54, 56, 57, 58, 59, 61, 62, 64, 66, 67, 68, 69, 70, 71, 72, 73, 81, 82, 83, 84, IS39, and CP6108). The procedure followed recommendations of the manufacturer with the following variations: 10 μl of template DNA was amplified and the amplified products were hybridized and detected using an automated Auto-LiPA staining system using 2.5 ml of each reagent per strip (compared to 3.0 ml in manual processing), as previously described (17). All specimens had adequate DNA, as indicated by internal control beta-globin measures above 250 for all samples included.
Visual evaluation and digitization of LA strips.
The LA results were initially evaluated by unmagnified examination of the strips by two independent observers. An unambiguous, continuous band was judged to indicate that biotinylated amplicons had hybridized to complementary probe sequences bound to the strips and was considered a positive result. A reference guide overlay provided by Roche was used to relate the location of the band(s) on the strip to the HPV genotype(s). The evaluators also subjectively graded the intensity of each band, as strong (S), moderate (M), weak (W), very weak (VW), or extremely weak (EW). A digitized record of hybridization signals on the LA strips was generated using an AutoChemi imaging system (UVP BioImaging Systems, Upland, CA) shortly after hybridization and detection were completed on the Auto-LiPA staining system. The strips were photographed while still wet. Pictures were obtained in a gray scale and stored using a 12-bit TIFF format as described previously (7).
Automated signal quantification of LA strips.
The digital images of LA results were evaluated at the Communications Engineering Branch, National Library of Medicine, Bethesda, MD, using a customized computer algorithm as described previously (7). Raw and background-corrected signal values were generated by an algorithm on a theoretical scale of 0 (white, no signal) to 1,000 (black, strongest possible signal). Only background-corrected values were used in the analyses. The algorithm was implemented in MATLAB, version 7.01, and executed on a Dell Optiplex GX270 (3.2 GHz, 2 GB of RAM) Windows XP computer.
Viral load.
Viral load reactions were carried out in 96-well, 0.2-ml PCR microplates with a 50-μl reaction mixture consisting of forward and reverse primers targeting the E6 gene, fluorescent probe, 1× Universal Mastermix with TaqGold (Applied Biosystems, Foster City, California), diethyl pyrocarbonate (DEPC)-treated water, and 5 μl of extracted DNA, as previously described (5, 11).
Using postdetection software linked to the ABI 7300, viral load was estimated by extrapolation from a standard curve of known concentrations of type-specific plasmid HPV DNA included on each reaction plate (five serial 1:10 dilutions of purified HPV 16, 18, 31, 33, 35, or 52 plasmid with an initial concentration of 250,000 copies in a background of 50 ng/μl human placental DNA [Sigma]). Human placental DNA at 50 ng/μl in low-salt Tris-EDTA buffer was used as a negative control on each plate. All standards and negative controls were run in duplicate. All specimens had adequate DNA, as indicated by internal control endogenous retrovirus 3 (ERV-3) loads above 100 for all samples included.
Statistical analysis.
All analyses were conducted on the infection level. To calculate percent agreement between LA and Q-PCR results, we dichotomized LA results based on the visual evaluation as negative (no signal) or positive (EW, VW, W, M, or S signal) and Q-PCR results as below or above the detection limit. Q-PCR results were log transformed for correlation analyses and for graphical display. We calculated the correlation coefficient (R) and the coefficient of determination (R2) with 95% confidence intervals as a measure of correlation between LA and Q-PCR for all five types separately and for all types combined. Correlation coefficients between 0.40 and 0.69 were interpreted as moderate correlation, and correlation coefficients of 0.70 and higher were interpreted as high correlation.
We created five disease groups based on histology and cytology information: group 1, CIN histology lower than grade 2 (<CIN2) and normal cytology; group 2, <CIN2 histology and observation of atypical squamous cells of undetermined significance (ASCUS) or low-grade squamous intraepithelial lesions (LSIL); group 3, CIN2 or <CIN2 and high-grade squamous intraepithelial lesions (HSIL); group 4, CIN3; group 5, cancer. We calculated mean signal intensity for LA and geometric means for Q-PCR to evaluate viral load by type and disease group. Q-PCR values below the detection limit were set to 1.
