Abstract
Human enteroviruses (HEV) are among the most common viruses infecting humans. Their circulation has been widely studied in most parts of the world but not in sub-Saharan Africa, where poliomyelitis remains prevalent. We report here the molecular characterization of 98 nonpoliovirus (non-PV) HEV strains isolated from 93 randomly selected cell culture-positive supernatants from stool samples collected from 1997 through 2006 from children with acute flaccid paralysis living in the Central African Republic (CAR). The isolates were typed by sequencing the VP1 coding region and sequenced further in the VP2 coding region, and phylogenetic studies were carried out. Among the 98 VP1 sequences, 3, 74, 18, and 3 were found to belong to the HEV-A, -B, -C, and -D species, respectively. Overall, 42 types were detected. In most cases, the VP2 type was correlated with that of the VP1 region. Some of the isolates belonged to lineages that also contain viruses isolated in distant countries, while others belonged to lineages containing viruses isolated only in Africa. In particular, one isolate (type EV-A71) did not fall into any of the genogroups already described, indicating the existence of a previously unknown genogroup for this type. These results illustrate the considerable diversity of HEV isolates from the stools of paralyzed children in the CAR. The presence of diverse HEV-C types makes recombination between poliovirus and other HEV-C species possible and could promote the emergence of recombinant vaccine-derived polioviruses similar to those that have been implicated in repeated poliomyelitis outbreaks in several developing countries.
INTRODUCTION
Human enteroviruses (HEV) (family Picornaviridae, genus Enterovirus) are among the most common viruses infecting humans. On the basis of the phylogenetic clustering of these viruses, they have been assigned to four species (HEV-A to -D) containing more than 100 types. The HEV-C species include the three types of poliovirus (PV) known to cause poliomyelitis.
HEV virions are icosahedral, have no envelope, and contain a single positively stranded RNA genome about 7,500 nucleotides (nt) in length. The genome contains only one functional open reading frame encoding a viral polyprotein that is cleaved to give rise to the functional proteins (48). The N-terminal part of the polyprotein contains the four structural proteins (VP1 to VP4) that are assembled to form the virion; the C-terminal part of the polyprotein contains the nonstructural viral proteins, including proteases and the RNA-dependent RNA polymerase.
Non-PV HEV-A to -D species cause a wide spectrum of diseases, with clinical signs ranging from mild febrile illness, such as the common cold, to severe forms, such as acute hemorrhagic conjunctivitis, myocarditis, encephalitis, and acute flaccid paralysis (47). Moreover, some HEV strains, such as HEV-C in particular, can recombine with the live attenuated vaccine PV strains to generate new neurovirulent viruses (22, 55); such vaccine-derived PV (VDPV) strains have already been implicated in many poliomyelitis outbreaks in various countries (31, 38). Pathogenic VDPV strains have never been isolated in the Central African Republic (CAR), but the widespread use of live attenuated PV vaccine in this country has raised fears about the possible emergence of pathogenic VDPV strains, which have already been found in several other sub-Saharan African countries (4), including the Democratic Republic of the Congo, which borders the CAR.
As a member of the worldwide network for poliomyelitis surveillance, the Institut Pasteur de Bangui, CAR, is responsible for virological investigations of all cases of human acute flaccid paralysis occurring in this country. These investigations involve the collection of stool samples from every patient with acute flaccid paralysis and the detection of PV by isolation in cell cultures (2). If cell cultures are found to be positive for HEV but negative for PV, the corresponding isolates are classified as non-PV HEV; no further investigations are conducted, but these isolates are added to the laboratory collection.
The circulation of HEV has been studied in many parts of the world, but little is known about the pattern of HEV circulation in sub-Saharan Africa, including the CAR. With the aim of improving knowledge of HEV circulation in this country, we retrospectively analyzed a panel of viruses classified as non-PV HEV, isolated from 1997 through 2006 in different regions of the CAR. These viruses were recovered from stool samples of children suffering from acute flaccid paralysis. Their genotypes were determined by sequencing the VP1 and VP2 regions, and phylogenetic analyses were conducted. These molecular analyses highlighted the tremendous diversity of HEV types and lineages circulating in the CAR, including at least 42 types from the 4 HEV species, and revealed the existence of a new genogroup within the type EV-A71.
MATERIALS AND METHODS
Virus samples.
Ninety-three cell culture supernatants positive for non-PV HEV were randomly selected from the virus collection of the Institut Pasteur de Bangui, CAR. All these strains were isolated on human rhabdomyosarcoma (RD) or human larynx epidermoid carcinoma (HEp-2c) cells infected with stool samples from children with acute flaccid paralysis (35). All these samples had been collected in the CAR from 1997 through 2006 for the purpose of diagnosis, in the context of poliomyelitis surveillance. All the supernatants considered in this study had previously tested negative for PV with WHO standard techniques (2).
Sequencing of the VP1 and VP2 coding regions.
