Abstract
We studied the clinical and epidemiological characteristics of Klebsiella oxytoca-associated diarrhea in hospitalized patients in Hong Kong. Between 1 November 2009 and 30 April 2011, all inositol-fermenting colonies found on Simmons citrate agar supplemented with inositol, tryptophan, and bile salts (SCITB agar) used for the culturing of diarrheal stool samples were screened by a spot indole test for K. oxytoca. The overall sensitivity of SCITB agar plus the spot indole test (93.3%) for the detection of K. oxytoca in stool samples was superior to that of MacConkey agar (63.3%), while the specificities were 100% and 60.4%, respectively. The former achieved a 23-fold reduction in the workload and cost of subsequent standard biochemical identifications. Cytotoxin production and the clonality of K. oxytoca were determined by a cell culture cytotoxicity neutralization assay using HEp-2 cells and pulsed-field gel electrophoresis (PFGE), respectively. Of 5,581 stool samples from 3,537 patients, K. oxytoca was cultured from 117/5,581 (2.1%) stool samples from 104/3,537 (2.9%) patients. Seventy-six of 104 (73.1%) patients with K. oxytoca had no copathogens in their diarrheal stool samples. Twenty-four (31.6%) of 76 patients carried cytotoxin-producing strains, which were significantly associated with antibiotic therapy after hospital admission (50% versus 21.2%; P = 0.01). Health care-associated diarrhea was found in 44 (42%) of 104 patients with K. oxytoca, but there was no epidemiological linkage suggestive of a nosocomial outbreak, and PFGE showed a diverse pattern. None of the patients with cytotoxin-producing K. oxytoca developed antibiotic-associated hemorrhagic colitis, suggesting that K. oxytoca can cause a mild disease manifesting as uncomplicated antibiotic-associated diarrhea with winter seasonality.
INTRODUCTION
Health care-associated diarrhea has posed a great challenge to infection control professionals, especially if it is caused by Clostridium difficile, norovirus, or rotavirus. These agents can lead to major outbreaks in hospitals, with high clinical attack rates (14, 16, 38). Proactive infection control measures may play a role in preventing nosocomial outbreaks due to norovirus (11), but only a few studies have demonstrated the successful control of nosocomial outbreaks due to C. difficile by a bundle approach (6, 27). While the increasing use of broad-spectrum antimicrobials, especially fluoroquinolones, poses a major risk for C. difficile-associated diarrhea and hemorrhagic colitis (22, 29), the intrinsic characteristic of hypersporulation of certain ribotypes of C. difficile, such as ribotypes 027 and 002, leading to the prolonged environmental survival of spores, may further contribute to the persistence of nosocomial outbreaks (2, 12).
In addition to toxigenic C. difficile, there is increasing evidence to suggest that cytotoxin-producing Klebsiella oxytoca is an important agent in patients with antibiotic-associated hemorrhagic colitis (8, 19, 21, 30, 40). K. oxytoca has fulfilled Koch's postulates for antibiotic-associated hemorrhagic colitis, and this has been demonstrated in a rat model (20). However, a recent study reported that bloody diarrhea occurred in fewer than 50% of patients who had cytotoxin-producing K. oxytoca isolated from their diarrheal stool samples (35).
K. oxytoca has been considered a part of the normal gut flora, colonizing up to 9% of healthy subjects (5, 20), and broad-spectrum antimicrobials have been increasingly used in response to the emergence of multidrug-resistant organisms in our locality (10, 17, 18). We conducted a prospective study to understand the epidemiology of these bacteria using homemade Simmons citrate agar supplemented with inositol, tryptophan, and bile salts (SCITB agar) as a differential and selective growth medium to screen for K. oxytoca in diarrheal stool samples. We investigated the clinical and epidemiological characteristics of patients with K. oxytoca isolated from diarrheal stool samples between 1 November 2009 and 30 April 2011. The findings and their potential implications for disease transmission and infection control practice are discussed.
MATERIALS AND METHODS
Setting.
