Abstract
A transcription-reverse transcription (RT) concerted reaction (TRCR) method was developed for rapid and specific detection of EV71 from clinical specimens. This method was validated with EV71 strains from all of the known genotypes (genotypes A, B1 to B5, and C1 to C5), with detection limits of 10 to 103 copies, and was useful for identification of EV71 from throat swabs of patients with hand, foot, and mouth disease (HFMD).
TEXT
Enterovirus 71 (EV71) is a major etiological agent for hand, foot, and mouth disease (HFMD) and herpangina, along with coxsackievirus A (CVA) types such as CVA10 and CVA16 (1, 10). In contrast to CVA infection (3), EV71 infection is also associated with severe neurological diseases, such as brain-stem encephalitis and poliomyelitis-like paralysis (4, 9, 14) with a high incidence (case severity rate of <0.3%) (6). Effective vaccines are not available at present, and only supportive care can be provided for the treatment of severe brain-stem encephalitis (reviewed in reference 15). Because of the rapid progression of symptoms in fatal cases of EV71 infection (≤12 h after hospitalization) (2), rapid diagnosis of EV71 infection is essential for severely EV71-infected patients to provide proper care among patients with or without HFMD caused by other EV infections.
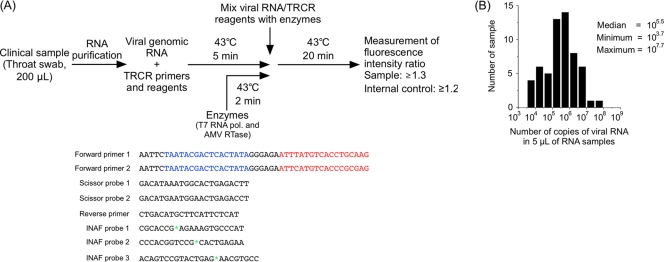
In this study, we aimed to develop a diagnostic test for EV71 by using a transcription-reverse transcription (RT) concerted reaction (TRCR) method (7, 13). The TRCR method is an isothermal RNA amplification method as well as an RT–loop-mediated isothermal amplification (LAMP) method, but it utilizes T7 RNA polymerase and avian myeloblastosis virus (AMV) reverse transcriptase instead of DNA polymerase (7, 13). The major advantages of the TRCR method are (i) rapid detection (20 min of reaction time), (ii) high specificity (by intercalation activating fluorescence [INAF] probes) (8), and (iii) high sensitivity (as low as 10 copies). For its rapid measurement of RNA, the TRCR method has been used as a diagnostic tool for Mycobacterium tuberculosis, Mycobacterium avium, and Mycobacterium intracellulare in some hospitals in Japan (11, 12). The procedure for using a TRCR method for EV71 detection is summarized in Fig. 1A.
Fig 1.
A TRCR method for the detection of EV71 from clinical specimens. (A) Procedure and a primer-probe set for the TRCR method. Viral RNA and TRCR reagents and enzymes were preincubated at 43°C for 5 min and 2 min, respectively, and then were mixed. EV71-targeting regions and T7 promoter regions of the forward primers are colored red and blue, respectively. The positions of oxazole yellow dye in INAF probes are indicated by green asterisks. (B) Histogram of RNA samples extracted from throat swabs of HFMD or herpangina patients in Japan (total 58 samples) grouped by the numbers of copies of viral RNA.
To determine the detection limits of the TRCR method, the numbers of copies of EV genomes in throat swabs from patients with HFMD or herpangina were determined by real-time PCR (5), with quantified viral genomic RNA of EV71(BrCr-TR) as the standard. The numbers of copies of EV genomes in 5 μl of purified viral RNA samples extracted from throat swabs were in a range of 103.7 to 107.7 copies, with a median of 105.5 copies (Fig. 1B). Therefore, we set the detection limit at 103.0 copies of EV viral genomes to develop a TRCR method for EV71 detection.
Primers and probes were designed in the VP1 coding region (nucleotides [nt] 3004 to 3214): there were two forward primers and two scissor probes, one reverse primer, and three INAF probes (Fig. 2). The specificity of the TRCR method to EV71 was mainly conferred by the INAF probes (Fig. 3), which require 100% nucleotide identity to the complementary target sequences for efficient emission of the fluorescence after hybridization (8). To validate the test for negative samples, we used an internal control (an artificial control RNA [103 copies] and INAF probe), the signals of which could validate the reaction in negative samples.
Fig 2.