The VP1 region was amplified by reverse transcription-PCR (RT-PCR) with generic primers amplifying the entire VP1 sequence (∼900 nt) of members of the species HEV-A to -D (11). If no amplicons were obtained, the samples were subjected to seminested PCR, leading to amplification of the first half of the VP1 sequence (∼470 nt). Sequencing was performed with a BigDye Terminator version 3.1 kit (Applied Biosystems) on an ABI Prism 3140 automated sequencer (Applied Biosystems). For some amplicons, the electropherograms showed superimposed peaks along the entire length of the sequence, suggesting the presence of a mixture of VP1 sequences. The corresponding amplicons were then inserted into the pCR-Blunt plasmid with a Zero Blunt PCR cloning kit (Invitrogen), and 10 clones were sequenced for each sample.
RT-PCR targeting the central part of VP2 (∼300 nt) was carried out on 89 of the 93 supernatants as previously described (40). Sequencing was performed with a GenomeLab Dye Terminator cycle sequencing Quick Start kit (Beckman Coulter) and a CEQ8000 automated DNA sequencer (Beckman Coulter).
Sequences were submitted to GenBank (see below). Isolate names are given in the following format: region of isolation-year of isolation-laboratory number.
Sequence analyses.
The sequences were compared with those of prototype and field strains available in GenBank (the respective accession numbers are shown here; see Fig. 2 to 5). Multiple sequence alignments were performed with CLC Main Workbench 6.0 software (CLC bio). Phylograms were constructed with the MEGA5 program (58), using the Jukes-Cantor algorithm for genetic distance determination and the neighbor-joining method. The robustness of the resulting trees was assessed with 1,000 bootstrap replications.
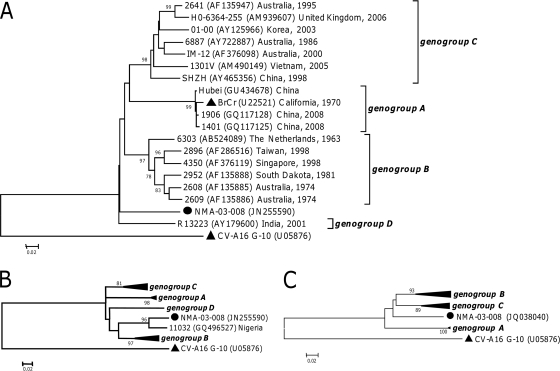
Fig 2.
Phylogenetic relationships between the Central African EV-A71 sequences and other EV-A71 sequences representing the different genogroups. (A) Phylogram based on VP1 sequences (nt 1 to 855 according to BrCr VP1 numbering). (B) Phylogram based on partial VP1 sequences (nt 157 to 458 according to BrCr VP1 numbering). (C) Phylogram based on partial VP2 sequences (nt 229 to 523 according to BrCr VP2 numbering). The Central African sequences are indicated by filled circles. For the other sequences, the location and year of isolation are indicated, if known. Filled triangles indicate the prototype strains. The percent bootstrap values are indicated if higher than 75. The CV-A16 G-10 sequence was introduced for correct rooting of the trees.
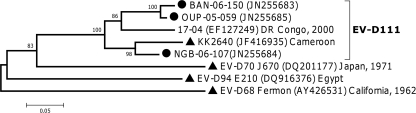
Fig 5.
Phylogenetic relationships between Central African EV-D111 sequences and other HEV-D sequences, based on partial VP1 sequences (nt 117 to 471 according to KK2640 VP1 numbering). The Central African sequences are indicated by filled circles. For the other sequences, the location and year of isolation are indicated, if known. Filled triangles indicate the prototype strains.
Nucleotide sequence accession numbers.
Sequences determined in this work have been submitted to GenBank (accession numbers JN255588 to JN255685 and JQ038038 to JQ038109 for the VP1 and VP2 regions, respectively).
RESULTS
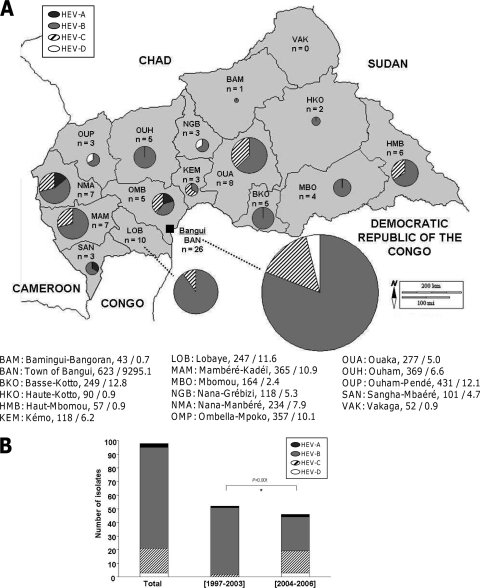
The geographic distribution of the stool samples reflected the heterogeneity of population density in the CAR (Fig. 1A), with the highest densities found in the western part of the country, including, in particular, the capital, Bangui.
Fig 1.
Distribution of the 98 HEV VP1 sequences. (A) Geographic distribution of the sequences in the 16 prefectures of the CAR and in the town of Bangui. Below the map, the number of inhabitants (in thousands) and the population density (in inhabitants per square kilometer) are indicated for each prefecture and for Bangui (data are from the 2003 census). (B) Distribution of the VP1 sequences within the 4 HEV species according to the period of isolation. *, chi-square analysis. The figure was drawn with an outline map provided by d-maps.com (http://d-maps.com/index.php?lang=fr).
