Abstract
Forty-one Mycobacterium abscessus complex isolates from 17 pediatric cystic fibrosis (CF) patients were typed using a novel variable-number tandem repeat (VNTR) scheme and an automated repetitive-PCR (rep-PCR) system. Both VNTR and rep-PCR typing methods differentiate between members of the M. abscessus complex. The isolates from individual patients are indistinguishable, and the data strongly suggest that individual CF patients are persistently infected with one strain and also suggests that different CF patients can harbor the same strain.
TEXT
The Mycobacterium abscessus complex comprises three closely related species (M. abscessus, M. bolletii, and M. massiliense) and can cause a wide range of infections in humans, including respiratory disease in hosts with compromised lung defenses, such as cystic fibrosis (CF) patients (5, 7). M. abscessus complex has an overall prevalence rate in CF patients of 5 to 6%, and it accounts for over half of all nontuberculous mycobacteria (NTM) isolated from this group (12, 15, 16). Associations have been made between M. abscessus and worse clinical outcomes (6, 8), but this association may not be straightforward, as other studies have failed to support this observation (11). Additionally, based on largely anecdotal evidence, CF patients infected with M. abscessus complex strains are often not considered for lung transplantation. In some CF patients, colonization appears to be transient, with isolation on a single occasion. However, others appear to be permanently infected with M. abscessus complex strains, with repeated isolation from sputum samples. Clinical outcome is variable, and it is unknown if this is associated with a particular species within the complex or strain variation. The members of the complex can be easily differentiated by sequencing of housekeeping genes, such as hsp65 and rpoB (1, 2, 3, 10, 13), but there is little published information on molecular typing beyond species level of the M. abscessus complex. One published study compared an automated repetitive-PCR (rep-PCR) typing system with multilocus sequence analysis and pulse-field gel electrophoresis (PFGE) to resolve ambiguous identification of M. abscessus isolates (18). The whole-genome sequence of M. abscessus was recently published (14), making it possible to design variable-number tandem-repeat (VNTR) typing schemes. One recent publication describes such an assay that can differentiate the individual members of the complex but not individual strains (4). In this study, we describe a variable-number tandem repeat (VNTR) typing scheme capable of strain discrimination within each species and compare it to an automated rep-PCR typing scheme (9). We have applied both methods to a collection of isolates from pediatric CF patients to investigate whether M. abscessus isolated repeatedly from CF patients is the same strain or if these patients are being reinfected by different strains. We also investigate whether there is a particular strain of M. abscessus that is associated with persistent infection in CF patients.
Forty-one M. abscessus complex isolates from 17 sequential pediatric CF patients at Great Ormond Street Hospital were subcultured from beaded, frozen stocks onto blood agar plates (Oxoid, Basingstoke, United Kingdom). All patients had positive cultures for M. abscessus complex between January 2004 and June 2010, and all available isolates were included in the study. However, for patients with a large number of stored isolates, we chose a representative selection of isolates. Three type strains were also included in the study: NCTC 13031 (ATCC 19977), NCTC 10882, and NCTC 10269. We had previously identified all isolates to subspecies level by hsp65 and rpoB gene sequencing (3).
