Abstract
This study describes the development and optimization of an immunomagnetic separation (IMS) method to isolate Mycobacterium bovis cells from lymph node tissues. Gamma-irradiated whole M. bovis AF2122/97 cells and ethanol-extracted surface antigens of such cells were used to produce M. bovis-specific polyclonal and monoclonal antibodies in rabbits and mice. They were also used to generate M. bovis-specific peptide ligands by phage display biopanning. The various antibodies and peptide ligands obtained were used to coat MyOne tosyl-activated Dynabeads (Life Technologies), singly or in combination, and evaluated for IMS. Initially, M. bovis capture from Middlebrook 7H9 broth suspensions (concentration range, 10 to 105 CFU/ml) was evaluated by IMS combined with an M. bovis-specific touchdown PCR. IMS-PCR results and, subsequently, IMS-culture results indicated that the beads with greatest immunocapture capability for M. bovis in broth were those coated simultaneously with a monoclonal antibody and a biotinylated 12-mer peptide. These dually coated beads exhibited minimal capture (mean of 0.36% recovery) of 12 other Mycobacterium spp. occasionally encountered in veterinary tuberculosis (TB) diagnostic laboratories. When the optimized IMS method was applied to various M. bovis-spiked lymph node matrices, it demonstrated excellent detection sensitivities (50% limits of detection of 3.16 and 57.7 CFU/ml of lymph node tissue homogenate for IMS-PCR and IMS-culture, respectively). The optimized IMS method therefore has the potential to improve isolation of M. bovis from lymph nodes and hence the diagnosis of bovine tuberculosis.
INTRODUCTION
Bovine tuberculosis (TB) caused by Mycobacterium bovis continues to be a significant animal health issue in some countries, including the United Kingdom and Ireland (21). Confirmation of infection with M. bovis by culture is the mainstay of disease diagnosis but can be problematic for various reasons. There may be small numbers of mycobacteria present in those tissues selected for culture, and the harsh decontamination procedures prior to culture methods can also significantly reduce M. bovis viability in vitro (3). The time taken to confirm the presence or absence of M. bovis in clinical samples by culture can be up to 8 weeks, and any delay in disease confirmation has consequences for disease control programs. Mycobacterial culture is also very expensive, placing a heavy financial burden on bovine tuberculosis control programs.
Other methods for the direct or indirect detection of M. bovis in bovines include the gamma interferon (IFN-γ) assay and PCR (21), but these diagnostic methods are also flawed to some degree. For example, detection of IFN-γ release from antigen-stimulated T cells has been identified as an auxiliary test for disease control programs by the European Union, but it does not have the specificity of the skin test in disclosing infected animals (22). PCR methods developed to demonstrate the presence of M. bovis DNA are not yet considered sensitive enough to replace the culture-based diagnostic methods in current use (5). The consequence of these facts is that expensive and time-consuming mycobacterial culture methods are still regarded as “gold standard” techniques for diagnosis of bovine tuberculosis (21), despite their shortcomings. More rapid, specific, and sensitive methods of isolating and detecting M. bovis cells are urgently needed, as these would greatly contribute to the success of bovine tuberculosis control programs.
Immunomagnetic separation (IMS) is a technique that has been used for the selective isolation of a range of bacterial genera from a variety of sample matrices (24). Previous studies describing IMS methods for detection of M. bovis (10, 27, 28) have employed polyclonal or monoclonal antibodies (PAbs or MAbs) to M. bovis, and little or no optimization of either magnetic beads or the antibody coating was undertaken. Sweeney et al. (27) reported capture efficiencies for M. bovis from spiked environmental samples (soil, badger feces, and urine) of 80.3 to 88.6%. The capture efficiency was assessed by quantitative PCR using secondary coated Dynabeads and a commercially available polyclonal antibody to M. bovis BCG (DakoCytomation, Glostrup, Denmark) in an indirect IMS approach (i.e., primary coated beads and antibody added separately to sample). The same researchers (28) subsequently altered their IMS method and used goat anti-mouse IgG Dynabeads secondarily coated with monoclonal antibody MBS43, which recognizes MPB83 (an M. bovis cell wall-associated protein), in a direct IMS approach (i.e., coated beads added to sample) for the same types of environmental samples. More recently, Garbaccio and Cataldi (10) reported the use of goat anti-rabbit IgG Dynabeads coated with anti-M. tuberculosis H37Rv rabbit antiserum for immunocapture of M. bovis from bovine tissue samples. In each of these previous studies, IMS was used in conjunction with either real-time PCR (27, 28) or touchdown PCR (10), commonly referred to as IMS-PCR, in order to obtain a rapid test result for the presence of M. bovis.
We recently optimized an IMS method for Mycobacterium avium subsp. paratuberculosis (9) and discovered that the use of two phage display-derived peptide ligands, aMp3 and aMptD (originally identified by Stratmann et al. [25, 26]), to coat MyOne tosyl-activated Dynabeads (Life Technologies) achieved maximal capture of M. avium subsp. paratuberculosis from broth and bovine milk and fecal samples. A range of bead types and antibody and peptide ligands were evaluated in the course of our study (9), and it was very evident from the results that different bead-ligand combinations achieved widely differing capture efficiencies. Hence, we hypothesized that the published IMS methods for M. bovis were not necessarily optimal IMS methods for this Mycobacterium species and that further improvements in capture capability could be achieved if a broader range of antibodies and alternative types of ligands were evaluated. Furthermore, we proposed the generation of novel peptide ligands for M. bovis by use of the phage display biopanning approach that Stratmann et al. (25) used to identify the M. avium subsp. paratuberculosis-specific peptide ligands. Phage display biopanning of M. bovis BCG (23) and M. tuberculosis (12) has been reported previously, but apparently not for M. bovis per se. Therefore, the objectives of the present study were to generate novel antibodies and phage display-derived peptide ligands for M. bovis surface antigens and to evaluate the performance of these, along with a monoclonal antibody specific for MPB83 (19), for IMS of M. bovis from veterinary diagnostic samples.
