Abstract
Although the presence of mecA is the genotypic determinant of methicillin-resistant Staphylococcus aureus (MRSA), certain MRSA strains, especially community-associated MRSA (C-MRSA), can display an oxacillin MIC in the Clinical and Laboratory Standards Institute susceptible breakpoint range (≤2 μg/ml). Among 91 and 180 isolates thought to be methicillin-susceptible S. aureus (MSSA) with oxacillin MICs of 2 and 1 μg/ml as determined by the Sensititre broth microdilution test initially, 52 (57.1%) and 6 (3.3%), respectively, were mecA positive. These mecA-positive low-oxacillin-MIC isolates belong to the dominant Taiwan C-MRSA clone (clonal complex [CC] 59), 56 of which carried SCCmec type V and were pvl positive, and 43 of which belonged to spa CC t437. All 271 isolates were retested by Sensititre, as well as by Vitek II and disk diffusion (DD). Based on the oxacillin results, the sensitivities of the Sensititre, Vitek II, and DD methods were 48.3% (28/58), 46.6% (27/58), and 89.6% (52/58), respectively. Although cefoxitin was better at detecting these isolates, 12.1, 10.4, and 5.2% of these isolates were still misidentified as MSSA by Sensititre, Vitek II, and DD, respectively. These results highlight the difficulty in the accurate identification of MRSA with borderline oxacillin MICs in the CC59:SCCmec V clone, which likely has contributed to its spread in the health care and community settings. Since this clone has now been detected in other countries, and since other C-MRSA lineages have also been found to have low-level β-lactam resistance, the findings of the present study may be relevant to other regions. Further studies are warranted to determine the extent and clinical impact of such misidentification.
INTRODUCTION
Methicillin-resistant Staphylococcus aureus (MRSA) arises when methicillin-susceptible S. aureus (MSSA) acquires the staphylococcal chromosome cassette mec (SCCmec) element, which contains the mecA gene encoding an altered penicillin-binding protein, PBP2′ (PBP2a) with lowered binding affinity for β-lactams (12). The emergence of community-associated MRSA (C-MRSA) infections in the late 1990s has changed the molecular epidemiology of MRSA worldwide (31). Several reports have indicated C-MRSAs now predominate in the health care setting as the cause of health care-associated MRSA (H-MRSA) infections as well (3, 16, 22, 25).
Why certain clones of C-MRSAs were able to establish a niche and spread rapidly in different geographic regions is unclear. Compared to H-MRSAs, C-MRSAs are less multidrug resistant to non-β-lactam agents, and some C-MRSAs display low-level oxacillin resistance (28, 30). ST59, the founder of clonal complex 59 (CC59), is prevalent in Taiwan (5, 14), and comprises two types: the predominant one carries SCCmec type V and is pvl positive, while the pvl-negative ST59 strains mostly carry SCCmec type IV (1, 5, 14). In addition to being the dominant C-MRSA in Taiwan, the CC59:SCCmec V clone has also been a significant cause of hospital-acquired bloodstream infections in a large medical center in Taiwan (4).
In addition to being highly prevalent in Taiwan, the ST59:SCCmec V “Taiwan clone” was recently reported as the most common CC59 C-MRSA in Western Australia (7). This clone also accounted for 13.1% of C-MRSA isolated from Chinese children in multiple hospitals in China (10). In Europe, the ST59:SCCmec V MRSA has also been detected in the Netherlands (15), while SCCmec V carrying ST338, a single-locus variant of ST59, has also been found in Poland (19). These reports indicate the potential for this clone to spread in other countries.
Accurate identification of methicillin (oxacillin) resistance in S. aureus is important for treatment and infection control purposes. Previously, while investigating genotypes of MSSA, we discovered several ST59 isolates thought to be MSSA with oxacillin MICs in the susceptible range (1 to 2 μg/ml) but contained type V SCCmec (4). The present study was carried out to compare methods for MRSA detection using a retrospective analysis of S. aureus isolates with oxacillin MICs of 1 to 2 μg/ml obtained through our national surveillance program. Our results confirmed that the pvl-positive ST59:SCCmec V clone is the major contributor of this low-level oxacillin-resistant mecA-positive phenotype. We also found fluctuation in the performance of the commonly used susceptibility testing methods in detecting this particular clone of MRSA.