VP1 typing.
The VP1 typing method was used to screen the 93 randomly selected cell culture supernatants. Strain mixing was detected in four samples, three of which had two different VP1 sequences, the remaining sample having three VP1 sequences. Overall, 98 VP1 sequences were fully or partly determined (Table 1). They belonged to 42 different types of the four HEV-A to -D species. More than 75% of the isolates belonged to species HEV-B, and about 20% belonged to species HEV-C. Only three isolates belonged to species HEV-A and three to species HEV-D. The most common types were echovirus 7 (E-7), coxsackievirus A 13 (CV-A13), E-13, E-11, and E-21, which together accounted for more than one-third (35 of 98) of the isolates.
Table 1.
Distribution of the Central African non-PV HEV into species and types
Only three of the isolates collected before 2004 belonged to species other than HEV-B, even though more than half the isolates (52 of 98) were collected during this period. The distributions of the four HEV species isolated before and after 2004 differed significantly (P < 0.001 in chi-square analysis) (Fig. 1B). Few HEV-C isolates were collected from 1997 through 2003, whereas HEV-C accounted for 34.8% of the isolates collected during 2004 to 2006. These differences may have resulted, at least in part, from the introduction in 2004 of the HEp-2c cell line for the routine culture of HEV in the Institut Pasteur de Bangui laboratory (100% of the HEV-A, 10% of the HEV-B, 90% of the HEV-C, and most of the HEV-D isolates in this study grew on the HEp-2c cell line).
VP2 typing.
In addition to the typing of VP1, we also typed 89 supernatants by sequencing part of the VP2 region, as described above. Seven of these samples could not be amplified by this method, and 11 gave results discordant with those for the VP1 sequence; those 18 samples, which merit additional detailed investigations (mixing of strains, recombinant strains, etc.), were discarded from this epidemiological study. We report in Table 1 the VP2 typing results for the 71 isolates that gave results concordant with those determined with the VP1 typing method.
Phylogenetic analyses of HEV-A isolates.
Only three HEV-A isolates were found in the panel. They belonged to types CV-A10 and EV-A71.
The two CV-A10 isolates were almost identical over the region of VP1 sequenced in this study. They belonged to lineage III (data not shown), which also includes isolates from Asia (26).
In the VP1 region, isolate NMA-03-008 displayed nucleotide and amino acid homologies of 79.9% and 93.6%, respectively, with the EV-A71 prototype strain BrCr. Nevertheless, it did not fall into any of the four known genogroups of EV-A71 (18, 23a, 56, 65) (Fig. 2A). The VP1 sequence of this isolate was very similar to a short VP1 sequence (∼300 nt) from a strain isolated in Nigeria in 2004 (GenBank accession number GQ496527). These two sequences displayed nucleotide and amino acid homologies of 92.7% and 97.0%, respectively. Compared with sequences representative of the four EV-A71 genogroups, these two sequences formed a separate cluster containing no other sequence (Fig. 2B). Despite the relatively small number of VP2 sequences from field viruses available from public databases, the phylogram for this region also supported the classification of isolate NMA-03-008 in a new genogroup (Fig. 2C). The analyses performed for both VP1 and VP2 indicated that isolate NMA-03-008 belonged to a previously unknown genogroup of EV-A71.
Phylogenetic analyses of HEV-B.
In VP1 analysis, 74 of the 89 sequences segregated with the HEV-B species and could be assigned to 32 different types. Seven HEV-B strains corresponded to recently described types (EV-B69, EV-B74, EV-B77, EV-B80, EV-B84, and EV-B101) for which only a few sequences are currently available in databases. The VP2 and VP1 methods gave the same type identification for 53 of these isolates (Table 1).
Based on the VP1 region, the Central African strains of the E-6, -11, -13, and -30 types belonged to several lineages that have already been described for these types and that include strains isolated from elsewhere around the world (9, 10, 27) (data not shown).
The E-7 type featured at least four main clusters (34), as supported by bootstrap values higher than 77% (Fig. 3); the prototype strain Wallace did not fall into any of these clusters and constituted a separate lineage. The A and C clusters contained isolates from different continents, whereas the B cluster contained only isolates from China. All eight E-7 isolates from the CAR grouped together in a new cluster (cluster D; bootstrap value of 96%) containing no previously reported sequences. A similar clustering was observed for the VP2 sequences of E-7 strains (data not shown).
Fig 3.
Phylogenetic relationships between the Central African E-7 sequences and other E-7 sequences available in GenBank (nt 1 to 413 according to Wallace VP1 numbering). The Central African sequences are indicated by filled circles. For the other sequences, the location and year of isolation are indicated, if known. The percent bootstrap values are indicated if higher than 70. Filled triangles indicate the prototype strains. The E-6 D'Amori sequence was introduced for correct rooting of the tree.
Phylogenetic analyses of HEV-C.