Comparison of the nucleotide sequences of the EV71 and CVA16 genomes examined for the primers and scissor probes. Genomic sequences of the EV71 and CVA16 strains are shown with their genotypes. Nucleotide differences in EV71 genomic sequences and in primers or probes are highlighted in red (genotype A), blue (genotype B), and green (genotype C).
Fig 3.
Comparison of the nucleotide sequences of the EV71 and CVA16 genomes for the INAF probes. Genomic sequences of EV71 and CVA16 strains are shown with their genotypes and names. Nucleotide differences in the EV71 genomic sequences and in INAF probes are highlighted in red (genotype A), blue (genotype B), and green (genotype C). Flanking nucleotides of oxazole yellow dye are colored in cyan in the INAF probe sequences. INAF probes (1, 2, and 3) and the target EV71 strains are colored in cyan, yellow, and pink, respectively. For detection of genotype C3 of EV71, INAF probes 2 and 3 were required. The amino acid sequences of EV71 and CVA16 in the INAF probe-binding region are shown below.
The conditions for the TRCR method are as follows. Briefly, 10 μl of the primer solution (330 mM potassium chloride, 0.24 μΜ scissor probe 1, 0.24 μΜ scissor probe 2, 1.5 μΜ forward primer 1, 1.5 μΜ forward primer 2, 1.5 μΜ reverse primer, 15 nΜ INAF probe 1, 15 nΜ INAF probe 2, 30 nΜ INAF probe 3, and 150 nΜ INAF probe for internal control), 10 μl of the TRCR substrate solution (51 mM magnesium chloride, 180 mM Tris-HCl [pH 8.6], 6 U RNase inhibitor, 3 mM dithiothreitol, 0.75 mM deoxynucleotide triphosphate, 8.4 mM nucleotide triphosphate, 10.8 mM ITP, and 103 copies of internal control RNA), and 5 μl of purified viral genomic RNA sample were mixed in a reaction tube (TRCR sample), and then the mixture was incubated at 43°C for 5 min. The TRCR enzyme mixture (6.4 U of avian myeloblastosis virus [AMV] reverse transcriptase and 142 U of T7 RNA polymerase) (5 μl) was incubated at 43°C for 2 min and then added to the TRCR sample. Fluorescence intensity ratios of samples (at 520 nm) or of an internal control (at 610 nm) were measured for 20 min or 30 min by using a TRCRapid-160 analyzer (Tosoh Corp.): samples with a reading of ≥1.3 of the fluorescence intensity ratio were judged as EV71 positive, and internal controls with >1.2 of the fluorescence intensity ratio were judged as valid in EV71-negative samples.
The detection limits and the specificity of this TRCR method were evaluated with the EV71 strains from all known genotypes (A, B1 to B5, and C1 to C5) (Table 1). The detection limits of this TRCR method varied among the genotypes: the highest sensitivity was found for genotypes A and C2 (10 copies), moderate sensitivity was found for genotypes B1, B2, B3, C1, and C5 (102 copies), and the lowest sensitivity was found for genotypes B4, B5, and C3 (103 copies). In contrast to positive signals for EV71 strains (104 copies), CVA16 (1010 copies) did not show positive signals at all, and only the signal of the internal control was detected (Table 1) (data not shown). We also analyzed the specificity of this TRCR method using viral RNAs of clinical isolates of CVA2, -4, -6, -10, -14, and -16. We could not detect these viral RNAs (>108 copies) by this TRCR method (data not shown).
Table 1.