MATERIALS AND METHODS
S. aureus isolates.
All S. aureus isolates from the Taiwan Surveillance of Antimicrobial Resistance (TSAR) program from years 2002, 2004, 2006, and 2008 with oxacillin MICs of 2 μg/ml were included in the present study. Isolates with oxacillin MICs of 1 μg/ml were also randomly selected for the study to include one to four isolates from each hospital (if present) each year. TSAR is a biennial national surveillance program of multiple hospitals throughout Taiwan (20). During July and September of the collection year, each hospital first collected 200 consecutive isolates to include 50 outpatient and 150 inpatient (30 intensive care unit [ICU] patients, 100 non-ICU adults, and 20 pediatric patients) isolates, after which 20 (2002 to 2006) to 50 (2008) isolates from blood and other sterile body sites were collected. In addition to the specimen source, the hospitals also provided the patient age and duplicate isolates were excluded. The same hospitals participated in TSAR between 2002 and 2008 except TSAR V (2006), in which one hospital did not participate. These hospitals included 11 medical centers and 15 regional hospitals located throughout Taiwan. All isolates were stored at −80°C. The species identification and purity of the isolates were confirmed as described previously (20).
Antimicrobial susceptibility testing (AST).
The reference broth microdilution (BMD) method was used for routine TSAR surveillance study. For isolates in 2002, 2004, and 2006, MICs of oxacillin and non-β-lactams were determined by the BMD method according to Clinical and Laboratory Standards Institute (CLSI) guidelines using custom-designed Sensititre panels (Trek Diagnostics, West Essex, England). Prior to 2006, only oxacillin MIC was used to determine methicillin resistance in staphylococci. For isolates in 2008, the Sensititre standard panel GPALL1F was used, which contained oxacillin and cefoxitin (6). For isolates between 2002 and 2006, an inoculum of 5 × 105 CFU/ml was used. However, for the GPALL1F panel, an inoculum of 3 × 105 CFU/ml was used according to the recommendation of the manufacturer. For the present study, 180 isolates with oxacillin MICs of 1 μg/ml and all 91 isolates with oxacillin MICs of 2 μg/ml regardless of mecA PCR results were tested by oxacillin and cefoxitin disk diffusion (DD) test, as well as by Vitek 2 AST-P583 according to the instructions of the manufacturer (bioMérieux, Marcy l'Etoile, France). These 271 isolates were also retested by broth microdilution method using the Sensititre standard panel GPALL1F. The Etest was performed on all mecA-positive isolates only. Test isolates were subcultured from −80°C twice prior to susceptibility testing. The BMD, Etest, and DD test samples were incubated at 35°C; the cefoxitin results were read after 18 to 20 h of incubation, and the oxacillin results were read after 24 h of incubation (6).
Molecular typing.
All isolates were first confirmed to be S. aureus by coagulase PCR (29). The mecA gene was determined by PCR by using the primers mA1 and mA2 according to a previously published protocol (17). SCCmec type was determined on all mecA-positive isolates. The genetic profiles of the mecA-positive strains were also determined based on protein A (spa) typing and Panton-Valentine leukocidin (pvl) gene detection. Multilocus sequence typing (MLST) was performed on strains selected from different spa types from each year. The protocols for MLST, pvl detection, spa typing, and SCCmec typing have been described previously (5, 8, 11, 17, 21).
Statistical analysis.
Susceptibility interpretation analysis was made using the Whonet software (http://www.who.int/drugresistance/whonetsoftware/en/). Univariate analysis was performed with the Statistical Package for the Social Sciences version 18.0 (SPSS, Chicago, IL). A P value of <0.05 was considered statistically significant.