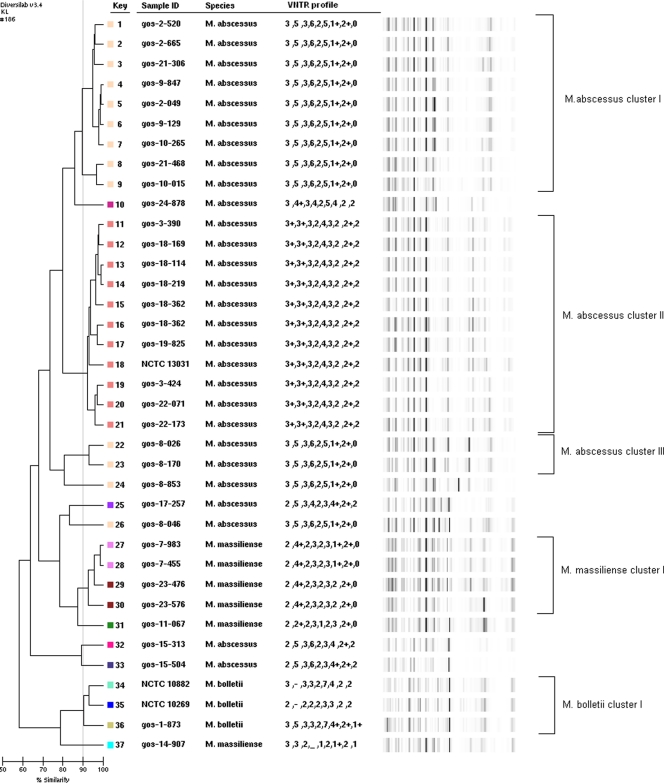
DNA was extracted with an UltraClean microbial DNA isolation kit (Mo-Bio, Cambridge, United Kingdom) as previously described (3). The DNA concentration was determined using a Nanodrop spectrophotometer (Fisher Scientific, Loughborough, United Kingdom). The DNA extract was then subjected to both typing methods. Tandem repeats were sought in the whole-genome sequence of M. abscessus CIP 104536T4 (ATCC 19977) using the Tandem Repeat Finder (http://tandem.bu.edu/). PCR primers were designed for the flanking regions of nine tandem repeat loci spread across the genome. PCR primers with similar melting points were chosen with the aim of ensuring that each locus could be amplified using the same thermal cycler conditions (Table 1). PCRs were performed by adding 3 μl of extracted DNA to 22 μl of PCR master mix containing 1× PCR buffer (Qiagen, Crawley, United Kingdom), a 200 μM concentration of each deoxynucleoside triphosphate, 10 pmol of each primer, and 1.5 U of Taq DNA polymerase. Thermal cycler conditions were as follows: 30 cycles of denaturation for 30 s at 94°C, annealing for 45 s at 55°C, and elongation for 60 s at 72°C, with a final elongation step of 72°C for 10 min. PCR amplicons were electrophoresed on a 1.5% 0.5× Tris-borate-EDTA (TBE) agarose gel and visualized after staining with GelRed alongside Hyperladder II (BioLine, London, United Kingdom) to estimate fragment size. Each locus was assigned a number based on its associated number of repeats, resulting in each isolate having a numerical code of nine numbers, with no amplification at a locus being scored with a dash and amplicons larger than expected (on the basis of whole repeat units) by a plus after the number. The rep-PCR was performed according to the Diversilab Mycobacterium kit protocol. The rep-PCR technology is based on the noncoding repetitive sequences that are interspersed throughout the genomes of all bacteria (17). Amplification of these sequences was performed with the rep-PCR primers provided in the kit, and multiple fragments throughout the bacterial genome were amplified simultaneously. These DNA fragments ranged in size from 50 bp to over 2,000 bp. The amplified PCR products were analyzed on a Diversilab microfluidic LabChip (BioMérieux, Hants, United Kingdom). Electrophoresis was performed automatically on a 2100 Bioanalyzer (Agilent Technologies, Stockport, United Kingdom), and a DNA fingerprint was produced for each isolate. These fingerprints were analyzed and compared to each other using the Diversilab online software (version 3.4) (9). Reports are generated based on different statistical methods; we used the Kullback-Leibler method. Isolates with 90% similarity were regarded as indistinguishable. The VNTR profile was added to the sample data and is also displayed in Fig. 1. Both the VNTR and rep-PCR typing methods were able to differentiate between the three members of the M. abscessus complex, as demonstrated by the distinct VNTR profiles and rep-PCR clusters of isolates previously identified as M. abscessus, M. bolletii, or M. massiliense by hsp65 and rpoB sequencing (3). Furthermore, the identical or almost identical VNTR profiles within groups of isolates were also supported by the rep-PCR clustering (Fig. 1). With the exception of two patients (gos-8 and gos-15), all isolates from a single patient are indistinguishable by rep-PCR. However, all isolates from patient gos-8 have identical VNTR profiles, and the two isolates from patient gos-15 have almost identical VNTR profiles (they differ at one locus). The isolates from four patients (gos-2, gos-9, gos-10, and gos-21) all have the VNTR profile 3,5,3,6,2,5,1+,2+,0 and fall into the rep-PCR cluster M. abscessus I. The isolates from another four patients (gos-3, gos-18, gos-19, and gos-22) and one of the type strains (NCTC 13031) all have the VNTR profile 3+,3+,3,2,4,3,2,2+,2 and fall into the rep-PCR cluster M. abscessus II. Patients gos-18 and gos-19 are siblings. Interestingly, for eight of these nine patients, M. abscessus was isolated on multiple occasions (Table 2). The isolates from patient gos-8 also had the VNTR profile 3,5,3,6,2,5,1+,2+,0, but they did not fall into the cluster M. abscessus I. The isolates from patients gos-7 and gos-23 fall into M. massiliense cluster I and have almost identical VNTR profiles (they differ at one locus). The isolate from patient gos-1 and two of the type strains (NCTC 10269 and NCTC 10882) fall into the rep-PCR cluster M. bolletii I, but they all have different VNTR profiles.
