Abstract
Fecal carriage of carbapenemase-producing Enterobacteriaceae (CPE) has not been extensively investigated, except in the cases of selected patients at risk, mostly during outbreaks. A total of 1,100 fecal samples randomly collected in our institution in two different periods in 2006 (n = 600) and 2009–2010 (n = 500) from hospitalized (26.8%) and nonhospitalized (73.2%) patients were screened for CPE. The first period coincided with an outbreak of VIM-1-producing Enterobacteriaceae, and the second one coincided with the emergence of KPC enzymes in our hospital. Diluted samples in saline were cultured in Luria-Bertani broth with 1 μg/ml imipenem and subcultured in MacConkey agar plates with 4 μg/ml ceftazidime. Growing colonies were screened for CPE (modified Hodge test and EDTA and boronic acid synergy tests). Carbapenemase genes, plasmids in which they are located, and clonal relatedness were determined. Individuals who exhibited fecal carriage of CPE (11/1,043, 1.1%; 95% confidence interval [CI], 0.53 to 1.88) included 8 hospitalized (carriage rate, 2.9%; 95% CI, 1.24 to 5.55) and 3 nonhospitalized patients (carriage rate, 0.4%; 95% CI, 0.08 to 1.14), the latter being identified in 2009. Eighty-two percent of colonized patients were not infected with CPE. Isolates harboring blaVIM-1 with or without blaSHV-12 were identified as Klebsiella pneumoniae (n = 8; ST39, ST688, ST253, and ST163), Enterobacter cloacae (n = 3; two pulsed-field gel electrophoresis [PFGE] types), Escherichia coli (n = 2; ST155 and ST2441), and Citrobacter freundii (n = 1). Some of these lineages had previously been detected in our institution. The blaVIM-1 gene was a member of the class 1 integrons In110 (blaVIM-1-aacA4-aadA1) and In113 (blaVIM-1-aacA4-dhfrII) located on plasmids IncN (n = 11; 30 to 50 kb) and IncHI2 (n = 3; 300 kb), respectively. Dissemination of blaVIM-1 class-1 integrons within highly transferable plasmids in a polyclonal population has potentially contributed to the maintenance and spread of CPE.
INTRODUCTION
Carbapenemase-producing Enterobacteriaceae (CPE) isolates have been increasingly reported in Europe (13, 20). They include enzymes belonging to the Ambler classes A (KPC types), B (VIM, IMP, and more recently NDM enzymes), and D (OXA-48). With the exception of OXA-48, these enzymes confer high-level resistance to most β-lactam compounds, such as penicillins and cephalosporins, but variably affect susceptibility to carbapenems (20, 32). Unlike in other South European countries such as Greece and Italy, the prevalence of CPE in Spain remains low and is mainly related to VIM-producing isolates (13, 28, 31, 35). Nevertheless, KPC-2-, KPC-3-, NDM-1-, and OXA-48-producing Enterobacteriaceae were recently detected in our country (6, 10, 24, 30).
Fecal carriage of CPE isolates has been investigated rarely compared with carriage of isolates producing extended-spectrum β-lactamases (ESBLs) (17, 18, 35, 36), particularly among patients not selected for their relation to outbreak cases. In addition, few surveillance culture recommendations are available for detecting CPE (1, 26, 36).
The objectives of the present study were to evaluate the prevalence of intestinal colonization with CPE in nonselected hospitalized and nonhospitalized patients from the same geographic area of Madrid during two different periods (2006 and 2009-2010), coinciding with CPE outbreak episodes in our institution (10, 31). Our study also assessed a microbiology protocol for screening these isolates in fecal material. Surveillance studies might serve for the implementation of control measures to curtail the spread of these isolates, both in the hospital and in the community. This approach has been recommended for vancomycin-resistant enterococci and ESBL-producing Enterobacteriaceae (7, 16).
MATERIALS AND METHODS
Epidemiological background.
The presence of CPE isolates was sought among 1,100 fecal samples randomly selected from those submitted (n = 11,474) to our laboratory for detection of enteropathogens (n = 9,842, 25.8% from hospitalized patients and 74.2% from nonhospitalized patients) or Clostridium difficile toxins (n = 1,632, 83.8% from hospitalized patients and 16.2% from nonhospitalized patients) during two different periods. The first period, January to April 2006, was coincidental with an outbreak of metallo-β-lactamase (MBL)-producing Enterobacteriaceae (31). The second, July 2009 to January 2010, corresponds to the months when KPC enzymes emerged in our institution and the ongoing presence of VIM isolates was detected (10). In the first period, fecal samples (n = 600) were recovered from 569 patients, 114 (20%) hospitalized and 455 (80%) nonhospitalized. In the second period, we studied samples (n = 500) from 474 patients, 166 (35%) hospitalized and 308 (65%) nonhospitalized. Patient clinical charts were reviewed to obtain epidemiological data. The study was approved by the ethical committee from our institution (reference number CEIC 210/06).
Screening protocol for CPE isolates in fecal samples and its assessment.