MATERIALS AND METHODS
Immunogen preparation.
Polyclonal and monoclonal antibodies were prepared using whole-cell suspensions or ethanol-extracted surface antigens. Mycobacterium bovis AF2122/97 (provided by M. Vordermeier, Veterinary Laboratory Agency, Weybridge, United Kingdom) was cultured in Middlebrook 7H9 broth containing 10% oleic acid-albumin-dextrose-catalase (both from Difco) (Middlebrook 7H9/OADC broth) to stationary phase, harvested by centrifugation, and washed in phosphate-buffered saline (PBS). Cells were then subjected to a 10-kGy dose of gamma radiation (Gammabeam 650 cobalt irradiator) to kill the bacteria with minimum damage to cell surface antigens and then were stored at −80°C. Irradiated whole-cell antigen (WCA) was used directly for antibody production and phage display biopanning. A second immunogen, ethanol-extracted antigen (EEA), was prepared by the method of Eda et al. (7). Briefly, the irradiated culture was centrifuged, pelleted, resuspended in a 1/10 volume of 80% ethanol, and vortexed for 30 s. Extracted M. bovis surface antigens were vacuum dried to remove ethanol, freeze-dried, and resuspended in water to a final protein concentration of 1 mg/ml.
Antibody production.
All animal procedures were carried out in accordance with the Animal Scientific Procedures Act (1986) and in accordance with the Agri-Food and Biosciences Institute (AFBI) Ethics Committee. Polyclonal antibodies were produced by inoculating rabbits with either WCA or EEA mixed with Montanide ISA 50V (Sepic, Paris, France) on six occasions. Initially, 500 μg of immunogen was used, with 400 μg used for subsequent immunizations given intramuscularly, at four distal sites. Ten days after each booster injection, blood samples were taken and tested by enzyme-linked immunosorbent assay (ELISA) for antibodies to irradiated M. bovis cells. Ten days after the final immunization, the rabbits were anesthetized, bled, and euthanized.
For monoclonal antibody production, BALB/c mice were immunized subcutaneously with 50 μl of either WCA or EEA mixed with Quil A (BrennTag, Superfoss, Denmark). Primary and secondary booster immunizations were injected at 3-week intervals, and the final booster injections were administered using Pam3Cys-Ser(Lys)4-OH (PCSL) adjuvant (EMC Microcollections, GmbH, Germany) by intraperitoneal injection. Serum antibody titers were monitored by ELISA 10 days after each booster injection. The most immunologically responsive mouse was given a final intraperitoneal injection of 100 μl of immunogen in PBS (pH 7.2) 4 days prior to B-cell fusion. A single-cell suspension was prepared from the spleen and fused with SP2 cells by use of polyethylene glycol according to a modification of the method of Kohler and Milstein (16). Monoclonal antibodies from cloned hybridoma cell lines were isotyped using an IsoStrip mouse monoclonal antibody isotyping kit (Roche Diagnostics, United Kingdom).
Antigen-coated ELISA.
Microtiter plates (Greiner, United Kingdom) were coated with WCA stock (optical density [OD], 0.46) (100 μl/well) diluted 1:20 in 0.1 M bicarbonate buffer (pH 9.4 to 9.7) and incubated at 4°C overnight. Plate wells were blocked with 200 μl of blocking solution (5 mg/ml bovine serum albumin [BSA] in 1 mM sodium acetate buffer [NaAc], pH 7.2) per well and incubated for 2 h at 37°C. The blocking buffer was discarded, and 100 μl hybridoma supernatant diluted 1:1 in NaAc buffer (pH 7.2) was added in duplicate to appropriate wells. An antibody-positive mouse serum was used as a positive control, and culture medium was used as the negative control. Plates were incubated in an incubator shaker for 1.5 h at 37°C, the solution was discarded, and then the plates were washed six times using wash buffer (9 g/liter NaCl, 0.0125% Tween 20). One hundred microliters of horseradish peroxidase-conjugated anti-mouse immunoglobulin (Dako, Cambridge, United Kingdom) diluted 1 in 2,000 in blocking buffer was added to each well and incubated at 37°C for 1 h. The plates were washed six times with wash buffer. One hundred microliters of 3,3′,5,5′-tetramethylbenzidine solution (TMB/E) was added to each well, and color development was stopped after 5 min by use of 25 μl of 2.5 M H2SO4. Optical density was measured at 450 nm using a spectrophotometer (Biotek). The optical density value was considered positive if the reading was >1.0.
Production of peptide ligands by phage display biopanning.
A commercially available phage display library (PhD-12; New England BioLabs Inc., Ipswich, MA) was used to biopan M. bovis WCA, EEA, and MPB83 (11, 30) per the manufacturer's instructions. Each antigen was used at 10 to 100 μg/ml to coat the surface of 60-mm petri dishes (Sarstedt) by incubation overnight at 4°C with shaking. Plates were blocked with 0.1 M sodium hydrogen carbonate, pH 8.6, containing 5 mg/ml BSA for 1 h at 4°C and washed 6 times with TBS-0.1% Tween 20 (50 mM Tris-hydrochloride, 150 mM sodium chloride, and 0.1% [vol/vol] Tween 20, pH 7.5). The PhD-12 phage library (2 × 1011 phage) was added to the petri dish, followed by gentle rocking for 1 h at room temperature to allow phage to bind to complementary antigen sequences. Plates were washed 10 times with TBS-0.1% Tween 20, and bound phage were released using 0.2 M glycine-HCl and 1 mg/ml BSA. Phage were amplified in Escherichia coli ER2738 and harvested from the culture supernatant by centrifugation and treatment with a 1/6 volume of 20% polyethylene glycol containing 2.5 M sodium chloride solution. These amplified phage were then used as the “new” phage library for two further rounds of biopanning with increased stringency of the wash steps, using TBS-0.5% Tween 20. After three rounds of biopanning, 10 phage plaques were randomly taken and amplified in E. coli ER2738 before DNA extraction and sequencing (DNA Sequencing and Services, University of Dundee, Dundee, Scotland). The amino acid sequence of the peptide was identified using the ExPASy translate tool (http://web.expasy.org/translate/; Swiss Institute of Bioinformatics). All peptide sequences were screened using the online SAROTUP tool (15) to identify and eliminate any previously recognized target-unrelated peptide sequences (29).