Table 1.
VNTR primer sequences and corresponding Mycobacterium abscessus genomic locations
| Locus no. | Locus name | Primer | Sequence | Repeat unit size (bp) | Flanking sequence (bp) | Genome locationa |
|---|---|---|---|---|---|---|
| 1 | 3416 | 3416F | 5′-CGT TCA TGG TCA GCA AGG AT-3′ | 30 | 228 | 3416798-3416904 |
| 3416R | 5′-TCC CAC GAG ACC ATC AGA AT-3′ | |||||
| 2 | 4356 | 4356F | 5′-CCA TCG AAG AAC CAA ACG AC-3′ | 126 | 180 | 4356558-4357020 |
| 4356R | 5′-AGT GGT GCC ATT GGT GGT A-3′ | |||||
| 3 | 3163 | 3163F | 5-CGC AAC ATC ATC GGA GAA C-3′ | 30 | 217 | 3163695-3163780 |
| 3163R | 5′-GCA TCC AGA TCC ACT GAT CC-3′ | |||||
| 4 | 4038 | 4038F | 5′-GAA GAC CCC CAC TCC AAT TT-3′ | 33 | 200 | 4038904-4038975 |
| 4038R | 5′-AAT AGA GCG TCG TGG TGG AT-3′ | |||||
| 5 | 4093 | 4093F | 5′-TCG TGT AGT CGC TTT GTG CT-3′ | 30 | 210 | 4093541-4093666 |
| 4093R | 5′-CCC GTA TAC GAG GAC GAT GT-3′ | |||||
| 6 | 3320 | 3320F | 5′-AAC CGT ATT CGT CGT CTG CT-3′ | 32 | 187 | 3320506-3320604 |
| 3320R | 5′-GTC AAC TGG ATC CGG AGA AA-3′ | |||||
| 7 | 2177 | 2177F | 5′-AAA ACG CGC GTA TCT TGA AC-3′ | 33 | 154 | 2177971-2178050 |
| 2177R | 5′-GTC GTA AAA GGC CCT CAT CA-3′ | |||||
| 8 | 3398 | 3398F | 5′-CGG TTT CAT GAC AAG CCA GT-3′ | 33 | 242 | 3398103-3398198 |
| 3398R | 5′-GTA GCT CTC GCC AAA AGT CG-3′ | |||||
| 9 | 2220 | 2220F | 5′-GGC AGA ATT ACC GAG TGC AG-3′ | 33 | 224 | 2220890-2220983 |
| 2220R | 5′-GAC ATG GGC ATC GAC AAA C-3′ |
Loci were selected using the Tandem Repeat Finder (http://tandem.bu.edu) using the whole-genome sequence of Mycobacterium abscessus CIP104536T (14).
Fig 1.
Diversilab rep-PCR dendrogram constructed using the Kullback-Leibler method. The VNTR profile is also displayed, and identical profiles are indicated by the color coding. The first part of the sample ID is the patient ID (e.g., gos-1). Clusters of isolates from different patients that are indistinguishable by both typing methods using a 90% similarity coefficient are indicated.
Table 2.