To evaluate whether viral load measurements had potential diagnostic utility, we calculated area under the curve (AUC) measures for two endpoints for detection of neoplasia of CIN2 or greater (combining disease groups 3, 4, and 5) and for detection of neoplasia of CIN3 or greater (combining disease groups 4 and 5). We calculated P values for the difference between the AUC measures and the diagonal line (AUC of 0.5), indicating no discriminative potential. For assays with AUC values significantly different from 0.5, we reported the sensitivity and specificity pair at the cutoff with the highest Youden's index (calculated as 1 − sensitivity + specificity), requiring at least 60% sensitivity and 40% specificity. We considered P values of <0.05 as statistically significant. All analyses were performed using SAS, version 9.1 (Cary, NC), and SPSS for Windows, version 19.0 (Chicago, IL).
RESULTS
Study population and HPV genotype prevalence.
A total of 104 women were included in the study; among them were 13 women with <CIN2 and normal cytology (mean age, 25 years), 19 women with <CIN2 and ASCUS or LSIL (mean age, 27 years), 42 women with CIN2 or <CIN2 and HSIL (mean age, 28 years), 17 women with CIN3 (mean age, 29 years), and 13 women with cervical cancer (mean age, 49 years) (Table 2). Seventy-four of 104 women (71.2%) had multiple infections according to LA, including other genotypes not analyzed by Q-PCR in this study. Based on the LA genotyping results, overall there were 51 infections with HPV16, 26 infections with HPV18, 17 infections with HPV31, 18 infections with HPV33, and 17 infections with HPV52 (Table 2). The Q-PCR assays gave very similar estimates, showing 49 infections with HPV16, 26 infections with HPV18, 17 infections with HPV31, 17 infections with HPV33, and 16 infections with HPV52. Of note, HPV52 is not directly measured in the LA assay, which uses a mixture of probes detecting HPVs 33, 35, 52, and 58 in combination. In addition, HPVs 33, 35, and 58 are detected separately. All 15 specimens that were positive for the mixed probe set and negative for HPVs 33, 35, and 58 in the LA were also positive for HPV52 by Q-PCR. The positive agreement between both assays using a dichotomous evaluation (positive versus negative) was very high, reaching 96% for HPV16, 100% for HPV18, 100% for HPV31, 94% for HPV33, and 94% for HPV52.
Table 2.
HPV prevalence measured by LA and Q-PCR in cervical disease categories
| Disease category | Total no. of subjects | Mean age (yrs [range]) | No. of subjects with multiple infectionsa | No. of infections by HPV type and assay method |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HPV16 |
HPV18 |
HPV31 |
HPV33 |
HPV52 |
|||||||||
| LA | Q-PCR | LA | Q-PCR | LA | Q-PCR | LA | Q-PCR | LA | Q-PCR | ||||
| <CIN2 and normal cytology | 13 | 25 (19-37) | 11 | 4 | 3 | 3 | 3 | 3 | 3 | 2 | 2 | 2 | 2 |
| <CIN2 and ASCUS or LSIL | 19 | 27 (19-41) | 14 | 9 | 9 | 3 | 3 | 5 | 5 | 2 | 2 | 4 | 4 |
| <CIN3 and HSIL | 42 | 28 (18-64) | 31 | 21 | 20 | 12 | 12 | 5 | 5 | 8 | 7 | 9 | 9 |
| CIN3 | 17 | 29 (19-52) | 13 | 8 | 8 | 5 | 5 | 4 | 4 | 5 | 5 | 1 | 1 |
| Cancer | 13 | 49 (38-69) | 5 | 9 | 9 | 3 | 3 | 0 | 0 | 1 | 1 | 1 | 0 |
| Total | 104 | 74 | 51 | 49 | 26 | 26 | 17 | 17 | 18 | 17 | 17 | 16 | |
Based on Linear Array (LA) genotyping. Disease categories are nonoverlapping.