Preliminary evaluation of antibody and peptide ligand specificity.
Antibody cross-reactivity was assessed using a range of Mycobacterium spp. which had been grown to stationary phase in Middlebrook 7H9/OADC broth (described above). ELISA microtiter plates were coated with WCA, EEA, and MPB83 and blocked with blocking buffer. A suspension of each Mycobacterium sp. (50 μl) was added to the appropriate wells prior to the antibody (50 μl). Inhibition of antibody-antigen binding was indicated if there was a reduction in the OD value compared to that in the absence of mycobacteria.
A phage ELISA binding assay (as described in the PhD-12 kit instructions) was used to assess if the consensus sequence phage clones derived from biopanning were capable of binding WCA. When antigen binding was confirmed, the 12-mer peptides were chemically synthesized in native form (Queen's University Belfast) or with N-terminal biotinylation (Cambridge Peptides Limited, Birmingham, United Kingdom) before further assessment of M. bovis specificity, cross-reactivity with other Mycobacterium spp., and application to IMS.
Detection sensitivity of magnetic beads coated with antibodies and peptide ligands.
All candidate antibodies and peptides produced during this study (Table 1) were used to coat MyOne tosyl-activated Dynabeads (Life Technologies) according to the manufacturer's instructions, following success with this bead type for M. avium subsp. paratuberculosis capture in a previous study (9). Assessment of the capture of M. bovis AF2122/97 by beads coated with each ligand was initially evaluated by IMS followed by M. bovis-specific PCR. Irradiated cultures were thoroughly declumped by vortexing twice with five 3-mm glass beads for 2 min, and then 10-fold dilutions (10−1 to 10−5) were prepared in PBS and subjected to automated IMS. After IMS, magnetic beads were resuspended in 100 μl Tris-EDTA buffer (pH 8) and heated to 99°C for 25 min to lyse attached M. bovis cells and release DNA. After a brief centrifugation, 5 μl supernatant was used as the DNA template in a touchdown PCR for M. bovis (32) employing INS1 and INS2 primers targeting the IS6110 element.
Table 1.
Details of coated beads evaluated for immunomagnetic separation of M. bovisa
| Bead no. | Ligand(s) |
|---|---|
| 1 | Peptide EEA308 (C-terminally amidated)b |
| 2 | Peptide EEA308 (N-terminal free amine)b |
| 3 | Peptide EEA302b |
| 4 | MAb 1F11c |
| 5 | PAb R43 |
| 6 | MAb 19G9 |
| 7 | MAb 11G3 |
| 8 | MAb 15E10 |
| 9 | Biotinylated peptide EEA302d |
| 10 | Biotinylated peptide EEA308d |
| 11 | MAb 11G3 + biotinylated peptide EEA302d |
All ligands evaluated were identified or produced during the course of this study, with the exception of MAb 1F11.
Synthesized in-house at Queen's University Belfast.
MPB83-specific monoclonal antibody available within the Agri-Food and Biosciences Institute (AFBI).
Synthesized in biotinylated form by Cambridge Peptides Limited.
Subsequently, once the optimal bead-ligand combination had been identified, the sensitivity of IMS capture of six other M. bovis strains with different variable-number-tandem-repeat (VNTR) profiles and spoligotypes (detailed in Table 2) by the dually coated MyOne tosyl-activated Dynabeads was assessed by IMS-PCR.
Table 2.
VNTR profiles and spoligotypes of all Mycobacterium bovis strains employed during this study
| Strain | Origina | VNTR profile | Spoligotype |
|---|---|---|---|
| AF2122/97 | GB field isolate | 4-4-5-3-4-8-10-8 | 140 |
| T/10/596 | NI field isolate | 3-4-5-2-4-6-9-8 | 129 |
| T/10/743 | NI field isolate | 4-4-4-3-2-7-6-8 | 273 |
| T/10/980 | NI field isolate | -44-5-3-4-7-11-9 | 140 |
| T/10/999 | NI field isolate | 4-4-4-1-4-7-10-7 | 142 |
| T/10/1009 | NI field isolate | 4-4-5-3-3-7-11-7 | 263 |
| T/10/1039 | NI field isolate | 3-3-3-3-4-7-11-8 | 145 |
GB, Great Britain; NI, Northern Ireland. The VNTR profile and spoligotype of NI field isolates were determined by the AFBI following isolation from clinical samples, with the exception of AF2122/97, which was analyzed from broth culture (Robin Skuce, AFBI, personal communication).
Nonspecific binding of other Mycobacterium spp. to coated beads.
The degree of nonspecific binding of the various antibody- and peptide-coated MyOne tosyl-activated Dynabeads was assessed by IMS experiments involving the following Mycobacterium strains: M. bovis BCG NCTC 5692, M. avium subsp. avium NCTC 13034, M. avium subsp. paratuberculosis ATCC 19698, M. fortuitum NCTC 10394, M. intracellulare NCTC 10425, M. kansasii NCTC 10268, M. xenopi NCTC 10042, M. terrae NCTC 10856, M. scrofulaceum NCTC 10803, M. marinum NCTC 2275, M. smegmatis mc2 155, and M. gordonae NCTC 10267. Stationary-phase broth cultures of each species were diluted to 103 to 104 CFU per ml in Middlebrook 7H9/OADC broth. One-hundred-microliter samples of the dilutions were spread onto Middlebrook 7H10/OADC agar plates to determine the accurate count (CFU/ml) before IMS, and 1 ml of each dilution was then subjected to automated IMS using each of the combinations of coated beads. Following IMS, beads were resuspended in 1 ml Middlebrook 7H9/OADC broth, and 100 μl was spread onto Middlebrook 7H10/OADC agar plates and incubated for an appropriate time at an appropriate temperature, depending on the species. Experiments were carried out in duplicate; colonies present after IMS were counted, and the mean was expressed as a percentage of the number of CFU present in the original suspension to indicate the degree of nonspecific binding by Mycobacterium spp. other than M. bovis.