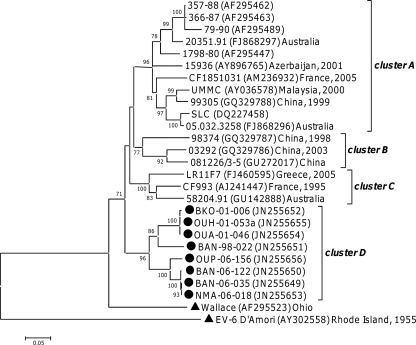
VP1 analysis assigned the 18 HEV-C isolates to seven different types, with almost half these sequences being of the CV-A13 type (Table 1). Previous phylogenetic analyses based on VP1 sequencing identified several clusters in this type (12). The eight Central African CV-A13 isolates fell into three of these clusters (Fig. 4): two fell into cluster A, which also contained strains isolated in North America and Asia between 1998 and 2000, and two others into cluster C, together with strains isolated in Madagascar. The four remaining sequences clustered with a strain isolated in the Democratic Republic of the Congo in 2000 (30); those five sequences together displayed homologies of at least 83.8% and 95.1% at the nucleotide and peptide levels, respectively. Together, they defined a new cluster, cluster D (bootstrap value of 100%). Those five sequences displayed a 6-nt-long deletion with respect to the other CV-A13 strains (VP1 nt 453 to 458, according to the numbering of prototype strain Flores). The phylogenetic tree based on the CV-A13 VP2 region displayed a similar pattern of clustering (data not shown).
Fig 4.
Phylogenetic relationships between partial VP1 Central African CV-A13 and CV-A17 sequences and sequences available in GenBank (nt 1 to 449 according to Flores VP1 numbering). The Central African sequences are indicated by filled circles. For the other sequences, the location and year of isolation are indicated in the tree. Filled triangles indicate the prototype strains. Previously reported clusters that do not contain any Central African sequences are collapsed. Percent bootstrap values are indicated if higher than 70.
Within the CV-A17 type, the VP1 sequence of isolate BAN-06-114 grouped with a strain isolated in the Democratic Republic of the Congo in 2000 (30), with a bootstrap value of 99% (Fig. 4). Within the EV-C99 type, the VP1 sequence of isolate MAM-06-060a fell into cluster B (12), which also contained sequences from Asia and North America (data not shown). Compared with VP1 sequences available in GenBank, the four CV-A20 and the two CV-A24 isolates constituted a unique lineage in their respective types (data not shown).
Phylogenetic analyses of HEV-D.
According to VP1 analysis, the three Central African HEV-D strains belonged to the recently described EV-D111 type, which previously included only two isolates (24, 30). As the five sequences depicted in Fig. 5 were obtained with primers targeting different regions of the genome, they displayed a match over only ∼350 nt (nt 117 to 471, according to KK2640 VP1 numbering). In this region, they featured homologies higher than 81.4% and 89.9% at the nucleotide and peptide levels, respectively. Despite the small number of sequences available for this type, two clusters were distinguishable in the VP1 region (Fig. 5).
No EV-D111 VP2 sequences were available from public databases. In comparison with other HEV-D VP2 sequences, the two Central African EV-D111 VP2 sequences clustered together (data not shown), displaying a nucleotide homology of 87.8% and an amino acid homology of 97.8%.
DISCUSSION
Our main goal in this study was to investigate the diversity of HEV strains circulating in the CAR by analyzing non-PV HEV strains isolated from the stools of children with acute flaccid paralysis of unknown origin. In addition to PV, other viruses, such as West Nile virus, Epstein-Barr virus, and non-PV HEV, may be implicated in acute flaccid paralysis (30, 33, 37, 54, 66). As this study was performed retrospectively, it was not possible to establish a causal link between the clinical symptoms and the presence of an HEV strain in the stools of the children. We have therefore a partial picture of the diversity of HEV in the CAR rather than a panel of the HEV strains actually responsible for acute flaccid paralysis.
We first subjected all the isolates to molecular typing on the basis of their VP1 sequences, which is the method most commonly used for HEV typing (11, 17, 20, 42–45, 62). An alternative typing method (40, 41), based on partial sequencing of the VP2 region, was used in parallel. A huge diversity of serotypes was observed. Most of the isolates belonged to species HEV-B, consistent with previous observations in many epidemiological investigations on HEV circulating in other countries (3, 6, 8, 13, 59, 63). This predominance could have been partly due to cell culture techniques, which may favor the detection of this species (19, 25), as suggested by the significant increase in the isolation of non-B HEV since 2004, when the systematic testing of stool samples on HEp-2c cells was introduced (Fig. 1B). The use of this cell line increased the frequency with which strains belonging to the HEV-C species were isolated in previous studies in Madagascar (49, 51). Similar results were also recently obtained in Cameroon (S. Sadeuh-Mba and F. Delpeyroux, unpublished data).
The VP1 method showed that the isolates belonged to 42 different types and to several lineages within some of these types; most of these typing results were confirmed by VP2 analysis (Table 1). Obtained from fewer than 100 stool samples, this result revealed the huge diversity of HEV strains cocirculating in the CAR. Some of the isolates belonged to lineages that also contained viruses isolated in distant countries, suggesting probable worldwide circulation of the viruses concerned. In contrast, some of the CAR isolates were found to belong to lineages containing viruses isolated only in Africa. Thus, the three HEV-D isolates of this study belonged to the recently described EV-D111 type (Fig. 5), which had previously been found only in Cameroon (24) and in the Democratic Republic of the Congo (30), both of which border the CAR. Similarly, some CV-A13 and CV-A17 isolates from the CAR defined new clusters within their own type with viruses isolated in the Democratic Republic of the Congo in 2000 to 2001 (30).