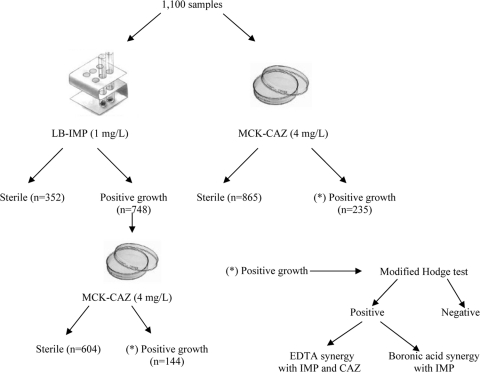
Figure 1 shows the working protocol for fecal samples. Briefly, 100 μl of saline fecal suspension (one full planting loop in 3 ml of saline) was cultured either in 2 ml tubes of Luria-Bertani broth (Pronadisa, Madrid, Spain) supplemented with imipenem (1 μg/ml) or in MacConkey agar plates (Oxoid Ltd., Basingstoke, England) supplemented with ceftazidime (4 μg/ml). Tubes and plates were incubated for 18 h and 48 h, respectively. Growth in tubes and each different colony that grew in ceftazidime-MacConkey plates, based on phenotypic morphology, were subcultured in selective ceftazidime-MacConkey plates for further analysis. Isolates from the same patient exhibiting different colonial morphotypes were also studied. The isolates from subcultured ceftazidime-MacConkey plates were identified using the WIDER system (Fco. Soria-Melguizo, Madrid, Spain) and/or MALDI TOF MS (Microflex LT instrument using the FlexControl 3.0 and MALDI BioTyper 2.0 software; Bruker Daltonics GmbH, Leipzig, Germany).
Fig 1.
Schematic representation of the working protocol. A total of 1,100 saline fecal suspensions were cultured in both MacConkey plates supplemented with 4 μg/ml of ceftazidime (MCK-CAZ) and Luria-Bertani broth with 1 μg/ml of imipenem (LB-IMP). Positive tubes of LB-IMP were subcultured in MCK-CAZ plates, and colonies from both media were tested for carbapenemase production (see Materials and Methods for further details).
All colonies were screened for carbapenemase production using the modified Hodge test (9). The positive ones were screened for MBL, KPC, or OXA-48 carbapenemase production. MBL production was assessed using the double-disk synergy test (MBL-DDST) consisting of imipenem (10 μg) and ceftazidime (30 μg) disks placed 20 mm (center to center) from a disk containing 1.9 mg of EDTA (31, 38). Enhancement of the inhibition zone in the area between the imipenem or ceftazidime disk and the EDTA disk was considered a positive result for MBL. KPC production was inferred by the increase in the inhibition zone of a meropenem disk compared with that of a meropenem disk supplemented with 0.3 mg of boronic acid (12). Presence of OXA-48 enzyme was searched for by PCR (see below). Phenotypic detection of ESBL was inferred by the ESBL-DDST using cefotaxime (30 μg), ceftazidime (30 μg), and amoxicillin-clavulanate (20 to 10 μg) disks (2).
In order to assess the sensitivity of the screening protocol, the detection limit (CFU/ml) was determined using CPE strains that had been characterized previously and that were representative of strains producing VIM-1 (Klebsiella pneumoniae and Enterobacter cloacae) (31), NDM-1 (K. pneumoniae NTCT 13443 strain), KPC-2 (Escherichia coli) and KPC-3 (K. pneumoniae) (reference 10 and this study), and OXA-48 (E. coli, kindly supplied by Patrice Nordmann, and E. cloacae, kindly supplied by Manuel Rodríguez-Iglesias and Luis Martinez-Martinez), as well as the susceptible reference strain E. coli ATCC 25922.
Serial dilutions (10−1, 10−2, 10−3, and 10−4) of a starting inoculum of ∼1 × 109 CFU/ml of each strain were made in 3 ml of saline and thoroughly mixed with one full planting loop of a fecal sample from a healthy volunteer negative for CPE. After an hour, 100 μl of each dilution was cultured on selective medium by using the protocol described above. The detection rate was calculated as the proportion of number of colonies of each strain growing on MacConkey-ceftazidime plates versus the initial inoculum size.
Susceptibility testing.
Antibiotic susceptibility testing was performed using the WIDER system (Fco. Soria-Melguizo). Carbapenem susceptibility was also performed by Etest (bioMérieux, Marcy l'Etoile, France), and results were interpreted according to EUCAST criteria (http://www.eucast.org).
Clonal relatedness.
Clonal relatedness was established by pulsed-field gel electrophoresis (PFGE) of XbaI (New England BioLabs, Inc.) digested genomic DNA (34). Multilocus sequencing typing (MLST) was also performed in K. pneumoniae (http://www.pasteur.fr/recherche/genopole/PF8/mlst/Kpneumoniae.html) and Escherichia coli (http://www.mlst.net) isolates. Clonal relatedness between the isolates recovered in this study and CPE isolates recovered in clinical samples from patients involved in the contemporary outbreaks due to MBL or KPC producing isolates in our institution was analyzed (10, 31).