Agreement between quantitative PCR and LA measurement.
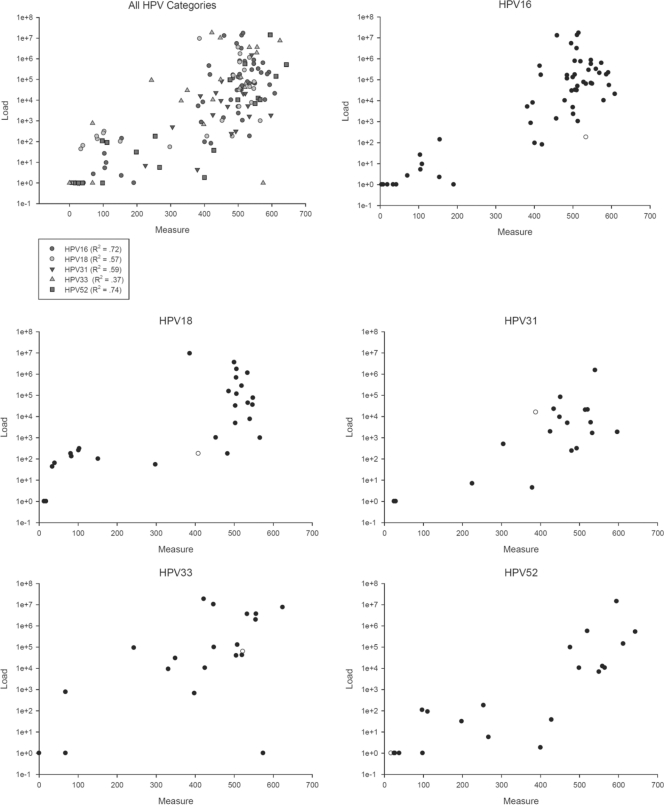
The range of LA measurements for HPV16 was from 4 to 609, and the range of viral load results was from 1 to 17,200,000. The ranges were very similar for the other types, with LA measurements between 1 and 600 and viral load measurements over a 7-log range. The LA measurement signal range and distribution were representative of our previous analysis of >1,000 women from this population (7). We compared the correlation between LA signal strength with log-transformed Q-PCR results. Overall, we observed strong correlation between the two quantifications except for HPV33, which showed moderate correlation between both measurements. The correlation coefficient, R, for HPV16 was 0.85 (R2 of 0.72; 95% confidence interval [CI], 0.58 to 0.80), for HPV18 it was 0.75 (R2 of 0.57; 95% CI, 0.29 to 0.72), for HPV31 it was 0.77 (R2 of 0.59; 95% CI, 0.22 to 0.74), for HPV33 it was 0.61 (R2 of 0.37; 95% CI, 0.05 to 0.60), and for HPV52 it was 0.86 (R2 of 0.74; 95% CI, 0.46 to 0.83) (Fig. 1A to F). When the analysis was restricted to women with multiple infections, we observed slightly, but insignificantly, stronger correlations between the two measurements for some types, with the correlation coefficient R for HPV16 of 0.85 (R2 of 0.73; 95% CI, 0.56 to 0.81), for HPV18 of 0.74 (R2 of 0.56; 95% CI, 0.22 to 0.74), for HPV31 of 0.78 (R2 of 0.61; 95% CI, 0.19 to 0.77), for HPV33 of 0.85 (R2 of 0.73; 95% CI, 0.34 to 0.84), and for HPV52 of 0.92 (R2 of 0.85; 95% CI, 0.57 to 0.91). The correlation coefficient R for all types combined was 0.78 (R2 of 0.61; 95% CI, 0.51 to 0.68), and 0.81 (R2 of 0.66; 95% CI, 0.55 to 0.75) when restricting to women with multiple infections.
Fig 1.