Through the use of both VP1 and VP2 analyses, this study also identified an EV-A71 VP1 sequence that did not belong to the genogroups already described within this type (Fig. 2). That VP1 sequence was close to a sequence of an EV-A71 isolate from Nigeria. Many EV-A71 strains collected worldwide have been sequenced, but only few were from Africa. Besides the two sequences analyzed here, only two other African strains, isolated in Kenya, have been partly sequenced in the VP1 region. Both these strains were shown to belong to genogroup C (21). Our findings indicate that there is an additional genogroup whose limited circulation, which is apparently confined to Africa, could explain the fact that it was not detected before. Since the occurrence of severe outbreaks in Europe in the 1970s, EV-A71 has been considered a major threat capable of causing poliomyelitis-like disease (1, 46, 47). It is known to be the major cause of hand, foot, and mouth disease (HFMD) and has been implicated in tens of thousands of cases in Asia, with hundreds of fatal cases of neurologic disorders, since the late 1990s. Over the same period, EV-A71 has been implicated in rare and limited outbreaks in developed countries (65). The rapid spread of HFMD in Asia, a region where the disease is endemic, appears to be related to the diversification of certain genogroups and the emergence of new subgenogroups (61). The factors underlying the evolution, diversification, and pathogenicity of this virus remain obscure. The discovery of a previously unknown genogroup circulating in at least some African regions may provide a useful additional piece of the puzzle. Given the highly pathogenic nature of some EV-A71 strains (46, 57), the pathogenicity and ability to cause epidemics of members of this new genogroup should be investigated in future studies.
With the VP1 method, five supernatants were found to contain at least two different types but this number probably represents an underestimation (30). The detection of mixed genomic sequences in field samples requires the cloning of RT-PCR products and the sequencing of several clones for each sample. This process is labor-intensive and not suitable for the screening of a large number of samples. We therefore chose to sequence the RT-PCR products directly, introducing a cloning step only when electropherograms showed superimposed peaks. The presence of two or more viruses would account for the discordance between VP1 and VP2 typing results for some samples. Alternatively, these discrepancies may have resulted from recombination events that had occurred within the capsid region. Such events are believed to be infrequent, probably because of the structural constraints imposed on virions, but previous studies have shown that they can occur in both PV species (14, 15, 23, 36, 39, 60, 67) and non-PV HEV species (16). Further analyses are under way to determine whether the discrepancies observed between the VP1 and VP2 methods for some samples can be attributed to such uncommon recombination events.
Given the bias due to the cell lines used for virus isolation mentioned above, the proportions of HEV-C strains detected in the CAR (approximately 18% for the whole period and 35% during the 2004 to 2006 period) are noteworthy. In most of the epidemiological studies that have been performed on HEV-infected patients in developed countries, the proportions of HEV-C isolates ranged from 0% to 4.7% (6, 8, 32, 52, 59, 63, 64). The high proportion of HEV-C species isolates in this study could have been due to the selection of stool samples from children with acute flaccid paralysis: in an investigation carried out over a 5-year period in China, in the same clinical context, 195 isolates were typed and 32 (16.4%) were found to be HEV-C (13). Similarly, in Cambodia, 40% of the non-PV HEV isolates from patients with acute flaccid paralysis were found to belong to the HEV-C species (7). The HEV-C species also include the three PV types. Intraspecies recombination events between wild-type or vaccine PV stains and non-PV HEV-C strains have been previously reported to have given rise to epidemic recombinant VDPV strains (22, 38). The cocirculation of PV and other HEV-C strains might provide a favorable context for the occurrence of such recombination events. Thus, the two outbreaks due to recombinant VDPV that occurred in Madagascar in the 2000s (50, 53) were linked to the high frequency of HEV-C on this island (12, 28, 29, 49, 51). A recombinant PV/non-PV HEV-C virus was also identified in Cambodia (7). The high frequency of HEV-C in the CAR, associated with the widespread use of oral PV vaccine and the sporadic importation of wild-type PV strains into the country (PV-3 in 2009 and PV-1 in 2011, according to reference 5), could promote the emergence of recombinant VDPV.
In conclusion, this retrospective report highlights the considerable diversity of HEV species circulating in the CAR. That population contains a number of lineages of viruses circulating worldwide but also several lineages that seem to be limited to Africa. In particular, the newly identified EV-A71 genogroup described here has never been observed outside central Africa and merits particular attention due to its potential pathogenicity. The high proportion of HEV-C isolates among those collected from children with acute flaccid paralysis is also a matter of concern, as it suggests that there may be a risk of emergence of recombinant PV/non-PV HEV-C strains with increased virulence.
ACKNOWLEDGMENTS
We are indebted to Coralie Tran, Jean-Michel Thiberge, Laure Diancourt, and Valérie Caro (Plateforme de génotypage des pathogènes et santé publique, Institut Pasteur, Paris, France) for virus sequencing, Matthew Pffloum (Emery University, Atlanta, GA) for initiating the VP1 amplification work, Alexandre Manirakiza (Institut Pasteur de Bangui) for the selection of isolates, and Jean-Luc Bailly (Université d'Auvergne) for fruitful advice.