This study was conducted in an acute-care university-affiliated teaching hospital with 1,400 beds and in 3 extended-care hospitals with 1,600 beds, serving a population of approximately 0.53 million. Between 1 November 2009 and 30 April 2011, K. oxytoca was screened for by an “added test” for all diarrheal stool specimens from four facilities. The stool specimens were sent to the laboratory of the teaching hospital for microbiological investigations, such as bacterial culture for stool pathogens, cytotoxin assays for C. difficile, reverse transcription-PCR (RT-PCR) for norovirus, and antigen detection for rotavirus. Culture and cytotoxin assays for K. oxytoca were performed.
Bacterial culture for K. oxytoca.
Besides the conventional MacConkey agar, a homemade SCITB agar was used in parallel for the isolation of K. oxytoca from stool samples. SCITB agar was prepared from Simmons citrate agar (with bromothymol blue as a color indicator) supplemented with 0.5% bile salts to inhibit the growth of Gram-positive organisms, 2% citrate and 1% inositol as the carbon sources (37), and 1% tryptophan as the nitrogen source, which favored the growth and indole production of K. oxytoca (28). Stool specimens were freshly inoculated onto MacConkey and SCITB agars, which were incubated for 48 h. As most Klebsiella spp. ferment inositol, a color change to yellow was observed for the bacterial colonies on SCITB agar. These colonies were then picked for a spot indole test (1 g p-dimethylaminocinnamaldehyde dissolved in a solution containing 10 ml of concentrated hydrochloric acid and 90 ml of distilled water). K. oxytoca appeared as large inositol-fermenting colonies when grown on SCITB agar and were positive by the spot indole test, whereas Klebsiella pneumoniae appeared as inositol-fermenting colonies but were negative by the spot indole test. Other common intestinal flora, such as Escherichia coli, Enterobacter spp., Citrobacter spp., Proteus spp., and Pseudomonas spp., showed up as tiny colonies, which were identified by conventional biochemical methods (Fig. 1). Indole-positive colonies were further confirmed to be K. oxytoca by standard biochemical methods (1) and by use of the Vitek II automated identification system (bioMérieux, Inc., Durham, NC). Suspected mucoid and lactose-fermenting colonies on MacConkey agar were also subjected to standard biochemical methods, and strains suspected of being K. oxytoca were then confirmed by the Vitek II automated identification system. The diagnostic gold standard for K. oxytoca was defined as suspected colonies on SCITB agar that were positive by the spot indole test or suspected colonies on MacConkey agar, with these suspected colonies from either agar being further confirmed by conventional biochemical methods and the Vitek II automated identification system.
Fig 1.
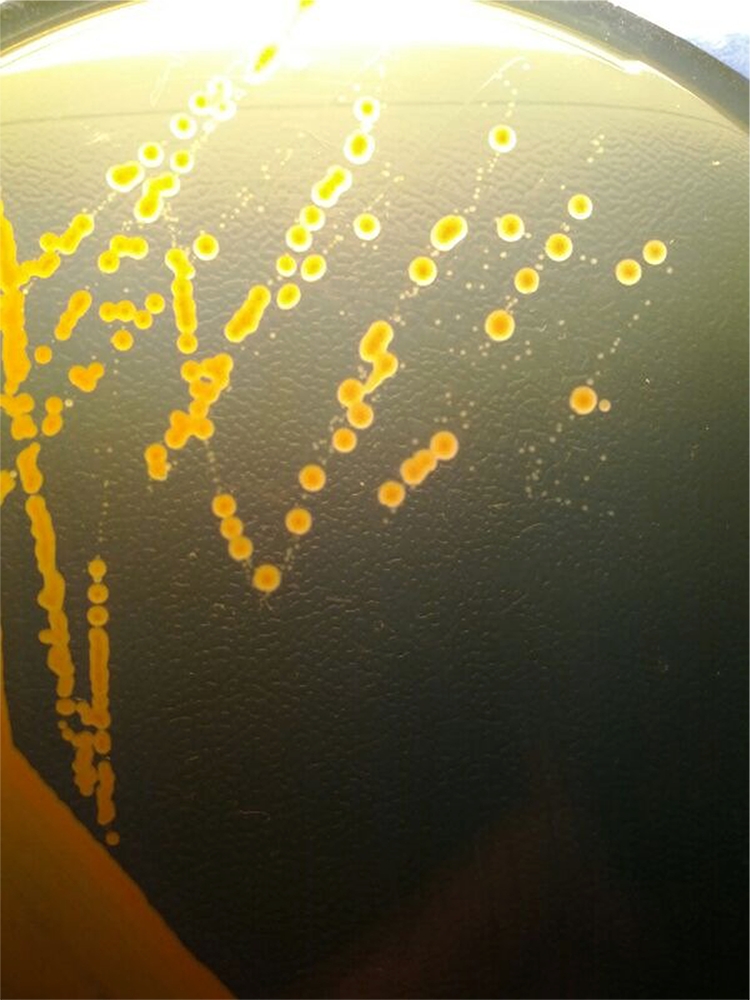
Fecal culture on a plate of Simmons citrate agar supplemented with inositol, tryptophan, and bile salts (SCITB agar) after 48 h of incubation at 37°C. Klebsiella spp. appear as large colonies, which ferment inositol with a color change to yellow, whereas other Enterobacteriaceae, such as Escherichia coli, appear as small colonies which do not ferment inositol in SCITB agar after 48 h of incubation at 37°C.
Cell culture cytotoxicity neutralization assay.