Correlation of LA signal measurement and Q-PCR. All 143 pairs of LA measurement and Q-PCR results are presented together and stratified by individual genotypes. Log-transformed Q-PCR results are plotted on the y axis, while LA measurement results are plotted on the x axis.
Viral load in cervical disease stages.
We analyzed the distribution of viral load estimates based on LA and Q-PCR in cervical disease categories (Table 3). We calculated mean LA signal intensities and geometric mean Q-PCR results for each type and disease category. For HPV16, the LA signal strength ranged from an average of 338 in women with <CIN2 and normal cytology to an average of 509 in women with CIN3. In agreement, the Q-PCR showed the lowest load in women with <CIN2 and normal cytology (274) and the highest in women with CIN3 (194, 387). For HPV18, both assays showed the lowest viral load among women with <CIN2 and normal cytology (LA measurement, 154; Q-PCR, 109) and the highest load among women with cancer (LA measurement, 518; Q-PCR, 92,167). The numbers for the other three types were lower, and estimates within disease categories were less stable. Still, both assays agreed on the disease category with the highest viral load, which was CIN3 for HPV31, cancer for HPV33, and CIN2 for HPV52.
Table 3.
Mean HPV viral load in cervical disease categories
| HPV type | Viral load by disease categorya |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| <CIN2 and normal cytology |
<CIN2 and ASCUS or LSIL |
CIN2 or <CIN2 and HSIL |
CIN3 |
Cancer |
|||||||||||
| n | Mean LA | Mean Q-PCR | n | Mean LA | Mean Q-PCR | n | Mean LA | Mean Q-PCR | n | Mean LA | Mean Q-PCR | n | Mean LA | Mean Q-PCR | |
| 16 | 4 | 338 | 274 | 9 | 395 | 3,635 | 21 | 442 | 10,712 | 8 | 509 | 194,387 | 9 | 457 | 80,749 |
| 18 | 3 | 154 | 109 | 3 | 363 | 55,617 | 12 | 429 | 12,108 | 5 | 332 | 1,747 | 3 | 518 | 92,167 |
| 31 | 3 | 411 | 206 | 5 | 473 | 3,462 | 5 | 440 | 4,007 | 4 | 483 | 9,221 | 0 | NAb | NA |
| 33 | 2 | 412 | 2,651 | 2 | 386 | 745,801 | 8 | 469 | 44,115 | 5 | 425 | 33,823 | 1 | 556 | 3,664,080 |
| 52 | 2 | 299 | 8 | 4 | 388 | 1,289 | 9 | 486 | 19,697 | 1 | 254 | 181 | 1 | 98 | NA |
Mean LA, arithmetic mean of measured LA signal intensities; mean PCR, geometric mean of Q-PCR results; n, number of women.
NA, not available.
To evaluate whether the viral load measurements for HPV16 and HPV18 could discriminate between women with neoplasia of CIN2 or greater and women with HPV infections but no confirmed lesions, we calculated the AUC for LA signal intensity and Q-PCR for two endpoints neoplasia of CIN2 and greater and neoplasia of CIN3 and greater (Table 4). For HPV16, the AUCs were higher for Q-PCR than for LA signal strength measurement (0.68 versus 0.62 for CIN2 and greater; 0.72 versus 0.55 for CIN3 and greater). In contrast, for HPV18, the AUCs were higher for the LA measurement (0.78 versus 0.60 for CIN2 and greater; 0.58 versus 0.53 for CIN3 and greater). Only two AUC estimates were significantly different from 0.5: the P value for HPV16 Q-PCR at the CIN3 cutoff was 0.01 (AUC 0.72), and the P value for the HPV18 LA measurement at the CIN2 cutoff was 0.04 (AUC 0.78). These AUCs correspond to a sensitivity of 82% and specificity of 47% for HPV16 Q-PCR at the CIN3 cutoff and to a sensitivity of 76% and specificity of 57% for HPV18 LA measurement at the CIN2 cutoff.
Table 4.