This study was funded by the Institut Pasteur (PTR276), the French Agence nationale de la recherche (ANR 09 MIEN 019), the Fondation pour la recherche médicale (DMI20091117313), and the WHO/CAR bureau office.
Footnotes
Published ahead of print 15 February 2012
REFERENCES
- 1. Abzug MJ. 2009. Enterovirus 71: emergence of the new poliomyelitis. S. Afr. J. Epidemiol. Infect. 24:5–8 [Google Scholar]
- 2. Anonymous 2004. Isolation and identification of polioviruses, p 87–100 In Polio laboratory manual. World Health Organization, Geneva, Switzerland [Google Scholar]
- 3. Anonymous 2010. Non-PV enterovirus and human parechovirus surveillance—United States, 2006-2008. MMWR Morb. Mortal. Wkly. Rep. 59:1577–1580 [PubMed] [Google Scholar]
- 4. Anonymous 2011. Update on vaccine-derived polioviruses—worldwide, July 2009-March 2011. MMWR Morb. Mortal. Wkly. Rep. 60:846–850 [PubMed] [Google Scholar]
- 5. Anonymous 2011. Wild poliovirus 2005–2011. World Health Organization, Geneva, Switzerland: http://www.polioeradication.org/Portals/0/Document/Data&Monitoring/Wild_poliovirus_list_2005_2011_08Nov.pdf [Google Scholar]
- 6. Antona D, et al. 2007. Surveillance of enteroviruses in France, 2000-2004. Eur. J. Clin. Microbiol. Infect. Dis. 26:403–412 [DOI] [PubMed] [Google Scholar]
- 7. Arita M, et al. 2005. A Sabin 3-derived poliovirus recombinant contained a sequence homologous with indigenous human enterovirus species C in the viral polymerase coding region. J. Virol. 79:12650–12657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bahri O, et al. 2005. Enteroviruses in Tunisia: virological surveillance over 12 years (1992-2003). J. Med. Microbiol. 54:63–69 [DOI] [PubMed] [Google Scholar]
- 9. Bailly JL, et al. 2009. Phylogeography of circulating populations of human echovirus 30 over 50 years: nucleotide polymorphism and signature of purifying selection in the VP1 capsid protein gene. Infect. Genet. Evol. 9:699–708 [DOI] [PubMed] [Google Scholar]
- 10. Bailly JL, et al. 2011. Repeated genomic transfers from echovirus 30 to echovirus 6 lineages indicate co-divergence between co-circulating populations of the two human enterovirus serotypes. Infect. Genet. Evol. 11:276–289 [DOI] [PubMed] [Google Scholar]
- 11. Bessaud M, et al. 2008. Characterization of the genome of human enteroviruses: design of generic primers for amplification and sequencing of different regions of the viral genome. J. Virol. Methods 149:277–284 [DOI] [PubMed] [Google Scholar]
- 12. Bessaud M, Joffret ML, Holmblat B, Razafindratsimandresy R, Delpeyroux F. 2011. Genetic relationship between cocirculating human enteroviruses species C. PLoS One 6:e24823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bingjun T, et al. 2008. Molecular typing and epidemiology of non-PV enteroviruses isolated from Yunnan Province, the People's Republic of China. J. Med. Virol. 80:670–679 [DOI] [PubMed] [Google Scholar]
- 14. Blomqvist S, Bruu AL, Stenvik M, Hovi T. 2003. Characterization of a recombinant type 3/type 2 poliovirus isolated from a healthy vaccinee and containing a chimeric capsid protein VP1. J. Gen. Virol. 84:573–580 [DOI] [PubMed] [Google Scholar]
- 15. Blomqvist S, et al. 2010. Recurrent isolation of poliovirus 3 strains with chimeric capsid protein Vp1 suggests a recombination hot-spot site in Vp1. Virus Res. 151:246–251 [DOI] [PubMed] [Google Scholar]
- 16. Bouslama L, et al. 2007. Natural recombination event within the capsid genomic region leading to a chimeric strain of human enterovirus B. J. Virol. 81:8944–8952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brown BA, et al. 2009. Resolving ambiguities in genetic typing of human enterovirus species C clinical isolates and identification of enterovirus 96, 99 and 102. J. Gen. Virol. 90:1713–1723 [DOI] [PubMed] [Google Scholar]
- 18. Brown BA, Oberste MS, Alexander JP, Jr, Kennett ML, Pallansch MA. 1999. Molecular epidemiology and evolution of enterovirus 71 strains isolated from 1970 to 1998. J. Virol. 73:9969–9975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bryden AS. 1992. Isolation of enteroviruses and adenoviruses in continuous simian cell lines. Med. Lab. Sci. 49:60–65 [PubMed] [Google Scholar]
- 20. Caro V, Guillot S, Delpeyroux F, Crainic R. 2001. Molecular strategy for ‘serotyping’ of human enteroviruses. J. Gen. Virol. 82:79–91 [DOI] [PubMed] [Google Scholar]
- 21. Chakraborty R, et al. 2004. An epidemic of enterovirus 71 infection among HIV-1-infected orphans in Nairobi. AIDS 18:1968–1970 [DOI] [PubMed] [Google Scholar]