Impact of tissue matrix on detection sensitivity of optimal IMS method.
A variety of normal bovine lymph node matrices (bronchial, retropharyngeal, and mesenteric lymph nodes) were collected from non-TB bovines and frozen at −20°C until required (<3 months), at which time they were ground with sand and PBS in a mortar and pestle and centrifuged briefly (300 × g for 3 min). The supernatant was then diluted 10-fold in PBS and divided into 900-μl aliquots. These were spiked with 100 μl irradiated M. bovis AF2122/97 culture at a range of cell concentrations (10 to 105 CFU/ml). IMS was performed using the dually coated Dynabeads with each spiked matrix, and recovery of M. bovis was compared by either touchdown PCR or culture.
The limits of detection of the IMS-PCR and IMS-culture methods were determined by spiking lymph node matrix with five replicate samples at four cell concentrations (102 to 103, 10 to 102, 1 to 10, and 0 CFU/ml), in duplicate. The 50% limit of detection (LOD50) and associated 95% confidence limits of IMS followed by touchdown PCR and culture were estimated using the generalized Spearman-Kärber LOD50 calculation for 4-level spiking protocols (1).
RESULTS
PAb production.
The EEA immunogen failed to generate an M. bovis-specific immune response in rabbits. Only rabbits immunized with WCA yielded polyclonal antibody capable of detecting the original M. bovis AF2122/97 cell suspension (WCA) by ELISA. Antibody R43 was both highly sensitive and specific, with a titer of 1:10,000 by ELISA and no cross-reactivity with the other Mycobacterium spp. tested (M. avium subsp. avium, M. fortuitum, M. intracellulare, M. kansasii, M. smegmatis, M. bovis BCG, and M. avium subsp. paratuberculosis).
MAb production.
Only mice immunized with WCA produced an M. bovis-specific immune response. A total of 774 hybridoma supernatants were assayed by ELISA for production of monoclonal antibody to M. bovis AF2122/97, and 71 of these were positive. The sensitivity of each MAb was determined by the degree of inhibition of binding to AF2122/97 cells, and specificity was characterized by determining cross-reactivity to M. intracellulare, M. avium subsp. paratuberculosis, and M. avium subsp. avium. Three cell lines (11G3, 19G9, and 15E10), found to be highly sensitive and specific for M. bovis, were cloned another two times, grown in bulk culture, and concentrated using Vivaspin columns. They were isotyped as IgM MAbs and purified by ammonium sulfate precipitation before use in IMS evaluations.
Phage display biopanning to generate peptide ligands.
Successful phage display biopanning should yield consensus amino acid sequences after at least three rounds. The outcomes of phage display biopanning of WCA, EEA, and MPB83 are summarized in Table 3. Biopanning with WCA did not yield any phage clones capable of binding M. bovis. After three rounds of biopanning with EEA, phage clones expressing three different peptide sequences were obtained, two of which were capable of binding whole M. bovis AF2122/97 cells. The corresponding peptides (designated peptide EEA302 and peptide EEA308) were chemically synthesized in native and N-terminally biotinylated forms and assessed for application to IMS. Biopanning of MPB83 yielded three phage clones capable of binding MPB83 but not whole M. bovis cells.
Table 3.
Outcomes of phage display biopanning of whole M. bovis cells (WCA), ethanol-extracted M. bovis cell surface antigens (EEA), and M. bovis surface antigen MPB83a
| Antigen | Clone identifier(s) | Amino acid sequence | Frequencyb | Ability to bind WCA |
|---|---|---|---|---|
| WCA | 302, 305, 309 | FHWTWQFPYTST | 3/10 | No |
| 303, 304, 307 | FHWNYYLYSQVS | 3/10 | No | |
| 301, 306 | WHWQWWLQTDAT | 2/10 | No | |
| 308 | FHWWPPSLANQP | 1/10 | No | |
| 310 | WHWNAWNWSSQQ | 1/10 | No | |
| EEA | 301, 303, 304, 306, 307, 308, 309 | SEFPRSWDMETN | 7/10 | Yes |
| 305 | FEFPRSWDMETN | 1/10 | No | |
| 302 | NFRVSIDVVKSR | 1/10 | Yes | |
| 310 | Unsuccessful sequencing | 1/10 | ||
| MPB83 | 304, 307 | GNLLHNHETYRH | 2/10 | Noc |
| 305 | HSLRWDWTARNS | 1/10 | Noc | |
| 306 | HSLRDDIRTMTA | 1/10 | Noc | |
| 309 | HKWGGNTLMAFR | 1/10 | Not tested | |
| 310 | HKPPTHIYLSWR | 1/10 | Not tested | |
| 308 | KVWPNMFANENI | 1/10 | Not tested | |
| 302 | KLWSIPKDLGPP | 1/10 | Not tested | |
| 303 | GYVHRELAWNMN | 1/10 | Not tested | |
| 301 | NMELHPHSLPRP | 1/10 | Not tested |
Two EEA-derived peptides (highlighted in bold) capable of binding WCA were chemically synthesized and subsequently evaluated for IMS purposes.
Number of clones with sequence/total number of clones tested after 3rd round of biopanning.
Did not bind WCA but did bind MPB83.
Evaluation of different ligands for M. bovis IMS.
