Abstract
A PCR-based assay was developed to discriminate the classical, El Tor, and Haitian types of ctxB alleles. Our retrospective study using this newly developed PCR showed that Haitian ctxB first appeared in Kolkata during April 2006, and 93.3% of strains isolated during 2011 carried the new allele. Dendrogram analysis showed a pulsed-field gel electrophoresis (PFGE) pattern of the new variant strains isolated recently that was distinct from the PFGE pattern of the strains carrying classical ctxB that closely matched the 2006 to 2007 variant strains.
TEXT
Cholera still continues to be an important cause of human infection, especially in developing countries that lack access to safe drinking water and proper sanitation. The recent devastating cholera outbreak in Haiti (13), for the first time in almost a century, placed this ancient disease at the forefront of the global public health agenda. In May 2011, the World Health Assembly recognized the reemergence of cholera as a significant global public health problem and called for the implementation of an integrated and comprehensive global approach to cholera control (17). This dreadful diarrheal disease is caused by the Gram-negative toxigenic bacterium Vibrio cholerae (7). To date, more than 200 serogroups of V. cholerae are known, but only serogroups O1 and O139 cause epidemic and pandemic cholera (7, 16). To date, the world has experienced seven pandemics of cholera. Among these, the first six were caused by the classical biotype strains, whereas the ongoing seventh pandemic has been caused by the El Tor biotype (16). In recent years, the emergence and dissemination of novel pathogenic variants of V. cholerae O1 throughout many Asian and African countries (1, 2, 3, 5, 9, 10, 11, 14, 15) indicated a cryptic change in cholera epidemiology. Our recent study showed that the El Tor variant strains of V. cholerae O1 have replaced the prototype El Tor biotype strains in Kolkata, India, since 1995 (15). This report, together with the recent massive cholera outbreak in Haiti, caused by V. cholerae organisms with a mutation in the 58th nucleotide of ctxB (3), motivated us to investigate the emergence and dissemination of this new variant of V. cholerae O1 biotype El Tor strains, if any, in Kolkata.
In this study, we have developed a double-mismatch-amplification mutation assay (DMAMA) to accurately discriminate the classical, El Tor, and Haitian type ctxB alleles through a rapid and simple PCR-based assay. A total of 142 V. cholerae O1 strains were included in this study. These strains were selected from the repository of the National Institute of Cholera and Enteric Diseases, Kolkata, India, covering different months of each year from 2004 to 2011. V. cholerae O1 strains O395 (serotype Ogawa), N16961 (serotype Inaba), and EL-1786 (Ogawa, El Tor) were used as standard strains for the classical, El Tor, and Haitian type, respectively.
Development of the DMAMA-PCR.
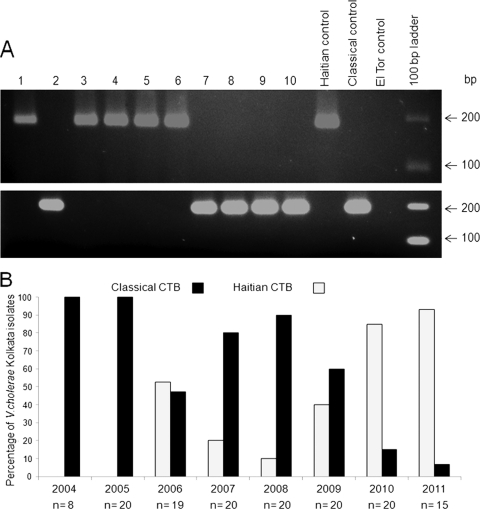
All 142 tested strains, along with the control strains, were grown in Luria-Bertani broth (Becton Dickinson, Sparks, MD) for 18 h and then streaked on Luria agar (LA) plates. In this study, we focused on ctxB in V. cholerae O1 strains to confirm the strains carrying Haitian, classical, and El Tor alleles in a simple PCR-based assay. Current methods for differentiating the biotype-specific cholera toxin B (CTB) subunit of V. cholerae O1 necessitate MAMA-PCR with biotype-specific primers, nucleotide sequencing of the ctxB allele, or an enzyme-linked immunosorbent assay (ELISA) using classical or El Tor CT-specific monoclonal antibodies. Among these, the first has been the method of choice as it is simple and less time consuming. However, reports on influxes of new variant strains of V. cholerae O1 with an additional mutation at the 20th amino acid position (58th nucleotide position) clearly point out its limitation in the discrimination of ctxB genotypes. The previously published MAMA-PCR (8) is based on two biotype-specific reverse primers, each bearing a mismatch at nucleotide position 