The HEp-2 cell line (ATCC CCL-23) was used for the cytotoxin-screening assay. Each well of a 96-well plate was seeded with 1 × 105 HEp-2 cells and incubated for 48 h in minimal essential medium (MEM) containing 10% fetal bovine serum (Gibco BRL), 100 U/ml penicillin (Gibco BRL), 100 μg/ml streptomycin (Gibco BRL), and 31.25 μg/ml gentamicin (Gibco BRL). A purified K. oxytoca colony was inoculated into 5 ml tryptone soy broth (TSB) and incubated for 20 h at 37°C with gentle agitation at 150 × g. After centrifugation at 20,000 × g for 10 min at 4°C, the supernatant was filtered through a membrane filter with a pore diameter of 0.2 μm (Millipore, MA). An equal volume of MEM was mixed with the filtrate (1:1 dilution with phosphate-buffered saline) and inoculated onto HEp-2 cells, followed by incubation in 5% carbon dioxide at 37°C for 72 h. A positive cytotoxic effect was recorded as cell rounding under light microscopy. The positive control, cytotoxin-producing K. oxytoca MH43-1, was a gift from Christoph Högenauer, Department of Internal Medicine, Medical University of Graz, Graz, Austria.
Molecular typing of cytotoxin-producing K. oxytoca by PFGE.
Cytotoxin-producing K. oxytoca isolates were grown at 37°C in brain heart infusion (BHI) broth with constant shaking for 2.5 h, and the turbidity was adjusted to an optical density at 610 nm (OD610) of 0.7. One milliliter of the cell suspension was centrifuged at 13,000 × g for 1 min, and the pellet was washed twice on ice with wash buffer (100 mM Tris, 100 mM EDTA [pH 8.0]). A 100-μl bacterial suspension was lysed with 10 μl of stock Lysozyme (10 mg/ml), 2 μl stock RNase (1 mg/ml), and 10 μl proteinase K (10 mg/ml) and incubated at 37°C for 20 min. Seven microliters of 20% sodium dodecyl sulfate and 140 μl of 1.2% InCert agarose were then mixed with the bacterial suspension. Agar plugs were allowed to solidify for 10 min at 4°C in plug molds. Plugs were then washed with ESP buffer (0.5 M EDTA [pH 9.0], 1% sodium lauryl sarcosine, 0.1 mg/ml proteinase K) at 55°C for 1 h twice. Further washing was performed with 8 to 10 ml of preheated (50°C) TE buffer (10 mM Tris [pH 8.0], 1 mM EDTA [pH 8.0]) in a 50°C shaking water bath for 15 min and repeated three times. Subsequently, a small portion of the plug (2 by 5 mm) was incubated with 50 U XbaI endonuclease (New England BioLabs, Beverly, MA) at 37°C with gentle shaking for 3 h. DNA fragments were resolved with 1% pulsed-field gel electrophoresis (PFGE) agarose in 250 ml 0.5× TBE buffer using a CHEF Mapper instrument (Bio-Rad). Electrophoresis was performed in 2 different sectors. In the first state, the initial switch time was 10 s, and the final switch time was 55 s for 12 h, with a linear ramp of 6 V/cm. In the second state, the initial switch time was 0.5 s, and the final switch time was 10 s for 12 h, with a linear ramp of 6 V/cm. Gel patterns were analyzed with the BioNumerics software package (version 6.0; Applied-Maths, Belgium). Cluster analysis was done by the unweighted-pair group method with average linkages (UPGMA) and the Dice coefficient, with 1% band position tolerance and 1% optimization settings. A cluster was defined as a similarity of 80% or more with the criteria of a difference of less than or equal to 6 bands, as described previously by Tenover et al. (36).