- 22. Combelas N, Holmblat B, Joffret ML, Colbere-Garapin F, Delpeyroux F. 2011. Recombination between poliovirus and coxsackie A viruses of species C: a model of viral genetic plasticity and emergence. Viruses 3:1460–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dedepsidis E, et al. 2008. Complete genomic characterization of an intertypic Sabin 3/Sabin 2 capsid recombinant. FEMS Immunol. Med. Microbiol. 52:343–351 [DOI] [PubMed] [Google Scholar]
- 23a. Deshpande JM, Nadkarni SS, Francis PP. 2003. Enterovirus 71 isolated from a case of acute flaccid paralysis in India represents a new genotype. Curr. Sci. 84:1350–1353 [Google Scholar]
- 24. Harvala H, et al. 2011. Detection and genetic characterization of enteroviruses circulating among wild populations of chimpanzees in Cameroon: relationship with human and simian enteroviruses. J. Virol. 85:4480–4486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Heim A. 2005. From poliovirus surveillance to enterovirus surveillance: a complete picture? J. Med. Microbiol. 54:1–2 [DOI] [PubMed] [Google Scholar]
- 26. Hu YF, et al. 2011. Complete genome analysis of coxsackievirus A2, a4, a5, and a10 strains isolated from hand, foot, and mouth disease patients in China revealing frequent recombination of human enterovirus A. J. Clin. Microbiol. 49:2426–2434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Iwai M, et al. 2006. Molecular epidemiology of echoviruses 11 and 13, based on an environmental surveillance conducted in Toyama Prefecture, 2002-2003. Appl. Environ. Microbiol. 72:6381–6387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jegouic S, et al. 2009. Recombination between polioviruses and co-circulating coxsackie A viruses: role in the emergence of pathogenic vaccine-derived polioviruses. PLoS Pathog. 5:e1000412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Joffret ML, et al. Common and diverse features of co-circulating type 2 and 3 recombinant vaccine-derived polioviruses isolated from patients with poliomyelitis and healthy children. J. Infect. Dis., in press [DOI] [PubMed] [Google Scholar]
- 30. Junttila N, et al. 2007. New enteroviruses, EV-93 and EV-94, associated with acute flaccid paralysis in the Democratic Republic of the Congo. J. Med. Virol. 79:393–400 [DOI] [PubMed] [Google Scholar]
- 31. Kew OM, Sutter RW, de Gourville EM, Dowdle WR, Pallansch MA. 2005. Vaccine-derived polioviruses and the endgame strategy for global polio eradication. Annu. Rev. Microbiol. 59:587–635 [DOI] [PubMed] [Google Scholar]
- 32. Khetsuriani N, Lamonte A, Oberste MS, Pallansch M. 2006. Neonatal enterovirus infections reported to the national enterovirus surveillance system in the United States, 1983-2003. Pediatr. Infect. Dis. J. 25:889–893 [DOI] [PubMed] [Google Scholar]
- 33. Kincaid O, Lipton HL. 2006. Viral myelitis: an update. Curr. Neurol. Neurosci. Rep. 6:469–474 [DOI] [PubMed] [Google Scholar]
- 34. Kyriakopoulou Z, et al. 2010. Molecular identification and full genome analysis of an echovirus 7 strain isolated from the environment in Greece. Virus Genes 40:183–192 [DOI] [PubMed] [Google Scholar]
- 35. Manirakiza A, Picard E, Ngbale R, Menard D, Gouandjika-Vasilache I. 2010. OPV strains circulation in HIV infected infants after National Immunisation Days in Bangui, Central African Republic. BMC Res. Notes 3:136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Martín J, et al. 2002. Isolation of an intertypic poliovirus capsid recombinant from a child with vaccine-associated paralytic poliomyelitis. J. Virol. 76:10921–10928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mehrabi Z, et al. 2011. Molecular detection of different types of non-PV enteroviruses in acute flaccid paralysis cases and healthy children, a pilot study. J. Clin. Virol. 50:181–182 [DOI] [PubMed] [Google Scholar]
- 38. Minor P. 2009. Vaccine-derived poliovirus (VDPV): impact on poliomyelitis eradication. Vaccine 27:2649–2652 [DOI] [PubMed] [Google Scholar]
- 39. Mueller JE, et al. 2009. Environmental poliovirus surveillance during oral poliovirus vaccine and inactivated poliovirus vaccine use in Cordoba Province, Argentina. Appl. Environ. Microbiol. 75:1395–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nasri D, et al. 2007. Typing of human enterovirus by partial sequencing of VP2. J. Clin. Microbiol. 45:2370–2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nasri D, et al. 2007. Basic rationale, current methods and future directions for molecular typing of human enterovirus. Expert Rev. Mol. Diagn. 7:419–434 [DOI] [PubMed] [Google Scholar]
- 42. Nix WA, Oberste MS, Pallansch MA. 2006. Sensitive, seminested PCR amplification of VP1 sequences for direct identification of all enterovirus serotypes from original clinical specimens. J. Clin. Microbiol. 44:2698–2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Norder H, Bjerregaard L, Magnius LO. 2001. Homotypic echoviruses share aminoterminal VP1 sequence homology applicable for typing. J. Med. Virol. 63:35–44 [PubMed] [Google Scholar]