A range of antibody- and peptide ligand-coated beads (Table 1) were evaluated for M. bovis capture potential. Preliminary testing was carried out using irradiated M. bovis AF2122/97 Middlebrook broth suspensions containing 104 CFU/ml and subjected to IMS followed by PCR. The PCR product band intensity obtained after IMS with each type of coated bead or bead mixture was compared to the PCR product band intensity from the original broth suspension before IMS (Fig. 1, lanes 1 and 11). Differing degrees of M. bovis recovery from each preparation were observed, as evidenced by different band intensities (Fig. 1). IMS using beads 1, 3, 6, 7, and 8 showed the highest level of M. bovis capture (Table 1). The most intense bands were obtained with peptide EEA302 (bead 3), MAb 15E10 (bead 8), and an equal mixture of beads containing MAb 19G9 (bead 6) and MAb 15E10 (bead 8). Further IMS trials were undertaken with these beads to determine their capability to capture M. bovis when smaller numbers of cells were present (101 to 105 CFU/ml). In addition, different combinations of beads were evaluated, either coated individually with antibodies or peptide ligand and used in mixtures of equal proportions or coated simultaneously with MAb 11G3 and biotinylated peptide EEA302.
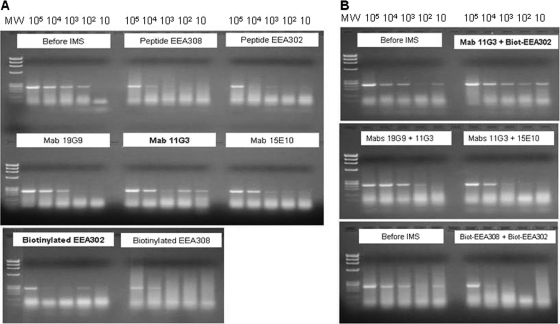
Fig 1.
Initial IMS-PCR evaluation of the potential of differently coated MyOne tosyl-activated Dynabeads to capture M. bovis AF2122/97 from broth suspensions containing approximately 104 CFU/ml. When used in 50:50 mixes, 5 μl of each type of coated bead was added separately to the sample. The expected IS6110 touchdown PCR product size was 245 bp. Lanes 1 and 11, M. bovis suspension before IMS; lanes 2 to 9, beads 1 to 8; lane 12, mix of beads 1 and 2; lane 13, mix of beads 6 and 8; lane 14, mix of beads 1 and 6; lane 15, mix of beads 1 and 8; lane 16, mix of beads 2 and 6; lane 17, mix of beads 2 and 8; lane 18, uncoated beads; lanes 10 and 19, negative control (water only). The DNA ladder shows X174 RF DNA/HaeIII fragments (Life Technologies). Bead details are given in Table 1.
Differences in M. bovis capture capability between bead types were evident when samples containing a range of M. bovis concentrations were subjected to IMS-PCR. Beads coated with peptide ligands EEA302, EEA308, and biotinylated EEA308 produced a PCR band only when the largest numbers of M. bovis cells were present (Fig. 2A), indicating relatively poor M. bovis capture capability. Beads coated singly with MAbs 19G9 and 15E10 (Fig. 2A) or with MAb mixtures (MAb 19G9 plus MAb 11G3 and MAb 11G3 plus MAb 15E10) (Fig. 2B) produced PCR bands of decreasing intensity for three or four of the five cell concentrations tested, indicating better M. bovis capture capability. MAb 11G3-coated beads and beads coated with a mixture of MAb 11G3 and biotinylated peptide EEA302 yielded PCR bands of decreasing intensity across all five cell concentrations tested (Fig. 2A and B) and thus demonstrated the best M. bovis capture capability (Fig. 2A and B). Beads coated separately with MAb 11G3 and biotinylated peptide EEA302 showed inconsistent PCR bands across the range of cell concentrations tested but did show capture of M. bovis at the lowest M. bovis concentrations tested (102 and 10 CFU/ml) (Fig. 2A). Inconsistent bands could be a result of nonspecific binding to the beads, as uncoated beads produced a faint band when tested by PCR after IMS. However, since the band representing 102 CFU/ml for biotinylated peptide EEA302 was more intense, it may have been the result of a clump of M. bovis cells being captured during IMS, as M. bovis tends to form clumps when cultured. Subsequently, beads coated simultaneously with MAb 11G3 and biotinylated peptide EEA302 were prepared and tested by IMS-PCR and IMS followed by culture on Middlebrook 7H10 agar. These dually coated beads proved to have even greater M. bovis capture capability than a mixture of beads coated individually with the same antibody and peptide ligand. Therefore, the dually coated beads were considered optimal for IMS analysis of M. bovis.
Fig 2.
Assessment of detection sensitivity of the “most promising” coated beads, used singly (A) or as 50:50 mixes (B), for capture of M. bovis AF2122/97 from spiked broth suspensions containing 10 to 105 CFU/ml by IMS-PCR. The expected IS6110 touchdown PCR product size was 245 bp. The DNA ladder shows X174 RF DNA/HaeIII fragments (Life Technologies).
Broth suspensions of six M. bovis field strains commonly encountered in Northern Ireland were tested by IMS-PCR. The results indicated that the MAb- and peptide-coated beads which had the best capture capability for M. bovis AF2122/97 also captured all six additional M. bovis strains across the full concentration range tested (10 to 105 CFU/ml) (Fig. 3). These results confirmed IMS capture capability for a range of M. bovis spoligotypes, not just the M. bovis strain originally used to generate antibodies and peptide ligands.
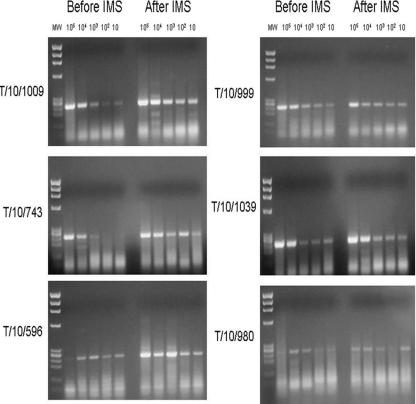
Fig 3.