Characterization of bla genes and analysis of the genetic environment of MBL genes.
Genes encoding carbapenemases (blaVIM, blaIMP, blaSPM, blaGES, and blaKPC) and ESBLs (blaTEM, blaSHV, and blaCTX-M) were sought by PCR using previously described primers and conditions and sequenced further (22, 31). blaNDM was detected by using primers NDM1-F (5′-AAGCTGAGCACCGCATTA-3′, positions 3186 to 3169) and NDM1-R (5′-GCTCATCACGATCATGCT-3′, positions 2473 to 2490), designed according to the sequence of the blaNDM-1 gene in the GenBank database under accession no. FN396876. For blaOXA-48, primers OXA-48-F (5′-GTGGCATCGATTATCGGAAT-3′, positions 2221 to 2240) and OXA-48-R (5′-AGCCCTAAACCATCCGATGT-3′, positions 2933 to 2914), designed according to the sequence of the blaOXA-48 gene in the GenBank database, under accession no. AY236073 were used. Characterization of class 1 integrons containing blaMBL was performed by PCR, sequencing, comparison of the restriction fragment length polymorphism (RFLP) patterns obtained by PCR, and further digestion with AluI enzyme. DNA of each integron showing a distinct RFLP type was sequenced using specific primers as previously described (33).
Transferability and location of blaMBL genes.
Mating experiments were performed using E. coli strain BM21, which is nalidixic acid and rifampin resistant, lactose fermentation positive, and plasmid free, as the recipient (2, 31). Transconjugants were selected on Luria-Bertani agar plates containing ceftazidime (4 mg/liter) and rifampin (300 mg/liter) and were incubated at 37°C for 24 h. Plasmid location of carbapenemase gene was assessed by hybridization of S1-digested genomic DNA with bla gene probes (15). Transfer and hybridization were performed by standard procedures. Labeling and detection were conducted by using a commercial AlkPhos kit (Amersham Life Sciences, Uppsala, Sweden) following the manufacturer's instructions.
Plasmid incompatibility groups were inferred according to the sequences obtained by using the PCR replicon-typing scheme described by Carattoli et al. (3). PCR results were confirmed by hybridization of S1 nuclease-digested genomic DNA from E. coli transconjugants (or wild-type strains in the absence of transfer) with appropriate probes (31).
RESULTS
Fecal carriage of CPE in hospitalized and nonhospitalized patients and carbapenemase characterization.
Fourteen of 1,100 fecal samples contained CPE. These samples were obtained from 11 of 1,043 (1.1%, exact 95% confidence interval [CI], 0.53 to 1.88) individual colonized patients; 8 were hospitalized patients (carriage rate, 2.9%; 95% CI, 1.24 to 5.55), and 3 were nonhospitalized patients (carriage rate, 0.4%; 95% CI, 0.08 to 1.14). There was no increase in the frequency of carbapenemase fecal carriers in hospitalized patients from 2006 [3.5% (4/114); CI, 0.96 to 8.74] to 2009–2010 [2.4% (4/166); CI, 0.66 to 6.05], but carbapenemase colonization in nonhospitalized patients emerged in the second period (1% [3/308]; CI, 0.20 to 2.82).
During the first period studied (January to April 2006), a total of 103 Gram-negative bacilli, including 40 nonfermentative isolates, were recovered in selective medium (Table 1). Four isolates displayed a consistently positive MBL-DDST with a positive blaVIM PCR result. These isolates were recovered from four hospitalized patients that were admitted to oncology and vascular surgery wards and general surgery and cardiovascular intensive care units. Two of these patients were also infected with an MBL-producing isolate, whereas the other two displayed only intestinal colonization. These isolates harbored blaVIM-1 and were identified as K. pneumoniae and E. cloacae (Table 2) isolates which showed PFGE patterns identical to those of VIM-1 epidemic clones (KPMBL-A-ST39 and ECMBL-A) that were previously detected in clinical samples in our institution (31). The epidemic K. pneumoniae clone also expressed SHV-12 and SHV-11.
Table 1.