Discrimination between transient infection and high-grade CIN using viral load
| HPV | Linear array result |
Q-PCR result |
||||||
|---|---|---|---|---|---|---|---|---|
| CIN2+ |
CIN3+ |
CIN2+ |
CIN3+ |
|||||
| AUC | P value | AUC | P value | AUC | P value | AUC | P value | |
| HPV16 | 0.62 | 0.20 | 0.55 | 0.56 | 0.68 | 0.06 | 0.72 | 0.01 |
| HPV18 | 0.78 | 0.04 | 0.58 | 0.52 | 0.60 | 0.50 | 0.47 | 0.82 |
AUC, area under the curve; CIN2+, neoplasia greater than CIN2; CIN3+, neoplasia greater than CIN3. The P value indicates the difference from the diagonal ROC curve (AUC = 0.5).
Visual quantification of LA signals and Q-PCR.
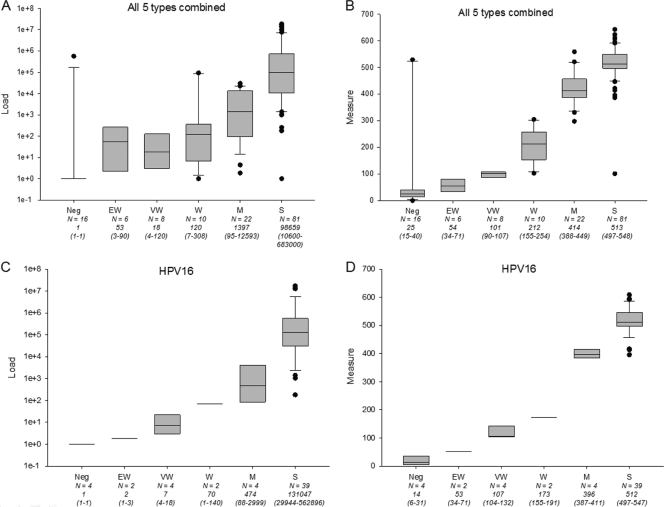
In addition to the quantitative comparison between LA and Q-PCR, we evaluated the distribution of Q-PCR results in six visual categories of LA signal strength, ranging from negative to strong. Since only HPV16 had representation of all visual signal strength categories, data are presented in aggregate for all five types (Fig. 2A and B) and for HPV16 separately (Fig. 2C and D). For HPV16 alone, the visual signal strength categories had increasing, almost discrete, viral load ranges measured by Q-PCR. Similarly, for the aggregated results of all five types, we observed an increase of Q-PCR-based viral load measures, albeit with less discrimination between the three weakest categories.
Fig 2.
Q-PCR and LA measurement by visual signal strength categories. A total of 143 viral load measures for all five HPV types (HPVs 16, 18, 31, 33, and 52) by Q-PCR (A) and LA densitometry (B) are shown stratified by visual signal strength categories (Neg, no signal; EW, extremely weak; VW, very weak; W, weak; M, moderate; S, strong). Similarly, viral load measures based on Q-PCR (C) and LA (D) are shown for HPV16 separately. Total numbers in each visual signal strength category are indicated together with median values and interquartile ranges (in parentheses).
DISCUSSION
We previously showed that densitometry measurement of signal strength obtained by LA HPV genotyping is more accurate than the visual evaluation of LA genotyping strips (7). Here, we demonstrate that the continuous viral load estimates obtained with this approach show high correlation with viral load estimates based on Q-PCR, the widely accepted gold standard for quantification of HPV in cervical specimens. For all five HPV genotypes analyzed, both measures agreed on the disease categories with the highest viral load, which varied from CIN2 (HPV52) to cancer (HPV18 and HPV33). The finding that high-grade CIN was associated with highest viral load for all types suggests that viral load may have clinical value. In this study, viral load based on Q-PCR provided modest differentiation between infection and neoplasia of CIN2 or greater for HPV16, while viral load based on signal intensity measurement provided modest differentiation between infection and neoplasia of CIN2 or greater for HPV18. Numbers were too low for the other types to evaluate clinical performance.