203 and, hence, incapable of identifying the Haitian type ctxB allele. Therefore, for discriminating the classical, El Tor, and Haitian type ctxB alleles, DMAMA-PCR was designed and validated in this study. We designed two allele-specific polymorphism detection forward primers, ctxB-F3 and ctxB-F4, each bearing a mismatch at its 3′ end (Table 1). These allele-specific primers each carry specific nucleotides, A and C, for the Haitian and classical allele, respectively, at the 3′ end. Furthermore, we enhanced the 3′ mismatch effect by introducing another nucleotide, G (instead of A), at the second nucleotide position (i.e., the 57th nucleotide) from the 3′ end of both primers. We used the ctxB reverse primer that is specific for the classical biotype (Rv-cla), as described by Morita et al. (8), as the conserved reverse primer. As shown in Fig. 1A, the DMAMA-PCR successfully discriminated the three different allelic subtypes of ctxB. V. cholerae O1 strains having the ctxB allele of genotype 7 yielded a 191-bp fragment of DNA with the primer pair ctxB-F3/Rv-cla but not with ctxB-F4/Rv-cla. The classical control strain (O395) produced just the opposite result with the same primer sets, and the El Tor strain (N16961) did not show any amplicon in either PCR assay due to the double mismatch in the forward and reverse primers (Fig. 1A).
Table 1.
Primer sequences, amplicon size, and annealing temperature used in PCR assays
Fig 1.
(A) DMAMA-PCR to detect the type of ctxB allele in representative Vibrio cholerae O1 strains of Kolkata using primers (ctxB-F3/Rv-cla) for the Haitian ctxB allele (top) and (ctxB-F4/Rv-cla) the classical ctxB allele (bottom). The extreme right lane contains a 100-bp size ladder. Lane 1, L19089 (V. cholerae O1, 2006); lane 2, L4706 (V. cholerae O1, 2006); lane 3, M12821(V. cholerae O1, 2007); lane 4, IDH00990 (V. cholerae O1, 2008); lane 5, IDH02003 (V. cholerae O1, 2009); lane 6, IDH03106 (V. cholerae O1, 2010); lane 7, IDH00504 (V. cholerae O1, 2008); lane 8, K16492 (V. cholerae O1, 2005); lane 9, J25916 (V. cholerae O1, 2004); lane 10, IDH03378 (V. cholerae O1, 2011); Haitian control, 2010EL-1786; Classical control, 0395; El Tor control, N16961. (B) Occurrence of ctxB allele type in Kolkata V. cholerae O1 strains from 2004 to 2011. A total of 142 strains were tested during the study period; “n” denotes the number of strains tested in each year. A V. cholerae O1 strain with Haitian type ctxB was isolated in Kolkata for the first time during April 2006.
Sequencing analysis to evaluate the PCR-based result.
To further confirm our PCR results, 14 representative strains, which yielded positive bands for the Haitian ctxB gene by DMAMA-PCR, were selected for DNA sequencing. For sequencing, a separate pair of primers (ctxB-F and ctxB-R) was used to provide the sequences of the whole ctxB genes. Nucleotide sequence analysis of the ctxB genes of the 14 representative strains of V. cholerae O1 revealed that the strains possessed DNA sequences identical to that of the classical type of ctxB but with an additional mutation at the 58th position (C to A). The deduced amino acid sequences of all 14 representative strains were aligned with the CTB sequences of the reference strains N16961 (El Tor) and O395 (classical). The amino acid sequences of all strains were found to be identical to the deduced amino acid sequence of the CTB of the O395 classical reference strain except for a histidine-to-asparagine substitution at the 20th position of the sequence encompassing the signal peptide (GenBank accession number JN806157-59). Thus, the result from DNA sequencing of the ctxB gene confirmed the results of DMAMA-PCR. We also sequenced the ctxB genes from three representative strains that yielded amplicons with the classical specific primers (ctxB-F4/Rv-cla). The deduced amino acid sequences of all three strains were found to be identical to that of the classical reference strain, with a histidine at position 39 and a threonine at position 68. Thus, the results from DNA sequencing of ctxB genes confirmed the results of DMAMA-PCR.