Identification, frequency, and phenotypic characterization of bacterial isolates recovered from 1,100 fecal samples in selective mediaa
| Organism (no. isolated in 2006/no. in 2010) | Number of isolates with: |
|||||||
|---|---|---|---|---|---|---|---|---|
| Positive MHT |
Positive MBL-DDST |
MBL (type) |
ESBL-DDST |
|||||
| 2006 | 2009-2010 | 2006 | 2009-2010 | 2006 | 2009-2010 | 2006 | 2009-2010 | |
| Escherichia coli (29/64) | 1 | 2 | 0 | 2 | 0 | 2 (VIM-1) | 21 | 30 |
| Enterobacter cloacae (8/18) | 2 | 3 | 2 | 1 | 2 (VIM-1) | 1 (VIM-1) | 1 | 0 |
| Citrobacter freundii (5/18) | 1 | 7 | 0 | 1 | 0 | 1 (VIM-1) | 0 | 1 |
| Morganella morganii (9/13) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Klebsiella pneumoniae (3/17) | 2 | 6 | 2 | 6 | 2 (VIM-1) | 6 (VIM-1) | 1 | 3 |
| Hafnia alvei (5/4) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Enterobacter aerogenes (1/2) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Citrobacter amalonaticus (1/0) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Enterobacter asburiae (1/0) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Klebsiella oxytoca (1/0) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Nonfermentative bacilli (40/40)b | 5 | 1 | 5 | 0 | 0 | 0 | 0 | 0 |
See Materials and Methods for further details. Abbreviations: MHT, modified Hodge test; MBL-DDST, double-disk synergy test for MBL detection; MBL, metallo-β-lactamase; ESBL-DDST, double-disk synergy test for extended-spectrum-β-lactamase detection.
Pseudomonas spp. (10/7), Stenotrophomonas maltophilia (7/2), Acinetobacter lwoffii (6/2), Sphingomonas paucimobilis (1/0), and others (16/29).
Table 2.
Epidemiological data for carbapenemase-producing Enterobacteriaceae isolatesa
| Species (no. of isolates) | PFGE type | Strain identification | MLST | Patient | Ward(s) and/or patient status | Mo and yr of isolation | Plasmid size(s) (kb)b | Inc group | Integron type | Coresistance(s) |
|---|---|---|---|---|---|---|---|---|---|---|
| Klebsiella pneumoniae (8) | KPMBL-A | RYC034268 | ST39 | A | Oncologyc | Mar-06 | 50, 100, 250 | N | B | Gm, Tb, Ak, Na, Fos, SXT |
| RYC042800 | ST39 | B | General surgeryd | Mar-06 | 30, 100, 250 | N | B | Gm, Tb, Ak, Na, Cip, Fos, SXT | ||
| KPMBL-B | RYC165502 | ST688 | C | Internal Medicine | Nov-09 | 50 | N | B | SXT, Fos | |
| KPMBL-C | RYC116749.1 | ST253 | D | Neurology | Aug-09 | 300 | HI2 | A | None | |
| KPMBL-D | RYC197897 | ST163 | E | Internal Medicine | Jan-10 | 50 | N | B | Na, SXT | |
| RYC197365.2e | ST163 | F | Outpatientf | Jan-10 | 50 | N | B | SXT, Fos | ||
| RYC197365.3e | ST163 | F | Outpatientf | Jan-10 | 50 | N | B | SXT | ||
| RYC197894 | ST163 | G | Outpatient | Jan-10 | 50 | N | B | Na, SXT | ||
| Enterobacter cloacae (3) | ECMBL-A | RYC023986 | H | Medical ICU | Feb-06 | 100, 300 | HI2 | A | Na, Cip, Fos, SXT | |
| RYC035509 | I | Cardiovascular ICU | Mar-06 | 100, 300 | HI2 | A | Na, Fos, SXT | |||
| ECMBL-B | RYC116749.2 | D | Neurology | Aug-09 | 50, 320 | N | B | SXT | ||
| Escherichia coli (2) | ECOMBL-A | RYC115509 | ST2441 | J | Outpatient | Aug-09 | 50 | N | B | Tb, SXT |
| ECOMBL-B | RYC165481 | ST155 | K | Nephrology | Nov-09 | 50 | N | B | Tb, Na, Cip, SXT | |
| Citrobacter freundii (1) | CFMBL-A | RYC197365.1 | F | Outpatientf | Jan-10 | 50 | N | B | Na, SXT |
Abbreviations: ICU, intensive care unit; Na, nalidixic acid; Cip, ciprofloxacin; SXT, trimethoprim-sulfonamide; Ak, amikacin; Gm, gentamicin; Tb, tobramycin.
Boldface indicates the size of the blaVIM-1-carrying plasmids, determined by hybridization; underlining indicates plasmids transferred by conjugation.
This patient was also infected with K. pneumoniae KPMBL-A (respiratory infection).
This patient was also infected with K. pneumoniae KPMBL-A (bacteremia and respiratory infection).
RYC197365.2 and RYC197365.3 exhibited a different morphotype.
This patient was previously admitted to a digestive disease ward.
In the second period studied (July 2009 to January 2010), a total of 176 Gram-negative bacilli were recovered. Ten isolates were CPE, later confirmed as MBL producers, and colonized 7 patients, 4 of them hospitalized in internal medicine (n = 2), nephrology (n = 1), and neurology (n = 1) wards (Tables 1 and 2). It should be noted that the remaining isolates were recovered from 3 nonhospitalized patients, only one of whom had been previously hospitalized (three months earlier in a internal medicine ward). None of these patients were infected with MBL-producing isolates, and contact with patients previously infected or colonized with these organisms was not demonstrated. All isolates harbored blaVIM-1 and corresponded to six K. pneumoniae isolates (KPMBL-B, -C, and -D clones), two E. coli isolates (ECOMBL-A and -B clones), one E. cloacae isolate and one Citrobacter freundii isolate (Table 2). One outpatient was simultaneously colonized with two K. pneumoniae isolates displaying different colonial morphotypes, both belonging to clone KPMBL-D-ST163, and one blaVIM-1 C. freundii isolate. In addition, one hospitalized patient was simultaneously colonized by a K. pneumoniae isolate (KPMBL-C) and an E. cloacae isolate (ECMBL-B clone) (Table 2).