RESULTS
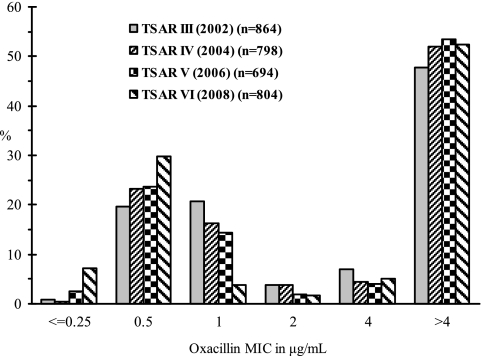
Totals of 864, 798, 694, and 804 nonduplicate S. aureus isolates were tested in 2002, 2004, 2006, and 2008, respectively, by the BMD method using the Sensititre panel. The oxacillin MIC distribution by year is show in Fig. 1. There were 180, 130, 100, and 31 isolates with oxacillin MICs of 1.0 μg/ml (OXA 1.0), while 34, 30, 13, and 14 isolates had oxacillin MICs of 2.0 μg/ml (OXA 2.0) in 2002, 2004, 2006, and 2008, respectively. Together, these OXA 1.0 and OXA 2.0 isolates represented 14.0% (441/3,160) and 2.9% (91/3,160) of the total 3,160 S. aureus isolates tested.
Fig 1.
Oxacillin MIC distribution for S. aureus from the Taiwan Surveillance of Antimicrobial Resistance (TSAR) tested between 2002 and 2008 based on data from the original testing results by broth microdilution method used for routine surveillance study.
All 91 OXA 2.0 isolates and 180 (40.8%) of the 441 randomly selected OXA 1.0 isolates were tested for the presence of mecA by PCR. Six of the OXA 1.0 isolates (3.3%, 6/180), while 52 (57.1%, 52/91) of the OXA 2.0 isolates tested mecA positive, and they were detected in all four study years (Table 1). A higher proportion of OXA 2.0 isolates from 2002 to 2004 were mecA positive compared to those from 2006 to 2008, but the difference was not statistically significant (59.4% [38/64] versus 51.8% [14/34]; P > 0.05). Among these 58 mecA-positive, low-oxacillin-MIC isolates, 56 carried SCCmec type V, the other 2 carried SCCmec type IV (Table 1). The majority (n = 43 [74.1%]) of the isolates belonged to spa clonal complex t437. MLST was performed on 16 strains, and all of them belonged to clonal complex 59 (13 ST59 and 3 ST338).
Table 1.
Presence of mecA in S. aureus with oxacillin MICs of 1 and 2 μg/ml as determined by broth microdilution and SCCmec type of mecA-positive isolates
| Yr | Oxacillin MIC of 1.0 μg/mla |
Oxacillin MIC of 2.0 μg/mlb |
SCCmec type(s) of mecA-positive isolates (no. of isolates)c | |||
|---|---|---|---|---|---|---|
| No of isolates | No. of isolates tested for mecA | No. (%) of mecA-positive isolates | No. of isolates tested | No. (%) of mecA-positive isolates | ||
| 2002 | 180 | 82 | 2 (2.4) | 34 | 20 (58.8) | IV (1), V (21) |
| 2004 | 130 | 50 | 1 (2.0) | 30 | 18 (60.0) | V (19) |
| 2006 | 100 | 35 | 1 (2.9) | 13 | 7 (53.8) | IV (1), V (7) |
| 2008 | 31 | 13 | 2 (15.4) | 14 | 7 (50.0) | V (9) |
| Total | 441 | 180 | 6 (3.3) | 91 | 52 (57.1) | IV (2), V (56) |
Due to the large number of isolates with an oxacillin MIC of 1.0 μg/ml (n = 441), only a portion (40.8%, 180/441) of randomly selected isolates were screened for the presence of mecA.
All isolates with an oxacillin MIC of 2 μg/ml were screened for the presence of mecA.
MLST was performed on 16 selected strains. Thirteen were ST59 and three were ST338, which is a single-locus variant of ST59.