Ability of dually coated MyOne tosyl-activated Dynabeads to capture a range of M. bovis strains (other than strain AF2122/97, which was used to generate capture ligands) from spiked broth suspensions containing 10 to 105 CFU/ml, as assessed by IMS-PCR. The expected IS6110 touchdown PCR product size was 245 bp. The DNA ladder shows X174 RF DNA/HaeIII fragments (Life Technologies). Spoligotype and VNTR information for the M. bovis strains is provided in Table 2.
The results from the IMS-PCR experiments on irradiated M. bovis cells were later verified by IMS analysis of viable M. bovis AF2122/97 cells by culture on Middlebrook 7H11 agar. IMS-culture results also confirmed that maximal capture of M. bovis cells was achieved using MyOne tosyl-activated Dynabeads coated simultaneously with MAb 11G3 and biotinylated peptide EEA302. Detection sensitivities of irradiation-killed and viable M. bovis cells for the dually coated beads were comparable.
Evaluation of IMS specificity with non-M. bovis mycobacteria.
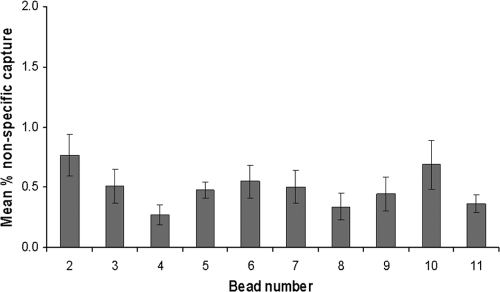
Broth suspensions of 12 Mycobacterium spp. other than M. bovis were subjected to IMS using all of the antibody- and peptide ligand-coated magnetic beads. In all cases, nonspecific binding after IMS, measured by culture, was determined to be less than 1% (Fig. 4). The bead-ligand combination which demonstrated maximal binding of M. bovis cells (tosyl-activated Dynabeads coated with both MAb 11G3 and biotinylated peptide EEA302) exhibited a mean % capture of non-M. bovis Mycobacterium spp. of 0.36% (range, 0.13 to 0.63%).
Fig 4.
Nonspecific capture of 12 Mycobacterium spp. other than M. bovis by differently coated MyOne tosyl-activated Dynabeads (bead details are given in Table 1). Data represent the mean % capture ± standard error of the mean for 12 different Mycobacterium spp. (listed in Materials and Methods) by each type of bead.
Effect of lymph node matrix on IMS detection sensitivity.
Spiking of various lymph node matrices with irradiated or live M. bovis AF2122/97 followed by IMS using the optimal dually coated beads and detection using either PCR or culture determined that there was no significant matrix effect on detection sensitivity. Subsequent LOD experiments using the Spearman-Karber calculation determined that the 50% limit of detection for M. bovis-spiked mesenteric lymph node matrix with IMS-culture was 57.7 CFU per ml of tissue homogenate (95% confidence interval [95% CI], 8.7 to 380.6 CFU/ml), whereas for IMS-PCR it was found to be 3.16 CFU/ml (95% CI, 0.57 to 17.63 CFU/ml), using spiked retropharyngeal lymph node matrix. The 10-fold greater detection sensitivity achieved by IMS-PCR than by IMS-culture was not unexpected, since all captured cells contribute DNA for PCR detection but both single cells and clumps of cells give rise to CFU values.
DISCUSSION
The objectives of this study were as follows: (i) to produce novel antibodies and peptide ligands for M. bovis surface antigens and (ii) to evaluate their efficacy for IMS analysis of M. bovis. The mycobacterial cell wall is complex, with many immunoreactive molecules thought to interact with host macrophages on first contact. These molecules comprise protein and nonprotein antigens, some of which are conformational in nature and whose three-dimensional structure is critical for immune recognition (23). Previously, M. bovis-derived immunogens used to produce antibodies have been whole cells prepared either by heat treatment and sonication or from culture filtrates (13, 18, 20, 31). Heat treatment is detrimental to native, surface epitopes and does not conserve heat-sensitive epitopes, which may partly explain the low diagnostic sensitivity and specificity of antibody tests developed using the antibodies generated during the studies cited above. Surface antigens of M. avium subsp. paratuberculosis are critical to high sensitivity and subspecies specificity for detection of this bacterium by flow cytometry (6). Similarly, antibodies or peptide ligands that bind M. bovis surface antigens are essential for IMS techniques. During the present study, M. bovis surface antigen characteristics were preserved by using gamma radiation prior to immunogen preparation, a technique successfully applied to polyclonal antibody preparation by Grant et al. (14). As a result, the immunogens, although nonviable, retained cellular morphology, could be stained and visualized by light or fluorescence microscopy, and were detectable by an appropriate PCR. It was therefore more likely that conformational cell surface epitopes were available for ligand production.
Carbohydrates are crucial molecules in pathogen-host interactions, and the outermost layer of M. tuberculosis is rich in saccharides and glycoconjugates, which are important in the pathogenesis of tuberculosis (2, 4, 8). In this study, surface antigens of M. bovis were extracted by ethanol to prepare EEA, an approach previously optimized to extract surface antigens from M. avium subsp. paratuberculosis cells (7). It was postulated that ethanol-extracted antigen increased the isolation of species-specific antigens from the mycobacterial surface, as opposed to antigens shared between mycobacterial species. Eda et al. (7) found a higher concentration of carbohydrate than protein in the ethanol extracts of M. avium subsp. paratuberculosis. Since M. bovis and M. avium subsp. paratuberculosis are related, with similar external compositions, antibodies or peptides obtained using EEA as an immunogen or biopanning target may bind to carbohydrate moieties on the M. bovis cell surface.