Overall, 13 isolates (4 in the first period and 9 in the second one) displayed positive results in the MHT but were not confirmed as carbapenemase producers (false-positive results). This could be due to ESBL production or hyperproduction of AmpC, as reported before (5, 25).
NDM-, KPC-, and OXA-type carbapenemase-producing isolates were not found in either of the studied periods.
Variability in antibiotic susceptibility.
All VIM-1 positive isolates were resistant to all penicillin inhibitor combinations and broad-spectrum cephalosporins. However, some VIM-positive isolates were susceptible to imipenem (MIC range, 0.25 to 4 μg/ml), meropenem (0.125 to 4 μg/ml), ertapenem (0.094 to 16 μg/ml), and doripenem (0.094 to 12 μg/ml) based on the Etest results. Using the EUCAST epidemiological cutoff (ECOFF) values, a number of isolates displayed MICs higher than the corresponding ECOFF values for imipenem (5 isolates), meropenem (13 isolates), doripenem (13 isolates) and ertapenem (14 isolates). As previously observed, differences in susceptibility levels to carbapenems among isolates belonging to the same PFGE clone were found (32). Although aztreonam is not hydrolyzed by MBLs, two K. pneumoniae isolates and two E. cloacae isolates recovered in 2006 were resistant to this antibiotic due to the expression of SHV-12 or AmpC hyperproduction, respectively. Non-β-lactam antibiotic resistances are shown in Table 2.
Clonal background.
An apparently high clonal diversity was observed among K. pneumoniae VIM-1 isolates (8 isolates corresponded to 4 PFGE types). Nevertheless, MLST studies revealed that most of them belonged to clonal complex 23 (CC23): ST163 and its single-locus variants ST253 and ST39 (http://eburst.mlst.net/). KPMBL-A-ST39 (n = 2) was recovered in 2006 from two unrelated patients, KPMBL-D-ST163 (n = 4) was recovered in 2010 from two outpatients and one hospitalized patient, and KPMBL-C-ST253 (n = 1) was recovered in 2009 from a hospitalized patient. The new type ST688, identified in a single patient, corresponded to the PFGE type KPMBL-B (Table 2). K. pneumoniae ST253 has already been associated with human hosts, while ST163 has been recovered from a horse in The Netherlands (http://www.mlst.net/).
The VIM-1 E. coli isolates belonged to the major complex CC155, which is usually associated with urinary tract infections (37), and to a new sequence type (ST2441). Different VIM-1 E. cloacae clones were detected in the two study periods, the ones from 2009-2010 being unrelated to those recovered during our institutional outbreak (31). A carbapenemase-producing C. freundii isolate was found for the first time in our hospital.
blaVIM-1 is part of the In110 and In113 integrons located on IncN and IncHI2 plasmids.
The blaVIM-1 genes were located within the class 1 integrons In110 and In113, which were identified in our institution during an outbreak and arbitrarily designated types A and B, respectively (31, 32). In110 (type A) (blaVIM-1-aacA4-aadA1, ca. 2.5 kb) was identified in two of three E. cloacae isolates and in only one K. pneumoniae isolate. In113 (type B) [blaVIM-1-aacA4-dhfrII (also called dfrB1)-aadA1-catB2, 4 kb] was detected in all but one K. pneumoniae, in one E. cloacae, and in all E. coli and C. freundii isolates. As previously observed, In110 was detected mainly in IncHI2 plasmids of 300 kb, whereas In113 was recovered from IncN plasmids between 30 and 50 kb (Table 2). Plasmids were transferred by conjugation in almost all strains (12 of 14).
Screening method assessment.
Not every CPE isolate recovered from the screening procedure was detected in both selective media. There were 5 carbapenemase-producing isolates that grew only in MacConkey-ceftazidime agar plates, not in LB-imipenem broth. It is also of note that 48 isolates grew only in LB-imipenem broth and were not later confirmed as carbapenemase producers.
The detection limit of the screening method when MacConkey-ceftazidime agar plates were used varied, with KPC and NDM variants showing the lowest values (≤10 CFU/ml) and VIM-1 producers showing the highest values (range, ≤10 to 2.1 × 102 CFU/ml). In the case of LB-imipenem broth plus MacConkey-ceftazidime agar plates, no better detection was observed (data not shown). Unlike the OXA-48-producing E. cloacae isolate (which also produced CTX-M-9), the OXA-48 producing E. coli isolate was able to grow only in LB-imipenem broth and not in MacConkey-ceftazidime agar plates.