Among the 271 isolates studied, 158 (58.3%) were from abscesses or wounds, and 43 (15.9%), 38 (14.0%), and 9 (3.3%) were from blood, respiratory, and urine specimens, respectively, with the remaining 23 (8.5%) from various other specimen types. Sixty isolates (22.1%) were from pediatric patients (≤18 years old), 204 (75.3%) were from adults, and the patient ages for the other 7 isolates are unknown. The 58 mecA-positive isolates were from 23 different hospitals in Taiwan; 54 (93.1%) were from wound specimens, 2 (3.4%) were from blood, 1 was from an ear, and the specimen source of one was unknown. The age of the patient was known for 55 of these isolates and included 19 (32.8%) pediatric and 36 (62.1%) adult patients. Twenty-six of the isolates were from inpatients, including 2 ICU and 24 non-ICU patients, and 32 were from outpatients. Thus, isolates from abscess or wound (odds ratio [OR], 11.11; 95% confidence interval [CI], 4.273 to 28.884; P < 0.001) and pediatric patients (OR, 2.163; 95% CI, 1.126 to 4.152, P = 0.02) were significantly more likely to be mecA positive with a low oxacillin MIC.
The performance of Sensititre (GPALL1F), Vitek II (AST-P583), and DD is summarized in the results presented in Table 2. Cefoxitin detected 87.9% (n = 51) of the 58 mecA-positive, low-oxacillin-MIC strains as MRSA by the Sensititre GPALL1F panel, while Vitek II detected 89.6% (n = 52). In contrast, by oxacillin MIC, only 48.3% (n = 28) of the Sensititre GPALL1F results and 46.6% (n = 27) of Vitek II results were in the resistant range (Table 2), and 19 isolates were missed by both methods (data not shown). Oxacillin and cefoxitin DD detected more isolates as MRSA: 89.6% (n = 52) and 94.8% (n = 55), respectively. However, among the 213 mecA-negative isolates, false-positive results by cefoxitin occurred in 1, 11, and 14 isolates by Sensititre, Vitek II, and DD, respectively. In addition, based on oxacillin results, 6 and 40 of the mecA-negative isolates were considered methicillin resistant by Vitek II and DD, respectively. Therefore, although oxacillin DD had higher sensitivity, it had the lowest specificity (81.2%) and PPV (56.5%) compared to Sensititre and Vitek II. By Etest, which was performed only on mecA-positive isolates, 93.1% (n = 54) and 65.5% (n = 38) of the isolates would be considered MRSA, but those included 17 isolates with oxacillin MIC 3.0 μg/ml (data not shown).
Table 2.
Performance of Sensititre, Vitek II, and oxacillin and cefoxitin disk diffusion in detecting mecA-positive, low-oxacillin-MIC S. aureus isolatesa
| Parameterb | Values (%) for all isolates (n = 271; mecA-positive isolates, n = 58; mecA-negative isolates, n = 213) |
|||||
|---|---|---|---|---|---|---|
| Sensititre broth microdilutionc |
Vitek II |
Disk diffusion |
||||
| Oxacillin | Cefoxitin | Oxacillin | Cefoxitin | Oxacillin | Cefoxitin | |
| Sensitivity | 48.3 (28/58) | 87.9 (51/58) | 46.6 (27/58) | 89.6 (52/58) | 89.6 (52/58) | 94.8 (55/58) |
| Specificity | 100 (213/213) | 99.5 (212/213) | 97.2 (207/213) | 94.8 (202/213) | 81.2 (173/213) | 93.4 (199/213) |
| PPV | 100 (28/28) | 98.1 (51/52) | 81.8 (27/33) | 82.5 (52/63) | 56.5 (52/92) | 79.7 (55/69) |
| NPV | 87.6 (213/243) | 96.8 (212/219) | 87.0 (207/238) | 97.1 (202/208) | 96.6 (173/179) | 98.5 (199/202) |
A total of 271 isolates were tested. These isolates tested as methicillin susceptible initially by Sensititre broth microdilution between 2002 and 2008, with oxacillin MICs of 1 (n = 180) or 2 (n = 91)μg/ml (see Table 1).