Screening of the Kolkata strains using the DMAMA-PCR.
After standardizing the DMAMA-PCR, we used this assay extensively to investigate the emergence and dissemination of the Haitian variant of V. cholerae strains in Kolkata. All the tested strains from 2004 through 2005 were positive for the classical type of ctxB, indicating that they are El Tor variant strains. The first appearance of Haitian type ctxB was noted in Kolkata during April 2006. There was an abrupt decrease in the isolation profile of V. cholerae O1 strains with the Haitian ctxB allele (CTB genotype 7) during 2007 and 2008. The percentage of the O1 isolates with CTB genotype 7 started to increase from 2009 (Fig. 1B), and more than 93% of Kolkata strains carried the Haitian ctxB allele in 2011.
Phylogenetic analysis based on PFGE.
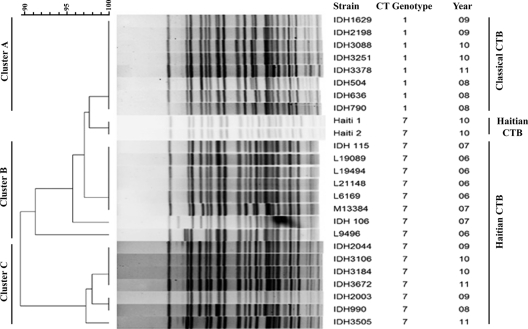
The results of DMAMA-PCR and the sequencing data clearly indicated the appearance of novel variant strains of V. cholerae O1 in Kolkata since 2006, which motivated us to take a closer look at the relatedness of these variants with the Haitian isolates. We also analyzed the NotI pulsed-field gel electrophoresis (PFGE) patterns with representative strains. The PFGE profiles of V. cholerae strains from Kolkata were compared using BioNumerics software (Applied Maths, Sint-Martens-Latem, Belgium) (Fig. 2). The similarity between strains was determined using the Dice coefficient, and cluster analysis was carried out using the unweighted-pair group method using average linkages (UPGMA). All the tested V. cholerae strains with classical ctxB (genotype 1) clustered together (Fig. 2, cluster A), with a similarity matrix of >98%. All the tested strains with Haitian ctxB (genotype 7) in 2006 and 2007 were also found to be closely related to each other, with a similarity matrix of >97% (Fig. 2, cluster B). Dendrogram analysis showed that clusters A and B were closely related to the Haitian V. cholerae strains. Interestingly, all the tested strains with Haitian ctxB in 2008 to 2011 formed a distinct cluster (Fig. 2, cluster C), suggesting considerable diversities in genomic content between strains containing Haitian ctxB in 2006 through 2007 and 2008 through 2011.
Fig 2.
PFGE patterns of the NotI-digested V. cholerae strains from Kolkata and Haitian control strains along with the dendrogram analysis using BioNumerics software (Applied Maths, Sint-Martens-Latem, Belgium). Analysis showed 3 distinct clusters, with all the V. cholerae strains having classical ctxB (genotype 1) clustered together. All the tested isolates with Haitian ctxB (genotype 7) from 2006 through 2007 and in 2008 through 2011, however, were found to form two distinct clusters, suggesting considerable diversities in genomic content between them.
Our results not only signify a cryptic change in the circulating strains in Kolkata but also raise questions about the origin of these variants of V. cholerae O1 El Tor. This new type of ctxB (genotype 7) was first reported by Goel et al. (5) in V. cholerae O1 strains isolated from a cholera outbreak in Kalahandi, Orissa, India, in 2007. But our results clearly show that in Kolkata, genotype 7 prevailed since April 2006. This finding tempted us to speculate that the Haitian type of ctxB may have originated from Kolkata and then disseminated to the neighboring regions like Orissa and other places, although confirmation of this hypothesis requires several other epidemiological and experimental validations, and then may have spread via Nepal to Haiti as reported from many investigations (6, 13). It has been hypothesized that the unique genetic composition of the variant type of strains increases their relative fitness, perhaps as a consequence of increased pathogenicity (4).