Viral load is a product of the number of HPV-infected cells and the level of viral particle production in the infected cells. Thus, in an extreme example, a widespread productive infection might be associated with high viral load, while a small incipient CIN3 with low-level virus production might be associated with low viral load. Furthermore, viral load in a cytological sample is subject to sampling variation in which there are varying proportions of lesional cells, normal epithelial cells, inflammatory exudate, and blood. A further complication in using viral load to predict neoplasia of CIN2 or greater is the high prevalence of multiple carcinogenic HPV infections detected in cervical samples. The current paradigm is that cervical lesions clonally expand following infection with one specific genotype (one virus-one lesion concept). On the cervical surface, multiple independent infections or lesions may occur that are caused by different genotypes. Without specific genotyping conducted in situ, assigning a causal HPV genotype to a specific lesion can only be based on assumptions (6, 15, 17). Currently, only HPV16 viral load has been shown to be associated with incident and prevalent cervical cancer precursors. This may be related to the fact that HPV16 is the most carcinogenic type and likely causal in most multiple-carcinogenic-type infections involving HPV16 and another carcinogenic type. For many other types, causal attribution is less clear when multiple carcinogenic types are present. Understanding the role of viral load for other carcinogenic types besides HPV16 will require multiple parallel evaluations of viral load for these types.
Interestingly, for HPV18, we observed a low viral load by both measures in the CIN3 category. This is in agreement with previous reports of a deficit of HPV18 detection in CIN3 compared to the prevalence in cervical cancer (8, 14). One explanation may be a high proportion of HPV18 infections associated with glandular lesions that are more difficult to sample and may be prone to false-negative results.
Recently, studies have focused on longitudinal observations of viral load to predict viral clearance or lesion progression (10, 12). Initial data indicate that repeated measurements can improve prediction of persistence or clearance, but these data are, so far, limited to HPV16 only. Our approach provides important opportunities for quantification of multiple parallel HPV genotype infections using a well-validated assay that is widely used in epidemiological studies. Furthermore, even without densitometry evaluation of LA signal strength, we observed that the visual evaluation of signal intensity using six categories provides a reasonable correlate of viral load as measured by Q-PCR, offering a simplified approach toward HPV viral load evaluation.
Our analysis was conducted in a large epidemiological study with highly standardized sample collection, uniform processing and genotyping of cervical samples, and excellent disease ascertainment by colposcopy and loop electrosurgical excision procedure (LEEP). The Q-PCR assay used for comparison in this analysis has been shown to be highly reproducible and to correlate well with other viral load assays (3, 5). A limitation of our study is the restriction to validation of signal strength measurements of only five HPV genotypes for which validated Q-PCR assays were available. However, based on the findings presented here, we can assume that the other types will show correlation to Q-PCR results in a similar range. While we validated the signal strength quantification only for LA genotyping in this study, the same approach is applicable to other strip-based HPV genotyping assays or to strip-based assays for other targets. While digitization of LA strips is not precisely reproducible due to inevitable variation in the digitization process, the resulting variation is expected to be minimal. Once linear array strips have been scanned, signal quantification is precisely reproducible when the parameters are unchanged.
In summary, measuring signal intensities on LA HPV genotyping strips provides quantitative information comparable to viral load measurements based on Q-PCR. We showed that signal strength measurement for HPV18 can distinguish between women without signs of high-grade CIN and women with CIN2 or higher-grade CIN. Extended analyses, accounting for multiple carcinogenic infections, are necessary to evaluate the role of viral load for other carcinogenic HPV types. Our approach offers the potential for viral load assessment for 37 types in parallel, simplifying conducting repeated measurements of viral load in epidemiologic studies and addressing the problems of multiple HPV genotype infections in studies of HPV load.
ACKNOWLEDGMENT
This work was supported by the Intramural Research Program of the National Cancer Institute.