PPV, positive predictive value; NPV, negative predictive value.
Sensititre results are based on retest on GPALL1F panels containing cefoxitin and oxacillin.
DISCUSSION
The factors contributing to the dissemination of C-MRSA are still unclear. However, low-level and heterogeneous oxacillin resistance has been reported as a main characteristic of C-MRSA (23, 28, 30). The present study found a high rate of mecA-positive strains in S. aureus isolates with oxacillin MIC in the susceptible range of the CLSI breakpoint (<2 μg/ml), including those with oxacillin MICs of 1 μg/ml (3.3%, 6/180), and especially in those with oxacillin MICs of 2 μg/ml (57.1%, 52/91). Nearly all of these isolates belong to the prevalent C-MRSA clone, CC59:SCCmec V:pvl-positive, in Taiwan (5, 14). Although the majority were from wound specimens, these isolates were from different years and adult and pediatric patients in more than 20 hospitals, indicating that they were not the result of an outbreak.
Several studies have found cefoxitin to be superior to oxacillin in detecting mecA-mediated resistance, especially in S. aureus strains with low-level resistance (9, 24, 26, 27), and cefoxitin MIC was added as a screening method for determining mecA-mediated resistance by the CLSI in 2008 (6). In the present study, without cefoxitin, both Sensititre and Vitek II would have missed more than half (51.7 to 53.4%) of these mecA-positive isolates because of their low oxacillin MICs. Our results confirm the importance of using cefoxitin as a marker for the detection of mecA-positive S. aureus and provided the genetic background for these isolates in our region. However, it is important to point out that even cefoxitin failed to identify several of these mecA-positive isolates (Sensititre missed 12.1% and Vitek II missed 10.4%) as MRSA, showing the difficulty in accurate identification of MRSA with borderline oxacillin MICs in the CC59:SCCmec V clone.
In the present study, upon retesting of the mecA-positive isolates with oxacillin MICs of 1 to 2 μg/ml by the broth microdilution method using the Sensititre standard GPALL1F panel, 28 of the 58 isolates had oxacillin MICs of >4 μg/ml, of which only 16 had oxacillin MICs of >4 μg/ml by Vitek II (data not shown). These results indicated fluctuation of the oxacillin MIC around the CLSI breakpoints; thus, oxacillin MIC is not a suitable method for detecting these mecA-positive isolates.
The reasons for the low-level oxacillin MIC in the CC59 genetic background are unknown at present. Previous studies have found VraSR and PBP4 to be associated with oxacillin resistance in C-MRSA of other lineages (18). Chromosomal factors affecting the expression of PBPs other than PBP2a, and several auxiliary genes affecting cell wall synthesis may also be involved (2, 18). If these mecA-positive low-oxacillin MIC isolates were misidentified as MSSA initially, they may emerge as highly oxacillin-resistant strains upon subsequent exposure to β-lactam agents (2). Further studies are warranted to determine the extent and clinical impact of such misidentification. In addition, since these isolates were from both inpatients and outpatients, they may have been missed by infection control measures.
In conclusion, there exists a subpopulation of mecA-positive MRSA in Taiwan that can escape detection by the commonly used susceptibility testing methods especially if cefoxitin was not included in the routine panel. These isolates belong to the dominant Taiwan C-MRSA clones (CC59:SCCmec V:pvl-positive and spa CC t437). Since CC59:SCCmec V isolates have now been reported in other countries (7, 10, 13, 15, 19) and since other C-MRSA lineages have also been found to have low-level β-lactam resistance (23, 28, 30), the findings and implications of our study may be relevant to predominant C-MRSA lineages on other continents. Our results indicate that as more clinical laboratories switch to using automated susceptibility testing instruments for determining antimicrobial susceptibility, there is a need for careful evaluation of the performance of these methods for the detection of mecA-positive S. aureus with a low-level oxacillin resistance to prevent their further spread in health care and community settings.