Detection of EV71 and CVA16 by the TRCR method
| No. of copies of RNA | Reaction time (min)a |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EV71-BrCr (A) | EV71-Nagoya (B1) | EV71-86-11316-NETH-86 (B2) | EV71-SK-EV006 (B3) | EV71-C7-Osaka (B4) | EV71-2972-Yamagata-2003 (B5) | EV71-0236 (C1) | EV71-1095 (C2) | EV71-003-KOR-00 (C3) | EV71-75-Yamagata-2003 (C4) | EV71-209 (C5) | CVA16-G-10 | |
| 10 | 18.2 (1.37) | ND (1.05) | ND (1.00) | NT | NT | ND (1.11) | ND (1.08) | 14.5 (1.60) | NT | NT | NT | NT |
| 102 | 12.5 (1.70) | 16.0 (1.49) | 18.9 (1.34) | 16.5 (1.45) | ND (1.12) | ND (1.05) | 19.4 (1.34) | 11.5 (1.88) | NT | ND (1.05) | 14.9 (1.57) | NT |
| 103 | 11.1 (1.61) | 10.7 (1.70) | 12.3 (1.91) | 11.1 (1.77) | 15.2 (1.63) | 10.6 (2.60) | 10.4 (1.98) | 9.7 (1.95) | 19.7 (1.30) | 12.4 (1.75) | 15.2 (1.52) | NT |
| 104 | 8.5 (16.7) | 10.4 (1.70) | 10.6 (2.03) | 9.9 (1.79) | 11.9 (1.80) | 9.0 (2.67) | 8.6 (2.06) | 8.6 (2.01) | 14.1 (1.36) | 10.2 (1.86) | 9.7 (1.77) | NT |
| 105 | 7.8 (1.74) | 9.1 (1.71) | 9.4 (2.10) | 8.8 (1.80) | 10.1 (1.88) | 8.1 (2.71) | 7.7 (2.05) | 7.6 (2.03) | 12.4 (1.39) | 9.2 (1.92) | 8.4 (1.85) | NT |
| 106 | 7.1 (1.74) | 8.0 (1.75) | NT | 8.0 (1.82) | 8.9 (1.92) | NT | 7.3 (1.99) | 6.8 (2.04) | 12.1 (1.38) | 8.0 (1.97) | 7.4 (1.89) | NT |
| 1010 | NT | NT | NT | NT | NT | NT | NT | NT | NT | NT | NT | ND (1.06) |
The reaction time that the fluorescence intensity ratio took to reach the threshold value (1.3) is shown. The fluorescence intensity ratio at 20 min of reaction is shown in parentheses. NT; not tested; ND, not detected.
Next, we tested the TRCR method with 58 throat swab samples from patients with HFMD or herpangina (Table 2). Clinical specimens (throat swabs) collected from HFMD or herpangina patients in 2009 and 2010 in Japan and those identified as EV positive by a cell culture method were used for the validation of this TRCR method. The experiments performed in the present study have been approved by the Committee for Ethical Regulation of the National Institute of Infectious Diseases, Japan. EV71 was successfully detected from all EV71-positive throat swab samples by this TRCR method, but not from samples positive for other EVs.
Table 2.
Detection of EV71 from throat swabs by the TRCR method
| Sample no. | No. of copies of viral RNA | Identified virus | EV71 detection (520 nm) |
Internal control (610 nm) |
||
|---|---|---|---|---|---|---|
| Reaction time (min)a | Fluorescence intensity ratio | Reaction time (min)a | Fluorescence intensity ratio | |||
| 1 | 1.4E+06 | EV71 | 10.6 | 1.54 | 19.4 | 1.46 |
| 2 | 2.3E+04 | CVA16 | NDb | 1.05 | 10.8 | 2.30 |
| 3 | 1.2E+05 | EV71 | 12.6 | 1.51 | 12.9 | 1.97 |
| 4 | 2.4E+05 | EV71 | 12.3 | 1.50 | 13.6 | 1.91 |
| 5 | 4.8E+05 | CVA16 | ND | 1.05 | 13.1 | 2.02 |
| 6 | 1.4E+04 | CVA16 | ND | 1.05 | 18.2 | 1.45 |
| 7 | 8.6E+05 | EV71 | 13.2 | 1.50 | 26.6 | 1.22 |