Epidemiology of K. oxytoca.
Patients with toxigenic K. oxytoca were clinically assessed by the infection control team. Case records were reviewed if the patients had been discharged. Patients with symptomatic diarrhea were classified as having a health care-associated infection if the symptoms started more than 48 h after admission to the hospital or within 4 weeks after discharge from the hospital or were classified as having community-associated diarrhea if the symptoms started within 48 h after admission to the hospital or more than 12 weeks after discharge from the hospital, as previously described for cases of C. difficile-associated diarrhea (15, 25). When symptoms developed 4 to 12 weeks after hospital discharge, the association was indeterminate. Infection control nurses would advise frontline health care workers on contact precautions for all diarrheal cases and conduct an outbreak investigation if there was epidemiological evidence of a nosocomial transmission of K. oxytoca, as previously described (11, 12). The demographic, clinical, and laboratory findings for patients with health care-associated and community-associated diarrhea with stool cultures that were positive for K. oxytoca were described. In addition, the risk factors for their acquisition of toxigenic K. oxytoca were also analyzed.
Statistical analysis.
A chi-square test, a t test, and the Mann-Whitney U test were used where appropriate. Screening performances between SCITB agar plus the spot indole test and MacConkey agar were compared by McNemar's test.
RESULTS
Between 1 November 2009 and 30 April 2011, a total of 5,581 diarrheal stool samples from 3,537 patients sent to our laboratory for bacterial and virological investigations were screened for the presence of K. oxytoca.
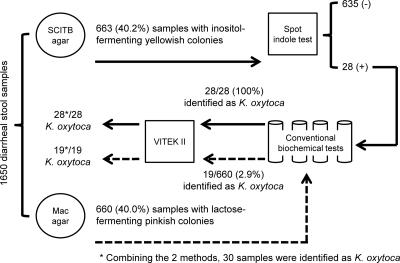
The first 1,650 diarrheal stool samples from 1,650 patients were inoculated onto both SCITB and MacConkey agars (Fig. 2). Characteristic large colonies which fermented inositol with a color change to yellow, suggestive of Klebsiella spp., were observed in SCITB agar for 663 samples from 663/1,650 (40%) patients. The spot indole test was performed for these 663 samples and was positive for 28 (4.2%) of them. All 28 strains were confirmed to be K. oxytoca by standard biochemical methods and the Vitek II automated identification system. For MacConkey agar, mucoid and lactose-fermenting colonies were found in 660 samples from 660/1,650 (40%) patients. Only 19 (2.9%) of 660 patients were found to have positive K. oxytoca samples by standard biochemical methods, confirmed by the Vitek II automated identification system. Combining the two methods, a total of 30 patients were confirmed to have K. oxytoca in the first 1,650 diarrheal stool samples. Two isolates identified on MacConkey agar were missed on SCITB agar. The overall performance of SCITB agar plus the spot indole test for the detection of K. oxytoca was superior to that of MacConkey agar by McNemar's test (P = 0.022). The sensitivities and specificities of the use of SCITB agar plus the spot indole test and MacConkey agar alone as a screening test were analyzed and are listed in Table 1. Furthermore, the number of patients requiring standard biochemical testing for the detection of K. oxytoca was significantly lower with SCITB agar plus the spot indole test (28/1,650; 1.7%) than with MacConkey agar (660/1,650; 40%) (P < 0.001). Because of these two major advantages, SCITB agar with the spot indole test was used as the single detection method after the first 1,650 samples.