DISCUSSION
Spread of CPE has been increasingly reported worldwide. Among them, MBL producers are prevalent in Asia and Europe, whereas KPC producers prevail in the United States (13, 25). CPE are still uncommon in Spain, although isolates producing MBL and, less frequently, KPC or OXA-48 enzymes have been reported (6, 10, 25, 31, 32, 36). A prevalence of 0.04% of MBL-producing Enterobacteriaceae isolates, which include VIM-1- and IMP-producing K. pneumoniae, Klebsiella oxytoca, E. coli, and E. cloacae isolates, was reported in a multicenter study performed in 2009, highlighting a nationwide distribution of these isolates in different Spanish regions (Madrid, Catalonia, Balearic Islands, and Andalusia) (21).
The frequency of CPE fecal colonization in our study is higher than the clinical prevalence previously found in Spanish hospitals (21), although it is still lower than that reported in another study in hospitalized patients in France during a nonoutbreak period (5.3%) (36). Colonization of patients from the community setting with CPE, all of them MBL producers, is also remarkable. These results are an alert for a hidden fecal carriage of MBL-producing isolates, as a high percentage of the isolates from positive fecal carriers (82%, 9 of 11 patients) were not infected with CPE isolates. Since the intestinal compartment could be a reservoir of resistant organisms, active surveillance studies can be implemented to detect fecal carriers. Such a surveillance system was established in Israel in the context of a nationwide epidemic and significantly decreased the spread of carbapenem-resistant K. pneumoniae isolates all over the country (29). Performing surveillance cultures when CPE prevalence remains low might avoid a trend toward endemicity, particularly in acute-care facilities (8).
Currently, there are no consensus recommendations on the detection of CPE fecal carriers. The combined use of an enriched broth supplemented with a carbapenem and a solid selective medium (MacConkey agar) supplemented with ceftazidime might increase the detection of carbapenemase fecal carriers. Nevertheless, the inclusion of a broth medium increases the laboratory workload. Some authors have proposed the use of chromogenic media for ESBL-producing isolates (1, 4). These ESBL-chromogenic media lack specificity for CPE, increasing the need for laboratory confirmation tests to discriminate CPE from ESBL producers. This is also the case in our method. Furthermore, the chromogenic media have also been shown to be inefficient for the detection of OXA-48 producers (4, 26). In the screening method assessment, we efficiently detected VIM, NDM, and KPC producers but not OXA-48 producers. Nevertheless, we were able to detect the OXA-48 E. cloacae isolate also coproducing an ESBL but not the E. coli control strain expressing only OXA-48. The problem of developing a method of detecting OXA-48 producers remains unsolved, although a phenotypic method for the detection of fecal carriage of OXA-48 Enterobacteriaceae was very recently proposed (26).
Despite the limitation of partial ineffective detection of OXA-48 producers, the method used was adequate for the detection of MBL and KPC carbapenemase producers for which carbapenem MICs were even in the susceptible clinical category. Likewise, in other studies that have been conducted for KPC-producing K. pneumoniae isolates (14), 1 μg/ml of imipenem was adequate for the recovery of MBL-producing Enterobacteriaceae from stool specimens. In fact, variations in carbapenem susceptibility of carbapenemase producers support the importance of basing the screening on ceftazidime rather than on imipenem. However, the use of imipenem is not always recommended due to its potential instability compared with other carbapenems.
Our study also revealed a polyclonal structure of VIM-1 producers regardless of the spread of some specific clones. K. pneumoniae isolates were distributed on different PFGE types and STs, although they belonged to the same complex, CC23, detected in hospitalized and nonhospitalized patients (37). This reflects the high risk of specific clones penetrating and disseminating in both settings. As far as we know, the K. pneumoniae STs found in our study have not yet been associated with blaVIM-1 (27) (http://www.pasteur.fr/recherche/genopole/PF8/mlst/Kpneumoniae.html). ST155 E. coli has been frequently associated with the spread of blaCTX-M-14 in healthy volunteers, which illustrates the role of widespread clones as vehicles of different bla genes.
Finally, the identification of the integrons In110 and In113, which were previously detected in our hospital and other Spanish institutions (31, 32), suggests endemicity of these elements and/or the IncN and IncHI2 plasmids in which they are located. IncN plasmids are increasingly associated with carbapenemase-encoding genes (both KPC and VIM types) in Enterobacteriaceae (11, 23). This reflects, as recently demonstrated (19), the high risk of intra- and interspecies horizontal transmission of carbapenemase genes among successful genetic platforms, enhancing their dissemination and persistence.
In conclusion, we describe fecal carriage of CPE, all of them MBL producers, mainly in patients not infected with these organisms in our hospital since 2006 and also in the surrounding geographic area since 2009. These results are an alert for hidden fecal carriage in patients with CPE that would not otherwise have been detected, as most of them were not infected with these organisms and had no demonstrated contact with infected or colonized patients. Moreover, the dissemination of highly transferable plasmids containing blaVIM-1-class 1 integrons among a variety of clones of different species has potentially contributed to the penetration, spread, and maintenance of CPE over time, leading to endemicity.