| 8 | 1.4E+04 | EV71 | 15.8 | 1.44 | 11.2 | 2.34 |
| 9 | 1.2E+05 | CVA16 | ND | 1.14 | 20.0 | 1.37 |
| 10 | 3.0E+05 | CVA16 | ND | 1.06 | 15.0 | 1.69 |
| 11 | 1.4E+05 | CVA16 | ND | 1.07 | 15.6 | 1.67 |
| 12 | 2.3E+05 | CVA16 | ND | 1.05 | 17.0 | 1.59 |
| 13 | 6.2E+03 | EV71 | 13.1 | 1.46 | 10.9 | 2.21 |
| 14 | 3.2E+05 | CVA16 | ND | 1.05 | 17.1 | 1.53 |
| 15 | 3.0E+05 | CVA16 | ND | 1.06 | 13.3 | 1.98 |
| 16 | 1.9E+07 | CVA4 | ND | 1.12 | 13.5 | 1.95 |
| 17 | 1.4E+05 | CVA16 | ND | 1.05 | 19.4 | 1.39 |
| 18 | 2.4E+06 | CVB4, CVA4 | ND | 1.05 | 29.1 | 1.20 |
| 19 | 6.0E+03 | CVA16 | ND | 1.05 | 12.4 | 2.00 |
| 20 | 1.4E+01 | Rhino | ND | 1.04 | 12.9 | 1.97 |
| 21 | 2.2E+05 | CVA4 | ND | 1.04 | 11.0 | 2.21 |
| 22 | 1.3E+05 | CVA16 | ND | 1.04 | 10.6 | 2.27 |
| 23 | 2.1E+05 | EV71 | 13.4 | 1.50 | 16.1 | 1.63 |
| 24 | 7.9E+05 | CVB4, CVA4 | ND | 1.05 | 11.0 | 2.20 |
| 25 | 1.9E+06 | CVA4 | ND | 1.12 | 10.6 | 2.32 |
| 26 | 4.6E+06 | CVA4 | ND | 1.06 | 15.4 | 1.61 |
| 27 | 5.2E+07 | CVA4 | ND | 1.07 | NDc | 1.10 |
| 28 | 3.9E+05 | EV71 | 13.5 | 1.51 | 18.3 | 1.40 |
| 29 | 1.6E+06 | CVB4 | ND | 1.06 | 12.4 | 1.97 |
| 30 | 6.6E+06 | CVB4 | ND | 1.05 | 12.4 | 2.08 |
| 31 | 9.1E+05 | CVA6 | ND | 1.06 | 11.0 | 2.27 |
| 32 | 1.2E+05 | CVB4 | ND | 1.04 | 10.5 | 2.24 |
| 33 | 5.9E+03 | CVA16 | ND | 1.05 | 23.0 | 1.23 |
| 34 | 1.9E+06 | CVA6 | ND | 1.07 | 15.5 | 1.75 |
| 35 | 3.8E+05 | CVB4 | ND | 1.04 | 12.1 | 2.23 |
| 36 | 2.3E+06 | CVA2 | ND | 1.05 | 14.8 | 1.75 |
| 37 | 9.9E + 04 | CVA10 | ND | 1.05 | 13.6 | 1.96 |
| 38 | 1.5E+06 | CVA6 | ND | 1.04 | 11.7 | 2.15 |
| 39 | 8.0E+06 | CVA10 | ND | 1.06 | 16.5 | 1.72 |
| 40 | 4.4E+04 | Echo11 | ND | 1.07 | 18.4 | 1.49 |
| 41 | 1.3E+06 | CVA10 | ND | 1.23 | 11.9 | 2.15 |
| 42 | 9.3E+06 | CVA10 | ND | 1.05 | 12.8 | 2.02 |
| 43 | 4.3E+04 | CVB3 | ND | 1.05 | 10.3 | 2.27 |
| 44 | 5.5E+03 | CVB3 | ND | 1.05 | 9.7 | 2.37 |
| 45 | 2.3E+04 | CVB3 | ND | 1.05 | 10.4 | 2.24 |
| 46 | 4.5E+06 | EV71 | 10.4 | 1.54 | 19.5 | 1.43 |
| 47 | 8.4E+05 | EV71 | 10.1 | 1.57 | 15.1 | 1.72 |
| 48 | 2.1E+04 | EV71 | 16.0 | 1.45 | 15.5 | 1.64 |
| 49 | 4.9E+05 | EV71 | 10.3 | 1.50 | 14.7 | 1.83 |
| 50 | 1.3E+05 | CVA16 | ND | 1.05 | 11.4 | 2.18 |
| 51 | 5.9E+05 | EV71 | 10.5 | 1.53 | 12.6 | 1.98 |
| 52 | 9.5E+06 | CVA4 | ND | 1.07 | 13.5 | 1.91 |
| 53 | 5.2E+04 | EV71 | 12.0 | 1.54 | 10.6 | 2.27 |
| 54 | 3.5E+05 | CVA3 | ND | 1.04 | 10.0 | 2.30 |
| 55 | 6.9E+05 | EV71 | 11.6 | 1.55 | 15.6 | 1.54 |
| 56 | 8.7E+05 | CVA2 | ND | 1.04 | 14.6 | 1.84 |
| 57 | 1.2E+04 | EV71 | 16.6 | 1.41 | 12.2 | 2.22 |
| 58 | 3.7E+05 | CVA16 | ND | 1.04 | 17.4 | 1.59 |
The reaction time that the fluorescence intensity ratio took to reach the threshold value (1.3) is shown.
ND, not detected.
For sample 27, a retest was performed with RNA sample diluted by 10-fold. Signal of the internal control (1.60), but not that of EV71 (1.05), was detected in the retest.