- 44. Oberste MS, et al. 1999. Typing of human enteroviruses by partial sequencing of VP1. J. Clin. Microbiol. 37:1288–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Oberste MS, Maher K, Kilpatrick DR, Pallansch MA. 1999. Molecular evolution of the human enteroviruses: correlation of serotype with VP1 sequence and application to picornavirus classification. J. Virol. 73:1941–1948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ooi MH, Wong SC, Lewthwaite P, Cardosa MJ, Solomon T. 2010. Clinical features, diagnosis, and management of enterovirus 71. Lancet Neurol. 9:1097–1105 [DOI] [PubMed] [Google Scholar]
- 47. Palacios G, Oberste MS. 2005. Enteroviruses as agents of emerging infectious diseases. J. Neurovirol. 11:424–433 [DOI] [PubMed] [Google Scholar]
- 48. Racaniello VR. 2007. Picornaviridae: the viruses and their replication, p 795–838 In Knipe DM. (ed), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 49. Rakoto-Andrianarivelo M, et al. 2007. Co-circulation and evolution of polioviruses and species C enteroviruses in a district of Madagascar. PLoS Pathog. 3:e191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rakoto-Andrianarivelo M, et al. 2008. Reemergence of recombinant vaccine-derived poliovirus outbreak in Madagascar. J. Infect. Dis. 197:1427–1435 [DOI] [PubMed] [Google Scholar]
- 51. Rakoto-Andrianarivelo M, et al. 2005. High frequency of human enterovirus species C circulation in Madagascar. J. Clin. Microbiol. 43:242–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Roth B, Enders M, Arents A, Pfitzner A, Terletskaia-Ladwig E. 2007. Epidemiologic aspects and laboratory features of enterovirus infections in Western Germany, 2000-2005. J. Med. Virol. 79:956–962 [DOI] [PubMed] [Google Scholar]
- 53. Rousset D, et al. 2003. Recombinant vaccine-derived poliovirus in Madagascar. Emerg. Infect. Dis. 9:885–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Saad M, et al. 2005. Acute flaccid paralysis: the spectrum of a newly recognized complication of West Nile virus infection. J. Infect. 51:120–127 [DOI] [PubMed] [Google Scholar]
- 55. Savolainen-Kopra C, Blomqvist S. 2010. Mechanisms of genetic variation in polioviruses. Rev. Med. Virol. 20:358–371 [DOI] [PubMed] [Google Scholar]
- 56. Schuffenecker I, et al. 2011. Epidemiology of human enterovirus 71 infections in France, 2000-2009. J. Clin. Virol. 50:50–56 [DOI] [PubMed] [Google Scholar]
- 57. Solomon T, et al. 2010. Virology, epidemiology, pathogenesis, and control of enterovirus 71. Lancet Infect. Dis. 10:778–790 [DOI] [PubMed] [Google Scholar]
- 58. Tamura K, et al. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tan CY, et al. 2011. A retrospective overview of enterovirus infection diagnosis and molecular epidemiology in the public hospitals of Marseille, France (1985-2005). PLoS One 6:e18022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tao Z, et al. 2010. Isolation of a recombinant type 3/type 2 poliovirus with a chimeric capsid VP1 from sewage in Shandong, China. Virus Res. 150:56–60 [DOI] [PubMed] [Google Scholar]
- 61. Tee KK, et al. 2010. Evolutionary genetics of human enterovirus 71: origin, population dynamics, natural selection, and seasonal periodicity of the VP1 gene. J. Virol. 84:3339–3350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Thoelen I, et al. 2004. Analysis of the serotype and genotype correlation of VP1 and the 5′ noncoding region in an epidemiological survey of the human enterovirus B species. J. Clin. Microbiol. 42:963–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Trallero G, et al. 2010. Enteroviruses in Spain over the decade 1998-2007: virological and epidemiological studies. J. Clin. Virol. 47:170–176 [DOI] [PubMed] [Google Scholar]
- 64. Tseng FC, et al. 2007. Epidemiological survey of enterovirus infections occurring in Taiwan between 2000 and 2005: analysis of sentinel physician surveillance data. J. Med. Virol. 79:1850–1860 [DOI] [PubMed] [Google Scholar]
- 65. van der Sanden S, van der Avoort H, Lemey P, Uslu G, Koopmans M. 2010. Evolutionary trajectory of the VP1 gene of human enterovirus 71 genogroup B and C viruses. J. Gen. Virol. 91:1949–1958 [DOI] [PubMed] [Google Scholar]
- 66. Wong M, Connolly AM, Noetzel MJ. 1999. Poliomyelitis-like syndrome associated with Epstein-Barr virus infection. Pediatr. Neurol. 20:235–237 [DOI] [PubMed] [Google Scholar]
- 67. Zhang Y, et al. 2010. Characterization of a rare natural intertypic type 2/type 3 penta-recombinant vaccine-derived poliovirus isolated from a child with acute flaccid paralysis. J. Gen. Virol. 91:421–429 [DOI] [PubMed] [Google Scholar]