ACKNOWLEDGMENTS
T.C. is supported by a fellow research contract from the Spanish Ministry of Science and Innovation (grant FI09/00901). This study was funded by research grants from the European Commission (LSHMCT-2008-223031 and FP7-HEALTH-2010-282512) and the CIBERESP Network for Biomedical Research in Epidemiology and Public Health (Instituto Carlos III, Spanish Ministry of Science and Innovation, reference number CB06/02/0053).
Footnotes
Published ahead of print 7 March 2012
REFERENCES
- 1. Adler A, et al. 2011. Laboratory and clinical evaluation of screening agar plates for detection of carbapenem-resistant Enterobacteriaceae from surveillance rectal swabs. J. Clin. Microbiol. 49:2239–2242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cantón R, et al. 2002. Epidemiology of extended-spectrum β-lactamase-producing Enterobacter isolates in a Spanish hospital during a 12-year period. J. Clin. Microbiol. 40:1237–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Carattoli A, et al. 2005. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63:219–228 [DOI] [PubMed] [Google Scholar]
- 4. Carrër A, Fortineau N, Nordmann P. 2010. Use of ChromID extended-spectrum β-lactamase medium for detecting carbapenemase-producing Enterobacteriaceae. J. Clin. Microbiol. 48:1913–1914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Carvalhaes CG, Picão RC, Nicoletti AG, Xavier DE, Gales AC. 2010. Cloverleaf test (modified Hodge test) for detecting carbapenemase production in Klebsiella pneumoniae: be aware of false positive results. J. Antimicrob. Chemother. 65:249–251 [DOI] [PubMed] [Google Scholar]
- 6. Cendejas E, Gómez-Gil R, Gómez-Sánchez P, Mingorance J. 2010. Detection and characterization of Enterobacteriaceae producing metallo-β-lactamases in a tertiary-care hospital in Spain. Clin. Microbiol. Infect. 16:181–183 [DOI] [PubMed] [Google Scholar]
- 7. Centers for Diseases Control and Prevention 1995. Recommendations for preventing the spread of vancomycin resistance. Recommendations of the Hospital Infection Control Practices Advisory Committee (HICPAC). Morb. Mortal. Wkly. Rep. 44:1–13 [PubMed] [Google Scholar]
- 8. Centers for Disease Control and Prevention 2009. Guidance for control of infections with carbapenem-resistant or carbapenemase-producing Enterobacteriaceae in acute care facilities. Morb. Mortal. Wkly. Rep. 58:256–260 [PubMed] [Google Scholar]
- 9. Clinical and Laboratory Standards Institute 2010. Performance standards for antimicrobial susceptibility testing: 19th informational supplement. CLSI document M100–S20. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 10. Curiao T, et al. 2010. Emergence of bla KPC-3-Tn4401a associated with a pKPN3/4-like plasmid within ST384 and ST388 Klebsiella pneumoniae clones in Spain. J. Antimicrob. Chemother. 65:1608–1614 [DOI] [PubMed] [Google Scholar]
- 11. García-Fernández A, et al. 2011. Multilocus sequence typing of IncN plasmids. J. Antimicrob. Chemother. 66:1987–1991 [DOI] [PubMed] [Google Scholar]
- 12. Giske CG, et al. 2011. A sensitive and specific phenotypic assay for detection of metallo-β-lactamases and KPC in Klebsiella pneumoniae with the use of meropenem disks supplemented with aminophenylboronic acid, dipicolinic acid and cloxacillin. Clin. Microbiol. Infect. 17:552–556 [DOI] [PubMed] [Google Scholar]
- 13. Grundmann H, et al. 2010. Carbapenem-non-susceptible Enterobacteriaceae in Europe: conclusions from a meeting of national experts. Euro Surveill. 15:19711. [DOI] [PubMed] [Google Scholar]
- 14. Landman D, Salvani JK, Bratu S, Quale J. 2005. Evaluation of techniques for detection of carbapenem-resistant Klebsiella pneumoniae in stool surveillance cultures. J. Clin. Microbiol. 43:5639–5641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu SL, Hessel A, Sanderson KE. 1993. Genomic mapping with I-Ceu I, an intron-encoded endonuclease specific for genes for ribosomal RNA, in Salmonella spp., Escherichia coli, and other bacteria. Proc. Natl. Acad. Sci. U. S. A. 90:6874–6878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lucet JC, et al. 1999. Control of a prolonged outbreak of extended-spectrum β-lactamase-producing Enterobacteriaceae in a university hospital. Clin. Infect. Dis. 29:1411–1418 [DOI] [PubMed] [Google Scholar]
- 17. Mammina C, et al. 2007. Surveillance of multidrug-resistant gram-negative bacilli in a neonatal intensive care unit: prominent role of cross transmission. Am. J. Infect. Control 35:222–230 [DOI] [PubMed] [Google Scholar]
- 18. March A, et al. 2010. Colonization of residents and staff of a long-term-care facility and adjacent acute-care hospital geriatric unit by multiresistant bacteria. Clin. Microbiol. Infect. 16:934–944 [DOI] [PubMed] [Google Scholar]