In summary, we developed a TRCR method for the specific detection of EV71 from clinical specimens. This TRCR method would be useful for simple, rapid, and specific diagnosis of EV71 infection and for rapid diagnosis of patients with severe EV71 infection, who need proper emergency care and need to be distinguished from among a large number of patients with HFMD caused by other EV infections.
ACKNOWLEDGMENTS
We are grateful to Junko Wada for excellent technical assistance. We also thank T. Itagaki, Yamanobe Pediatric Clinic, and F. Katsushima and Y. Katsushima, Katsushima Pediatric Clinic, for collecting the clinical specimens.
This study was supported by Research on Emerging and Re-emerging Infectious Diseases from the Ministry of Health, Labor and Welfare.
Footnotes
Published ahead of print 22 February 2012
REFERENCES
- 1. Brown BA, Pallansch MA. 1995. Complete nucleotide sequence of enterovirus 71 is distinct from poliovirus. Virus Res. 39:195–205 [DOI] [PubMed] [Google Scholar]
- 2. Chang LY, et al. 1999. Clinical features and risk factors of pulmonary oedema after enterovirus-71-related hand, foot, and mouth disease. Lancet 354:1682–1686 [DOI] [PubMed] [Google Scholar]
- 3. Chang LY, et al. 1999. Comparison of enterovirus 71 and coxsackie-virus A16 clinical illnesses during the Taiwan enterovirus epidemic, 1998. Pediatr. Infect. Dis. J. 18:1092–1096 [DOI] [PubMed] [Google Scholar]
- 4. Chumakov M, et al. 1979. Enterovirus 71 isolated from cases of epidemic poliomyelitis-like disease in Bulgaria. Arch. Virol. 60:329–340 [DOI] [PubMed] [Google Scholar]
- 5. Dierssen U, Rehren F, Henke-Gendo C, Harste G, Heim A. 2008. Rapid routine detection of enterovirus RNA in cerebrospinal fluid by a one-step real-time RT-PCR assay. J. Clin. Virol. 42:58–64 [DOI] [PubMed] [Google Scholar]
- 6. Ho M, et al. 1999. An epidemic of enterovirus 71 infection in Taiwan. N. Engl. J. Med. 341:929–935 [DOI] [PubMed] [Google Scholar]
- 7. Ishiguro T, et al. 2003. Intercalation activating fluorescence DNA probe and its application to homogeneous quantification of a target sequence by isothermal sequence amplification in a closed vessel. Anal. Biochem. 314:77–86 [DOI] [PubMed] [Google Scholar]
- 8. Ishiguro T, et al. 1996. Fluorescence detection of specific sequence of nucleic acids by oxazole yellow-linked oligonucleotides. Homogeneous quantitative monitoring of in vitro transcription. Nucleic Acids Res. 24:4992–4997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McMinn PC. 2002. An overview of the evolution of enterovirus 71 and its clinical and public health significance. FEMS Microbiol. Rev. 26:91–107 [DOI] [PubMed] [Google Scholar]
- 10. Pulli T, Koskimies P, Hyypia T. 1995. Molecular comparison of coxsackie A virus serotypes. Virology 212:30–38 [DOI] [PubMed] [Google Scholar]
- 11. Takakura S, et al. 2005. Rapid detection of Mycobacterium tuberculosis in respiratory samples by transcription-reverse transcription concerted reaction with an automated system. J. Clin. Microbiol. 43:5435–5439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tanaka H, Hirose H, Kato Y, Kida S, Miyajima E. 2010. Clinical evaluation of TRCRapid M.TB for detection of Mycobacterium tuberculosis complex in respiratory and nonrespiratory specimens. J. Clin. Microbiol. 48:1536–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Taya T, Saitoh J, Ishizuka T, Matsubayashi K, Ishiguro T. 1999. Homogeneous detection of a target nucleic acid sequence by combination of the intercalation activating fluorescence DNA probe and the isothermal sequence amplification. Nucleic Acids Symp. Ser. 42:51–52 [DOI] [PubMed] [Google Scholar]
- 14. Wang SM, et al. 2003. Pathogenesis of enterovirus 71 brainstem encephalitis in pediatric patients: roles of cytokines and cellular immune activation in patients with pulmonary edema. J. Infect. Dis. 188:564–570 [DOI] [PubMed] [Google Scholar]
- 15. Wang SM, Liu CC. 2009. Enterovirus 71: epidemiology, pathogenesis and management. Expert Rev. Anti Infect. Ther. 7:735–742 [DOI] [PubMed] [Google Scholar]