Footnotes
Published ahead of print 15 February 2012
REFERENCES
- 1. Boulet GA, et al. 2009. Human papillomavirus 16 load and E2/E6 ratio in HPV16-positive women: biomarkers for cervical intraepithelial neoplasia ≥2 in a liquid-based cytology setting? Cancer Epidemiol. Biomarkers Prev. 18:2992–2999 [DOI] [PubMed] [Google Scholar]
- 2. Doorbar J. 2007. Papillomavirus life cycle organization and biomarker selection. Dis. Markers. 23:297–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fontaine J, et al. 2005. High level of correlation of human papillomavirus-16 DNA viral load estimates generated by three real-time PCR assays applied on genital specimens. Cancer Epidemiol. Biomarkers Prev. 14:2200–2207 [DOI] [PubMed] [Google Scholar]
- 4. Gravitt PE, et al. 2007. High load for most high risk human papillomavirus genotypes is associated with prevalent cervical cancer precursors but only HPV16 load predicts the development of incident disease. Int. J. Cancer 121:2787–2793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gravitt PE, et al. 2003. Reproducibility of HPV 16 and HPV 18 viral load quantitation using TaqMan real-time PCR assays. J. Virol. Methods 112:23–33 [DOI] [PubMed] [Google Scholar]
- 6. Gravitt PE, et al. 2007. Human papillomavirus (HPV) genotyping using paired exfoliated cervicovaginal cells and paraffin-embedded tissues to highlight difficulties in attributing HPV types to specific lesions. J. Clin. Microbiol. 45:3245–3250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jeronimo J, et al. 2008. Evaluation of linear array human papillomavirus genotyping using automatic optical imaging software. J. Clin. Microbiol. 46:2759–2765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kovacic MB, et al. 2006. Relationships of human papillomavirus type, qualitative viral load, and age with cytologic abnormality. Cancer Res. 66:10112–10119 [DOI] [PubMed] [Google Scholar]
- 9. Lorincz AT, et al. 2002. Viral load of human papillomavirus and risk of CIN3 or cervical cancer. Lancet 360:228–229 [DOI] [PubMed] [Google Scholar]
- 10. Marks M, et al. 2011. Kinetics of DNA load predict HPV 16 viral clearance. J. Clin. Virol. 51:44–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Marks M, et al. 2009. Confirmation and quantitation of human papillomavirus type 52 by Roche linear array using HPV52-specific TaqMan E6/E7 quantitative real-time PCR. J. Virol. Methods 156:152–156 [DOI] [PubMed] [Google Scholar]
- 12. Monnier-Benoit S, et al. 2006. Dynamics of HPV16 DNA load reflect the natural history of cervical HPV-associated lesions. J. Clin. Virol. 35:270–277 [DOI] [PubMed] [Google Scholar]
- 13. Pretet JL, Dalstein V, Monnier-Benoit S, Delpeut S, Mougin C. 2004. High risk HPV load estimated by Hybrid Capture II correlates with HPV16 load measured by real-time PCR in cervical smears of HPV16-infected women. J. Clin. Virol. 31:140–147 [DOI] [PubMed] [Google Scholar]
- 14. Schiffman M, et al. 2011. Human papillomavirus testing in the prevention of cervical cancer. J. Natl. Cancer Inst. 103:368–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sherman ME, et al. 2003. Determinants of human papillomavirus load among women with histological cervical intraepithelial neoplasia 3: dominant impact of surrounding low-grade lesions. Cancer Epidemiol. Biomarkers Prev. 12:1038–1044 [PubMed] [Google Scholar]
- 16. Wang SS, et al. 2009. Human papillomavirus cofactors by disease progression and human papillomavirus types in the study to understand cervical cancer early endpoints and determinants. Cancer Epidemiol. Biomarkers Prev. 18:113–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wentzensen N, et al. 2009. Multiple human papillomavirus genotype infections in cervical cancer progression in the study to understand cervical cancer early endpoints and determinants. Int. J. Cancer 125:2151–2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xi LF, et al. 2011. Viral load in the natural history of human papillomavirus type 16 infection: a nested case-control study. J. Infect. Dis. 203:1425–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]