VNTR profile, rep-PCR cluster, and frequency of M. abscessus isolation
| Patient IDa or strain | Total no. of M. abscessus isolates | No. used in this study | No. of weeks between first and last isolates | Identity by hsp65/rpoB sequence | Rep-PCR cluster | VNTR profileb |
|---|---|---|---|---|---|---|
| Gos-2 | 11 | 3 | 176 | M. abscessus | M. abscessus I | 3,5,3,6,2,5,1+,2+,0 |
| Gos-9 | 6 | 3 | 102 | M. abscessus | M. abscessus I | 3,5,3,6,2,5,1+,2+,0 |
| Gos-21 | 4 | 3 | 52 | M. abscessus | M. abscessus I | 3,5,3,6,2,5,1+,2+,0 |
| Gos-10 | 4 | 2 | 114 | M. abscessus | M. abscessus I | 3,5,3,6,2,5,1+,2+,0 |
| Gos-8 | 7 | 3 | 121 | M. abscessus | M. abscessus III | 3,5,3,6,2,5,1+,2+,0 |
| 1 | None | 3,5,3,6,2,5,1+,2+,0 | ||||
| Gos-18 | 32 | 6 | 287 | M. abscessus | M. abscessus II | 3+,3+,3,2,4,3,2,2+,2 |
| Gos-19 | 1 | 1 | M. abscessus | M. abscessus II | 3+,3+,3,2,4,3,2,2+,2 | |
| Gos-22 | 15 | 4 | 99 | M. abscessus | M. abscessus II | 3+,3+,3,2,4,3,2,2+,2 |
| Gos-3 | 8 | 3 | 49 | M. abscessus | M. abscessus II | 3+,3+,3,2,4,3,3,2+,2 |
| NCTC 13031 | M. abscessus | M. abscessus II | 3+,3+,3,2,4,3,3,2+,2 | |||
| Gos-17 | 1 | 1 | M. abscessus | None | 2,5,3,4,2,3,4+,2+,2 | |
| Gos-15 | 3 | 1 | 191 | M. abscessus | None | 2,5,3,6,2,3,4+,2+,2 |
| 1 | None | 2,5,3,6,2,3,4,2+,2 | ||||
| Gos-24 | 1 | 1 | M. abscessus | None | 3,4+,3,4,2,5,4,2,2 | |
| Gos-14 | 1 | 1 | M. massiliense | None | 3,3,2,-,1,2,1+,2,1 | |
| Gos-11 | 1 | 1 | M. massiliense | None | 2,2+,2,3,1,2,3,2+,0 | |
| Gos-7 | 3 | 2 | 71 | M. massiliense | M. massiliense I | 2,4+,2,3,2,3,1+,2+,0 |
| Gos-23 | 4 | 4 | 61 | M. massiliense | M. massiliense I | 2,4+,2,3,2,3,2,2+,0 |
| Gos-1 | 2 | 1 | 26 | M. bolletii | M. bolletii I | 3,5,3,3,2,7,4+,2+,1+ |
| NCTC 10882 | M. bolletii | M. bolletii I | 3,−,3,3,2,7,4,2,2 | |||
| NCTC 10269 | M. bolletii | M. bolletii I | 2,−,2,2,2,3,3,2,2 |
Patients gos-18 and gos-19 are siblings.
A dash indicates no amplification at a locus.
The two typing methods we used in this study are both molecular methods that look at targets that are interspersed throughout the genome, either by looking at regions of the genome that exhibit length variation due to variable-number tandem repeats (VNTR) or by targeting PCR primers to noncoding repetitive sequences (rep-PCR) (4, 9). The M. abscessus complex isolates from the majority of individual patients are indistinguishable by both the VNTR and rep-PCR typing methods. Moreover, several CF patients in this study share M. abscessus complex strains that are indistinguishable by both methods. It is not yet clear if these distinct VNTR profiles and rep-PCR clusters represent a single clonal group, but these data strongly suggest that individual patients are persistently infected with one strain and appear to also suggest that some patients harbor the same strain. Whether the typing methods used in this study have sufficient resolution to identify individual strains of M. abscessus complex is uncertain, but finding six distinct profiles among 12 patients with M. abscessus complex suggests that they do. Whole-genome sequencing of a selection of isolates from this study would unequivocally establish this. Intriguingly, the apparent association between two of the VNTR profiles and rep-PCR clusters M. abscessus I and II with multiple isolations of M. abscessus from individual CF patients suggests a possible link between strain and chronic infection. This requires further investigation, as it could be a factor in disease severity, decline in lung function, and clinical outcome.