ACKNOWLEDGMENTS
We express our sincere appreciation to the following hospitals for their participation in the Taiwan Surveillance of Antimicrobial Resistance (TSAR): Buddhist Tzu Chi General Hospital; Cathay General Hospital; Changhua Christian Hospital; Cheng-Ching Hospital; Chung Shan Medical University Hospital; Ditmanson Medical Foundation Chia-Yi Christian Hospital; Far Eastern Memorial Hospital; Hua-Lien Hospital; Jen-Ai Hospital; Kaohsiung Armed Forces General Hospital; Kaohsiung Chang Gung Memorial Hospital of the C.G.M.F.; Kaohsiung Medical University Chung-Ho Memorial Hospital; Kaohsiung Veterans General Hospital; Kuang Tien General Hospital; Lo-Hsu Foundation, Inc., Lotung Poh-Ai Hospital; Mennonite Christian Hospital; Min-Sheng Healthcare; National Cheng Kung University Hospital; Saint Mary's Hospital Luodong; Show Chwan Memorial Hospital; Tungs' Taichung MetroHarbor Hospital; Taichung Veterans General Hospital; Tainan Sin-Lau Hospital, the Presbyterian Church in Taiwan; Taipei City Hospital Heping Fuyou Branch; Taipei City Hospital Zhongxiao Branch; and Tri-Service General Hospital. We thank Li-Yun Hsieh for assistance in statistical analysis.
This project was supported by intramural grants from the National Health Research Institutes (97A1-CLPP01, 97A1-CLPP07, ID-099-PP-01, and ID-099-PP-07).
Footnotes
Published ahead of print 29 February 2012
REFERENCES
- 1. Boyle-Vavra S, Ereshefsky B, Wang CC, Daum RS. 2005. Successful multiresistant community-associated methicillin-resistant Staphylococcus aureus lineage from Taipei, Taiwan, that carries either the novel staphylococcal chromosome cassette mec (SCCmec) type VT or SCCmec type IV. J. Clin. Microbiol. 43:4719–4730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chambers HF. 1997. Methicillin resistance in staphylococci: molecular and biochemical basis and clinical implications. Clin. Microbiol. Rev. 10:781–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chambers HF, Deleo FR. 2009. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 7:629–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen CJ, et al. 2009. Change in the molecular epidemiology of methicillin-resistant Staphylococcus aureus bloodstream infections in Taiwan. Diagn. Microbiol. Infect. Dis. 65:199–201 [DOI] [PubMed] [Google Scholar]
- 5. Chen FJ, et al. 2005. Methicillin-resistant Staphylococcus aureus in Taiwan. Emerg. Infect. Dis. 11:1761–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Clinical and Laboratory Standards Institute 2008. Performance standards for antimicrobial susceptibility testing; 18th informational supplement M100-S18. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 7. Coombs GW, et al. 2010. Differentiation of clonal complex 59 community-associated methicillin-resistant Staphylococcus aureus in Western Australia. Antimicrob. Agents Chemother. 54:1914–1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Felten A, Grandry B, Lagrange PH, Casin I. 2002. Evaluation of three techniques for detection of low-level methicillin-resistant Staphylococcus aureus (MRSA): a disk diffusion method with cefoxitin and moxalactam, the Vitek 2 system, and the MRSA-screen latex agglutination test. J. Clin. Microbiol. 40:2766–2771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Geng W, et al. 2010. Molecular characteristics of community-acquired, methicillin-resistant Staphylococcus aureus isolated from Chinese children. FEMS Immunol. Med. Microbiol. 58:356–362 [DOI] [PubMed] [Google Scholar]
- 11. Harmsen D, et al. 2003. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 41:5442–5448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hiramatsu K, Cui L, Kuroda M, Ito T. 2001. The emergence and evolution of methicillin-resistant Staphylococcus aureus. Trends Microbiol. 9:486–493 [DOI] [PubMed] [Google Scholar]
- 13. Ho PL, et al. 2007. Molecular epidemiology and household transmission of community-associated methicillin-resistant Staphylococcus aureus in Hong Kong. Diagn. Microbiol. Infect. Dis. 57:145–151 [DOI] [PubMed] [Google Scholar]
