LETTER
Human enterovirus 71 (EV71) is an important pathogen and it can cause hand, foot, and mouth disease (HFMD), herpangina, and severe neurological symptoms. There was no intensive surveillance of EV71 until May 2008, when HFMD was classified as a category C notifiable infectious disease by the Ministry of Health of China (6). Acute flaccid paralysis (AFP) surveillance, developed for the polio eradication program, can obtain nonpolio enterovirus (NPEV) isolates as a side benefit. Here, we report the identification of EV71 isolated from AFP cases in 1990 to 2010.
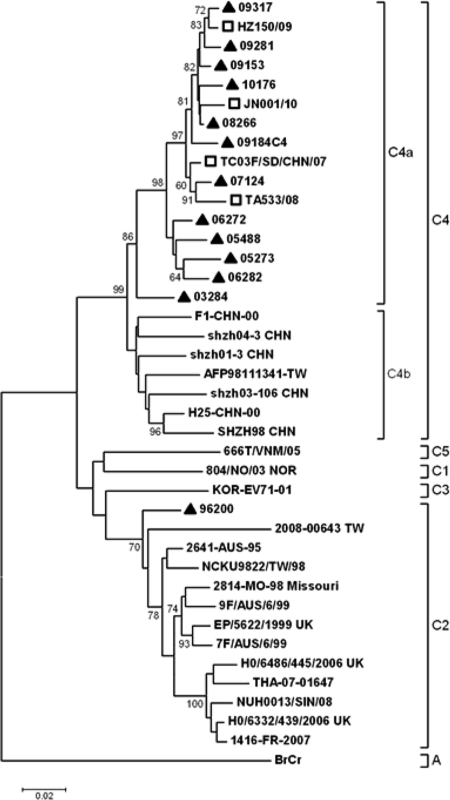
All together, 768 NPEVs were isolated from AFP cases in Shandong Province in this period. Primer pairs 486/488 and 040/011 were used to amplify partial VP1 sequences, and the combination of the two sequences yielded the entire VP1 coding region (2, 3). After molecular typing, 13 (1.7%) EV71 strains were identified, including 1 isolate in 1996, 1 in 2003, 2 in 2005, 2 in 2006, 1 in 2007, 1 in 2008, 4 in 2009, and 1 in 2010. The VP1 sequences were deposited in GenBank under accession numbers GQ253420 to GQ253423, GQ253395, GQ253401, and JQ326297 to JQ326306. Only the isolate in 1996, named 96200, was typed as C2 subgenogroup, and the others were C4 strains. According to the AFP surveillance data, strain 96200 was isolated from an 8-year-old female. The final diagnosis was Guillain-Barre syndrome (GBS), and the patient had residual paralysis at the 60-day clinical follow-up. Phylogenetic analysis based on VP1 entire coding regions of genogroup C strains of EV71 was conducted (Fig. 1). Shandong C4 strains all belonged to C4a, closely related to local HFMD strains in 2007 to 2010. The mean p distance within the Shandong C4 branch was 8.7%. C2 strain 96200 showed a relative remote relationship with other C2 strains. Homologous comparison revealed it had 92.5% to 97.4% VP1 similarities with all C2 members available in GenBank, with the highest homology with Australia strain 2641 (AF135947) in 1995.
Fig 1.
Phylogenetic tree based on complete VP1 sequences of EV71 isolates in Shandong Province and subgenogroup C reference strains. Black triangles indicate strains from local AFP surveillance (represented as YY+serial number), and squares indicate Shandong HFMD reference strains in 2007 to 2010.
In China, large-scale outbreaks of HFMD occurred repeatedly since 2007, when the EV71-associated HFMD outbreak in Linyi City of Shandong Province triggered a nationwide epidemic. Subgenogroup C4 was thought to be the sole viral genetic lineage circulating in mainland China since 1998 (6). This study reveals that the C2 subgenogroup had existed in mainland China previously. So, more retrospective study of EV71 strains in other provinces of mainland China is needed to comprehend the EV71 circulation before 1998.
C2 had a global distribution. Since the 1998 HFMD outbreak in Taiwan, C2 had been frequently reported in Asian, European, and American countries (1, 4, 5). Cocirculation of various genetic subgenogroups could be frequently observed in a given region (4). And the reemergence of C2 in Taiwan in 2008 (1) implied we need to closely monitor if the subgenogroup C2 outbreaks happen or not in mainland China in the next few years.
To our knowledge, this is this first report of the C2 subgenogroup of EV71 in mainland China, and this study reveals that not a single EV71 subgenogroup had existed previously in China.
Nucleotide sequence accession numbers.
VP1 sequences determined in this study were deposited in GenBank under accession numbers JQ326297 to JQ326306.
ACKNOWLEDGMENTS
This study was supported by grants from the Health Department of Shandong Province, China (2011QZ013), and the Science and Technology Development Planning Project of Shandong Province (2009GG10002055).
Footnotes
Published ahead of print 22 February 2012
REFERENCES
- 1. Huang Y, et al. 2010. Genetic diversity and C2-like subgenogroup strains of enterovirus 71, Taiwan, 2008. Virol. J. 7:277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Oberste MS, et al. 1999. Typing of human enteroviruses by partial sequencing of VP1. J. Clin. Microbiol. 37:1288–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Oberste MS, et al. 2006. Species-specific RT-PCR amplification of human enteroviruses: a tool for rapid species identification of uncharacterized enteroviruses. J. Gen. Virol. 87:119–128 [DOI] [PubMed] [Google Scholar]
- 4. van der Sanden S, Koopmans M, Uslu G, van der Avoort H. 2009. Epidemiology of enterovirus 71 in the Netherlands, 1963 to 2008. J. Clin. Microbiol. 47:2826–2833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang J, et al. 2002. Change of major genotype of enterovirus 71 in outbreaks of hand-foot-and-mouth disease in Taiwan. J. Clin. Microbiol. 40:10–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang Y, et al. 2011. Emergence and transmission pathways of rapidly evolving evolutionary branch C4a strains of human enterovirus 71 in the central plain of China. PLoS One 6:e27895. [DOI] [PMC free article] [PubMed] [Google Scholar]