Results from this study indicated that M. bovis WCA was very immunogenic in rabbits and mice, producing high-titer polyclonal antibodies (diluted 1 in 10,000 in an ELISA) and highly specific and sensitive monoclonal antibodies for M. bovis. In contrast, EEA failed to stimulate antibody production in either rabbits or mice. Using the PhD-12 phage display random peptide library to produce peptide ligands, biopanning with WCA failed to produce any phage clones that bound whole M. bovis cells, while biopanning with MPB83 produced phage clones recognizing only the protein, not whole M. bovis cells. In contrast, biopanning of EEA generated two phage clones that bound whole M. bovis cells. These were subsequently chemically synthesized to provide peptide ligands for IMS evaluation. Since Eda et al. (7) showed that EEA from M. avium subsp. paratuberculosis cells was predominantly carbohydrate, it is possible that the EEA peptide ligands produced in this study are specific for nonprotein components, possibly lipopolysaccharide surface antigens of M. bovis.
In total, four monoclonal antibodies, one polyclonal antibody, and two peptide ligands (in both biotinylated and nonbiotinylated forms) were used to coat MyOne tosyl-activated Dynabeads, and the beads were tested for the ability to recover M. bovis from various matrices by IMS followed by culture and PCR. They were also tested for cross-reactivity to a range of other Mycobacterium spp., using culture only. The results indicated that a combination of MAb 11G3 and biotinylated peptide EEA302, used simultaneously to coat MyOne tosyl-activated Dynabeads, provided high specificity and the most recovery of M. bovis from samples. IMS using two or more binding reagents has been shown to achieve more (synergistic) bacterial capture than that with beads coated with a single reagent (9, 17). With M. bovis IMS, both a polyclonal antibody and a monoclonal antibody were used previously to capture this bacterium from sample matrices, including badger feces and urine, soil, and slurry (27, 28), and from spiked cattle tissue samples (10). However, we are not aware of any previous reports of phage display-derived peptide ligands being used in combination with a monoclonal antibody to achieve immunocapture of any bacterium, so this ligand combination represents a key finding of this study. The peptide, derived by phage display biopanning of an ethanol extract of M. bovis surface antigens, and the monoclonal antibody, derived from a whole M. bovis cell preparation, combined synergistically to enhance capture, enabling sensitive detection.
Characterization of the specificity of the ligands by ELISA and culture confirmed that the dually coated tosyl-activated Dynabeads did not significantly cross-react with any of the environmental Mycobacterium spp. tested. These nontuberculous Mycobacterium spp. are occasionally encountered during culture of clinical specimens, but the high specificity of the M. bovis-specific IMS developed during this study will minimize false-positive results when other mycobacterial contaminants are present in tissue samples. The sensitivity of the optimized IMS method used with PCR (IMS-PCR) on homogenized lymph node samples spiked with M. bovis was 3.16 CFU/ml, compared to 57.7 CFU/ml for IMS with culture (IMS-culture), indicating the potential to detect M. bovis in clinical samples. The difference in sensitivity of the two methods is likely due to the ease with which DNA can be detected by PCR, whereas enumeration of M. bovis by culture is not as accurate due to clumping or cording of cells.
In conclusion, we report here the production of several novel antibodies and phage display-derived peptide ligands specific to M. bovis and their evaluation for use in IMS. The optimal bead-ligand combination proved to be MyOne tosyl-activated Dynabeads coated simultaneously with MAb 11G3 and peptide EEA302. This optimized IMS method has considerable potential to improve the isolation of M. bovis from infected tissues and to reduce the time taken for accurate diagnosis. For example, it could potentially circumvent the need for chemical decontamination prior to MGIT culture. Chemical decontamination of clinical samples is currently employed prior to mycobacterial culture to inactivate nonmycobacterial contaminants, but it is known to have a significant deleterious effect on the viability of mycobacteria and may result in false-negative culture results (3). Using IMS, small numbers of M. bovis cells may be captured selectively and concentrated from tissue samples, and subsequent washing steps should effectively remove sample components or contaminating bacteria loosely adhering to the beads, leaving just specifically bound M. bovis cells. Alternatively, IMS used in combination with M. bovis-specific PCR methods would enable rapid and quantitative detection of M. bovis without the need for culture. These possibilities are the subjects of ongoing research employing the new IMS method to process lymph node samples from naturally infected cattle. Preliminary results indicate that both the IMS-culture and IMS-PCR approaches have considerable potential to improve the efficacy and speed of bovine TB diagnosis.
ACKNOWLEDGMENTS
This research was funded by the Department of Environment, Food and Rural Affairs, United Kingdom (project SE3262).
We thank Nuala Greer for her expertise in the production of polyclonal antibody.