- 19. Mathers AJ, et al. 2011. Molecular dissection of an outbreak of carbapenem-resistant Enterobacteriaceae reveals intergenus KPC carbapenemase transmission through a promiscuous plasmid. mBio 2:e00204–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Miriagou V, et al. 2010. Acquired carbapenemases in Gram-negative bacterial pathogens: detection and surveillance issues. Clin. Microbiol. Infect. 16:112–122 [DOI] [PubMed] [Google Scholar]
- 21. Miró E, et al. 2010. Estudio de la prevalencia de β-lactamasas AmpC plasmídicas y carbapenemasas en Enterobacterias en España, abstr. 706, p 321 XIV Congreso SEIMC [Google Scholar]
- 22. Monstein HJ, et al. 2007. Multiplex PCR amplification assay for the detection of blaSHV, blaTEM and blaCTX-M genes in Enterobacteriaceae. APMIS 115:1400–1408 [DOI] [PubMed] [Google Scholar]
- 23. Novais A, et al. 2007. Emergence and dissemination of Enterobacteriaceae isolates producing CTX-M-1-like enzymes in Spain are associated with IncFII (CTX-M-15) and broad-host-range (CTX-M-1, -3, and -32) plasmids. Antimicrob. Agents Chemother. 51:796–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pitart C, et al. 2011. First outbreak of a plasmid-mediated carbapenem-hydrolyzing OXA-48 β-lactamase in Klebsiella pneumoniae in Spain. Antimicrob. Agents Chemother. 55:4398–4401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Queenan AM, Bush K. 2007. Carbapenemases: the versatile β-lactamases. Clin. Microbiol. Rev. 20:440–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ruppé E, et al. 2011. Development of a phenotypic method for detection of fecal carriage of OXA-48-producing Enterobacteriaceae after incidental detection from clinical specimen. J. Clin. Microbiol. 49:2761–2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Samuelsen O, et al. 2011. Molecular characterization of VIM-producing Klebsiella pneumoniae from Scandinavia reveals genetic relatedness with international clonal complexes encoding transferable multidrug resistance. Clin. Microbiol. Infect. 17:1811–1816 [DOI] [PubMed] [Google Scholar]
- 28. Sánchez-Romero I, et al. 2012. Nosocomial outbreak of VIM-1-producing Klebsiella pneumoniae isolates of multilocus sequence type 15: molecular basis, clinical risk factors, and outcome. Antimicrob. Agents Chemother. 56:420–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schwaber MJ, et al. 2011. Israel Carbapenem-Resistant Enterobacteriaceae Working Group. Containment of a country-wide outbreak of carbapenem-resistant Klebsiella pneumoniae in Israeli hospitals via a nationally implemented intervention. Clin. Infect. Dis. 52:848–855 [DOI] [PubMed] [Google Scholar]
- 30. Solé M, et al. 2011. First description of an Escherichia coli strain producing NDM-1 carbapenemase in Spain. Antimicrob. Agents Chemother. 55:4402–4404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tato M, et al. 2007. Complex clonal and plasmid epidemiology in the first outbreak of Enterobacteriaceae infection involving VIM-1 metallo-β-lactamase in Spain: toward endemicity? Clin. Infect. Dis. 45:1171–1178 [DOI] [PubMed] [Google Scholar]
- 32. Tato M, et al. 2010. Carbapenem heteroresistance in VIM-1-producing Klebsiella pneumoniae isolates belonging to the same clone: consequences for routine susceptibility testing. J. Clin. Microbiol. 48:4089–4093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tato M, Coque TM, Baquero F, Cantón R. 2010. Dispersal of carbapenemase blaVIM-1 gene associated with different Tn402 variants, mercury transposons, and conjugative plasmids in Enterobacteriaceae and Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 54:320–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tenover FC, et al. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tórtola MT, et al. 2005. First detection of a carbapenem-hydrolyzing metalloenzyme in two Enterobacteriaceae isolates in Spain. Antimicrob. Agents Chemother. 49:3492–3494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vidal-Navarro L, Pfeiffer C, Bouziges N, Sotto A, Lavigne JP. 2010. Faecal carriage of multidrug-resistant Gram-negative bacilli during a non-outbreak situation in a French university hospital. J. Antimicrob. Chemother. 65:2455–2458 [DOI] [PubMed] [Google Scholar]
- 37. Woodford N, Turton JF, Livermore DM. 2011. Multiresistant Gram-negative bacteria: the role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol. Rev. 35:736–755 [DOI] [PubMed] [Google Scholar]
- 38. Yong D, et al. 2002. Imipenem-EDTA disk method for differentiation of metallo-β-lactamase-producing clinical isolates of Pseudomonas spp. and Acinetobacter spp. J. Clin. Microbiol. 40:3798–3801 [DOI] [PMC free article] [PubMed] [Google Scholar]