- 14. Huang YC, Chen CJ. 2011. Community-associated methicillin-resistant Staphylococcus aureus in children in Taiwan, 2000s. Int. J. Antimicrob. Agents 38:2–8 [DOI] [PubMed] [Google Scholar]
- 15. Huijsdens XW, et al. 2006. Multiple cases of familial transmission of community-acquired methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 44:2994–2996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Klevens RM, et al. 2007. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA: the journal of the American Medical Association. 298:1763–1771 [DOI] [PubMed] [Google Scholar]
- 17. Kondo Y, et al. 2007. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob. Agents Chemother. 51:264–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Llarrull LI, Fisher JF, Mobashery S. 2009. Molecular basis and phenotype of methicillin resistance in Staphylococcus aureus and insights into new β-lactams that meet the challenge. Antimicrob. Agents Chemother. 53:4051–4063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Luczak-Kadlubowska A, et al. 2008. Countrywide molecular survey of methicillin-resistant Staphylococcus aureus strains in Poland. J. Clin. Microbiol. 46:2930–2937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McDonald LC, et al. 2004. The status of antimicrobial resistance in Taiwan among Gram-positive pathogens: the Taiwan Surveillance of Antimicrobial Resistance (TSAR) programme, 2000. Int. J. Antimicrob. Agents 23:362–370 [DOI] [PubMed] [Google Scholar]
- 21. Mellmann A, et al. 2007. Based Upon Repeat Pattern (BURP): an algorithm to characterize the long-term evolution of Staphylococcus aureus populations based on spa polymorphisms. BMC Microbiol. 7:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Otter JA, French GL. 2011. Community-associated methicillin-resistant Staphylococcus aureus strains as a cause of healthcare-associated infection. J. Hosp. Infect. 79:189–193 [DOI] [PubMed] [Google Scholar]
- 23. Qi W, et al. 2005. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in Zurich, Switzerland (2003): prevalence of type IV SCCmec and a new SCCmec element associated with isolates from intravenous drug users. J. Clin. Microbiol. 43:5164–5170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Roisin S, Nonhoff C, Denis O, Struelens MJ. 2008. Evaluation of new Vitek 2 card and disk diffusion method for determining susceptibility of Staphylococcus aureus to oxacillin. J. Clin. Microbiol. 46:2525–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Seybold U, et al. 2006. Emergence of community-associated methicillin-resistant Staphylococcus aureus USA300 genotype as a major cause of health care-associated blood stream infections. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 42:647–656 [DOI] [PubMed] [Google Scholar]
- 26. Swenson JM, et al. 2007. Detection of mecA-mediated resistance using reference and commercial testing methods in a collection of Staphylococcus aureus expressing borderline oxacillin MICs. Diagn. Microbiol. Infect. Dis. 58:33–39 [DOI] [PubMed] [Google Scholar]
- 27. Swenson JM, Tenover FC. 2005. The results of disk diffusion testing with cefoxitin correlate with presence of mecA in Staphylococcus spp. J. Clin. Microbiol. 43:3818–3823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wannet WJ, et al. 2004. Widespread dissemination in The Netherlands of the epidemic Berlin methicillin-resistant Staphylococcus aureus clone with low-level resistance to oxacillin. J. Clin. Microbiol. 42:3077–3082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Watanabe S, et al. 2005. Structural comparison of ten serotypes of staphylocoagulases in Staphylococcus aureus. J. Bacteriol. 187:3698–3707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Witte W, Pasemann B, Cuny C. 2007. Detection of low-level oxacillin resistance in mecA-positive Staphylococcus aureus. Clin. Microbiol. Infect. 13:408–412 [DOI] [PubMed] [Google Scholar]
- 31. Zetola N, Francis JS, Nuermberger EL, Bishai WR. 2005. Community-acquired methicillin-resistant Staphylococcus aureus: an emerging threat. Lancet Infect. Dis. 5:275–286 [DOI] [PubMed] [Google Scholar]