Footnotes
Published ahead of print 8 February 2012
REFERENCES
- 1. AOAC International Presidential Task Force on Best Practices in Microbiological Methodology 2006. Final report and executive summaries. Appendix K. Proposed use of a 50% limit of detection value in defining uncertainty limits in the validation of presence-absence microbial detection methods. http://www.fda.gov/ucm/groups/fdagov-public/@fdagov-foods-gen/documents/document/ucm088702.pdf
- 2. Brennan PJ. 2003. Structure, function and biogenesis of the cell wall of Mycobacterium tuberculosis. Tuberculosis (Edinburgh) 83:91–97 [DOI] [PubMed] [Google Scholar]
- 3. Corner LA, Trajstman AC, Lund K. 1995. Determination of optimum concentration of decontaminants for primary isolation. N. Z. Vet. J. 43:129–133 [DOI] [PubMed] [Google Scholar]
- 4. Daffe M, Draper P. 1998. The envelope layers of mycobacteria with reference to their pathogenicity. Adv. Microb. Physiol. 39:131–203 [DOI] [PubMed] [Google Scholar]
- 5. Department of Environment Food and Rural Affairs 2010. Bovine TB and the use of PCR: summary of 12 July meeting. http://archive.defra.gov.uk/foodfarm/farmanimal/diseases/atoz/tb/documents/pcr-meeting100712.pdf Accessed 3 October 2011
- 6. Eda S, et al. 2005. New method of serological testing for Mycobacterium avium subsp. paratuberculosis (Johne's disease) by flow cytometry. Foodborne Pathog. Dis. 2:250–262 [DOI] [PubMed] [Google Scholar]
- 7. Eda S, et al. 2006. A highly sensitive and subspecies-specific surface antigen enzyme-linked immunosorbent assay for diagnosis of Johne's disease. Clin. Vaccine Immunol. 13:837–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ehlers MRW, Daffe M. 1998. Interactions between Mycobacterium tuberculosis and host cells: are mycobacterial sugars the key? Trends Microbiol. 6:328–335 [DOI] [PubMed] [Google Scholar]
- 9. Foddai A, Elliott CT, Grant IR. 2010. Maximising capture efficiency and specificity of magnetic separation for Mycobacterium avium subsp. paratuberculosis cells. Appl. Environ. Microbiol. 76:7550–7558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Garbaccio SG, Cataldi AA. 2010. Evaluation of an immunomagnetic capture method followed by PCR to detect Mycobacterium bovis in tissue samples from cattle. Rev. Argent. Microbiol. 42:247–253 [DOI] [PubMed] [Google Scholar]
- 11. Garnier T, et al. 2003. The complete genome sequence of Mycobacterium bovis. Proc. Natl. Acad. Sci. U. S. A. 100:7877–7882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gevorkian G, et al. 2005. Peptide mimotopes of Mycobacterium tuberculosis carbohydrate immunodeterminants. Biochem. J. 387:411–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Glatman-Freedman A, Martin JM, Riska PF, Bloom BR, Casadevall A. 1996. Monoclonal antibodies to surface antigens of Mycobacterium tuberculosis and their use in a modified enzyme-linked immunosorbent spot assay for detection of mycobacteria. J. Clin. Microbiol. 34:2795–2802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Grant IR, Ball HJ, Rowe MT. 1998. Isolation of Mycobacterium paratuberculosis from milk by immunomagnetic separation. Appl. Environ. Microbiol. 64:3153–3158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Huang J, Ru B, Li S, Lin H, Guo F-B. 2010. SAROTUP: scanner and reporter of target-unrelated peptides. J. Biomed. Biotechnol. 2010:101932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kohler G, Milstein C. 1975. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 256:495–497 [DOI] [PubMed] [Google Scholar]
- 17. Kretzer JW, et al. 2007. Use of high-affinity cell wall-binding domains of bacteriophage endolysins for immobilization and separation of bacterial cells. Appl. Environ. Microbiol. 73:1992–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lyashchenko KP, et al. 2001. Novel monoclonal antibodies against major antigens of Mycobacterium bovis. Scand. J. Immunol. 53:498–502 [DOI] [PubMed] [Google Scholar]
- 19. McNair J, Corbett DM, Girvin RM, Mackie DP, Pollock JM. 2001. Characterisation of the early antibody response in bovine tuberculosis: MPB83 is an early target with diagnostic potential. Scand. J. Immunol. 53:365–371 [DOI] [PubMed] [Google Scholar]
- 20. Morris JA, Thorns CJ, Woolley J. 1985. The identification of antigenic determinants on Mycobacterium bovis using monoclonal antibodies. J. Gen. Microbiol. 131:1825–1831 [DOI] [PubMed] [Google Scholar]
- 21. Office International des Epizooties 2011. Chapter 2.4.7. Bovine tuberculosis. Manual of diagnostic tests and vaccines for terrestrial animals 2009. http://www.oie.int/fileadmin/Home/eng/Health_standards/tahm/2.04.07_BOVINE_TB.pdf Accessed 3 October 2011
- 22. Schiller I, et al. 2010. Bovine tuberculosis: a review of current and emerging diagnostic techniques in view of their relevance for disease control and eradication. Transbound. Emerg. Dis. 57:205–220 [DOI] [PubMed] [Google Scholar]
- 23. Sharma A, Saha A, Bhattacharjee S, Majumdar S, Das Gupta SK. 2006. Specific and randomly derived immunoactive peptide mimotopes of mycobacterial antigens. Clin. Vaccine Immunol. 13:1143–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stevens KA, Jaykus LA. 2004. Bacterial separation and concentration from complex sample matrices: a review. Crit. Rev. Microbiol. 30:7–24 [DOI] [PubMed] [Google Scholar]
- 25. Stratmann J, Strommenger B, Stevenson K, Gerlach GF. 2002. Development of a peptide-mediated capture PCR for detection of Mycobacterium avium subsp. paratuberculosis in milk. J. Clin. Microbiol. 40:4244–4250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stratmann J, Dohmann K, Heinzmann J, Gerlach GF. 2006. Peptide aMptD-mediated capture PCR for detection of Mycobacterium avium subsp. paratuberculosis in bulk milk samples. Appl. Environ. Microbiol. 72:5150–5158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sweeney FP, et al. 2006. Immunomagnetic recovery of Mycobacterium bovis from naturally infected environmental samples. Lett. Appl. Microbiol. 2:460–462 [DOI] [PubMed] [Google Scholar]
- 28. Sweeney FP, et al. 2007. Environmental monitoring of Mycobacterium bovis in badger feces and badger sett soil by real-time PCR, as confirmed by immunofluorescence, immunocapture and cultivation. Appl. Environ. Microbiol. 73:7471–7473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vodnik M, Zager U, Strukelj B, Lunder M. 2011. Phage display: selecting straws instead of a needle from a haystack. Molecules 16:790–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wiker HG, et al. 1998. Immunochemical characterization of the MPB70/80 and MPB83 proteins of Mycobacterium bovis. Infect. Immun. 66:1445–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wood PR, et al. 1988. Production and characterisation of monoclonal antibodies specific for Mycobacterium bovis. J. Gen. Microbiol. 134:2599–2604 [DOI] [PubMed] [Google Scholar]
- 32. Zumárraga MJ, et al. 2005. Use of touch-down polymerase chain reaction to enhance the sensitivity of Mycobacterium bovis detection. J. Vet. Diagn. Invest. 17:232–238 [DOI] [PubMed] [Google Scholar]