Recent reports by several research groups showed a putative link between the strains associated with cholera in Haiti and in Nepal (6, 13), underscoring the speed at which infectious diseases can be transferred globally even to other countries where they are not endemic. Implementing a coordinated, integrated multidisciplinary approach is the only effective way to prevent and contain outbreaks among vulnerable populations living in high-risk areas. Prevention, preparedness, and response depend upon an effective and holistic surveillance system and are linked and interdependent. We strongly believe that the DMAMA-PCR will be an easy and accurate tool for tracking the emergence and dissemination of Haitian variant ctxB in V. cholerae O1 isolates and, therefore, will help in understanding cholera epidemiology around the globe.
ACKNOWLEDGMENTS
This work was supported in part by the Japan Initiative for Global Research Network on Infectious Diseases (J-GRID) of the Ministry of Education, Culture, Sports, Science and Technology of Japan and by the Indian Council of Medical Research, Government of India.
Footnotes
Published ahead of print 22 February 2012
REFERENCES
- 1. Ang GY, et al. 2010. Molecular evidence of cholera outbreak caused by a toxigenic Vibrio cholerae O1 El Tor variant strain in Kelantan, Malaysia. J. Clin. Microbiol. 48:3963–3969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ansaruzzaman M, et al. 2004. Cholera in Mozambique, variant of Vibrio cholerae. Emerg. Infect. Dis. 10:2057–2059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chin CS, et al. 2011. The origin of the Haitian cholera outbreak strain. N. Engl. J. Med. 364:33–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ghosh-Banerjee J, et al. 2010. Cholera toxin production by the El Tor variant of Vibrio cholera O1 compared to prototype El Tor and classical biotypes. J. Clin. Microbiol. 48:4283–4286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Goel AK, et al. 2008. A new variant of Vibrio cholerae O1 El Tor causing cholera in India. J. Infect. 57:280–281 [DOI] [PubMed] [Google Scholar]
- 6. Hendriksen RS, et al. 2011. Population genetics of Vibrio cholerae from Nepal in 2010: evidence on the origin of the Haitian outbreak. mBio 2(4):e00157–11 doi:10.1128/mBio.00157-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kaper JB, Morris JJ, Jr, Levine MM. 1995. Cholera. Clin. Microbiol. Rev. 8:48–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Morita M, et al. 2008. Development and validation of a mismatch amplification mutation PCR assay to monitor the dissemination of an emerging variant of Vibrio cholerae O1 biotype El Tor. Microbiol. Immunol. 52:314–317 [DOI] [PubMed] [Google Scholar]
- 9. Nair GB, et al. 2002. New variants of Vibrio cholerae O1 biotype El Tor with attributes of the classical biotype from hospitalized patients with acute diarrhea in Bangladesh. J. Clin. Microbiol. 40:3296–3299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nair GB, et al. 2006. Cholera due to altered El Tor strains of Vibrio cholerae O1 in Bangladesh. J. Clin. Microbiol. 44:4211–4213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nguyen BM, et al. 2009. Cholera outbreaks caused by an altered Vibrio cholerae O1 El Tor strain producing classical cholera toxin B in Vietnam in 2007 to 2008. J. Clin. Microbiol. 47:1568–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Olsvik O, et al. 1993. Use of automated sequencing of polymerase chain reaction-generated amplicons to identify three types of cholera toxin subunit B in Vibrio cholerae O1 strains. J. Clin. Microbiol. 31:22–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Piarroux R, et al. 2011. Understanding the cholera epidemic, Haiti. Emerg. Infect. Dis. 17:1161–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Raychoudhuri A, et al. 2008. Biotyping of Vibrio cholerae O1: time to redefine the scheme. Indian J. Med. Res. 128:695–698 [PubMed] [Google Scholar]
- 15. Raychoudhuri A, et al. 2009. Classical ctxB in Vibrio cholerae O1, Kolkata, India. Emerg. Infect. Dis. 15:131–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Safa A, Nair GB, Kong RYC. 2010. Evolution of new variants of Vibrio cholerae O1. Trends Microbiol. 18:46–54 [DOI] [PubMed] [Google Scholar]
- 17. World Health Organization 2011. Cholera 2010. Wkly. Epidemiol. Rec. 86:325–340 [PubMed] [Google Scholar]