Footnotes
Published ahead of print 7 March 2012
REFERENCES
- 1. Adekambi T, Berger P, Raoult D, Drancourt M. 2006. Rpo gene sequence-based characterization of emerging non-tuberculous mycobacteria with descriptions of Mycobacterium bolletii sp. nov., Mycobacterium phocaicum sp. nov. and Mycobacterium aubagnense sp. nov. Int. J. Syst. Evol. Microbiol. 56:133–143 [DOI] [PubMed] [Google Scholar]
- 2. Adekambi T, Colson P, Drancourt M. 2003. RpoB-based identification of nonpigmented and late-pigmenting rapidly growing mycobacteria. J. Clin. Microbiol. 41:5699–5708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Blauwendraat C, Dixon G, Hartley JC, Foweraker J, Harris KA. 2012. The use of a two-gene sequencing approach to accurately distinguish between the species within the Mycobacterium abscessus complex and Mycobacterium chelonae. Eur. J. Clin. Microbiol. Infect. Dis. [Epub ahead of print.] doi:10.1007/s10096-011-1510-9 [DOI] [PubMed] [Google Scholar]
- 4. Choi GE, et al. 2011. Efficient differentiation of Mycobacterium abscessus complex isolates to the species level by a novel PCR-based variable-number tandem-repeat assay. J. Clin. Microbiol. 49:1107–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Esther CR, Jr, Esserman DA, Gilligan P, Kerr A, Noone PG. 2010. Chronic Mycobacterium abscessus infection and lung function decline in cystic fibrosis. J. Cystic Fibrosis 9:117–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Esther CR, Jr, Henry MM, Molina PL, Leigh MW. 2005. Nontuberculous mycobacterial infection in young children with cystic fibrosis. Paediatr. Pulmonol. 40:39–44 [DOI] [PubMed] [Google Scholar]
- 7. Glassroth J. 2008. Pulmonary disease due to nontuberculous mycobacteria. Chest 133:243–251 [DOI] [PubMed] [Google Scholar]
- 8. Hayes D. 2005. Mycobacterium abscessus and other nontuberculous mycobacteria: evolving respiratory pathogens in cystic fibrosis: a case report and review. South Med. J. 98:657–661 [DOI] [PubMed] [Google Scholar]
- 9. Healy M, et al. 2005. Microbial DNA typing by automated repetitive-sequence-based PCR. J. Clin. Microbiol. 43:199–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kim HY, et al. 2007. Outbreak of Mycobacterium massiliense infection associated with intramuscular injections. J. Clin. Microbiol. 45:3127–3313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Olivier KN, et al. 2003. Nontuberculous mycobacteria. II: nested-cohort study of impact on cystic fibrosis lung disease. Am. J. Respir. Crit. Care Med. 167:835–840 [DOI] [PubMed] [Google Scholar]
- 12. Pierre-Audigier C, et al. 2005. Age-related prevalence and distribution of nontuberculous mycobacterial species among patients with cystic fibrosis. J. Clin. Microbiol. 43:3467–3470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ringuet H, et al. 1999. Hsp65 sequencing for identification of rapidly growing mycobacteria. J. Clin. Microbiol. 37:852–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ripoll F, et al. 2009. Non-mycobacterial virulence genes in the genome of the emerging pathogen Mycobacterium abscessus. PLoS One 4:e5660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Roux AL, et al. 2009. Multicenter study of prevalence of nontuberculous mycobacteria in patients with cystic fibrosis in France. J. Clin. Microbiol. 47: 4124–4128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sermet-Gaudelus I, et al. 2003. Mycobacterium abscessus and children with cystic fibrosis. Emerg. Infect. Dis. 9:1587–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Versalovic J, Schneider M, de Bruijn F, Lupski J. 1994. Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods Mol. Cell. Biol. 5:25–40 [Google Scholar]
- 18. Zelazny AM, et al. 2009. Cohort study of molecular identification and typing of Mycobacterium abscessus, Mycobacterium massiliense and Mycobacterium bolletii. J. Clin. Microbiol. 47:1985–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]