Fig 2.
Workflow and diagnostic performance of SCITB agar plus the spot indole test and MacConkey (Mac) agar for the detection of Klebsiella oxytoca in the first 1,650 diarrheal stool samples.
Table 1.
Diagnostic performances of SCITB agar plus the spot indole test and MacConkey agar as a screening test for Klebsiella oxytoca in the first 1,650 diarrheal stool samplesa
| Test | No. of samples |
|
|---|---|---|
| Positive for K. oxytoca | Negative for K. oxytoca | |
| SCITB agar plus spot indole test, K. oxytoca | 28 | 0 |
| SCITB agar plus spot indole test, non-K. oxytoca | 2 | 1,620 |
| MacConkey agar, K. oxytoca | 19 | 641 |
| MacConkey agar, non-K. oxytoca | 11 | 979 |
The diagnostic gold standard for K. oxytoca was defined as suspected colonies on SCITB agar that were positive by the spot indole test or suspected colonies on MacConkey agar, with these suspected colonies from either agar being further confirmed by conventional biochemical methods and the Vitek II automated identification system. The sensitivity and specificity of SCITB agar plus the spot indole test were 93.3% (95% confidence interval [CI], 76.5 to 98.8%) (28/30) and 100% (95% CI, 99.7 to 100%) (1,620/1,620), respectively, and those of MacConkey agar were 63.3% (95% CI, 43.9 to 79.5%) (19/30) and 60.4% (95% CI, 58.0 to 62.8%) (979/1,620), respectively. The screening performance of SCITB agar plus the spot indole test was found to be significantly better than that of MacConkey agar by McNemar's test (P = 0.022). The number of patients requiring standard biochemical identification was 23 times [(19 + 641)/28] higher if MacConkey agar was used as a screening test than if SCITB agar with the spot indole test was used.
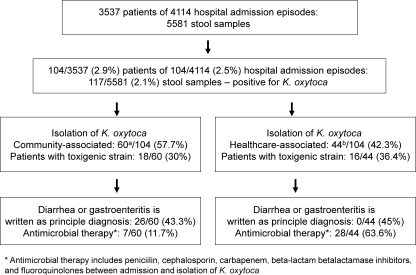
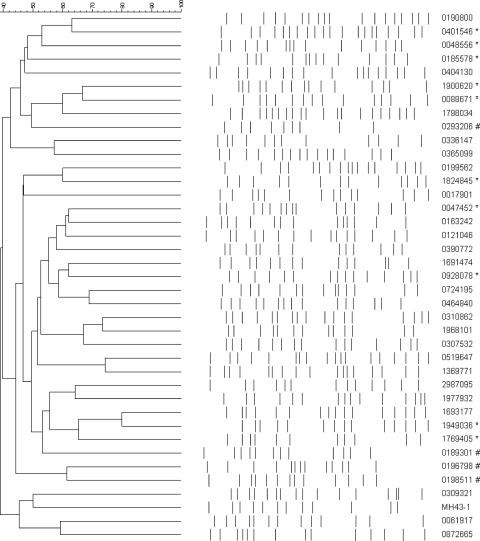
Among the 5,581 stool samples from 3,537 patients, a total of 117 (2.1%) samples from 104 (2.9%) patients were positive for K. oxytoca (Fig. 3). Cytotoxin-producing strains of K. oxytoca were detected in 34 (29.1%) samples from 34 (32.7%) patients by a cell culture cytotoxicity neutralization assay. PFGE patterns of 34 strains of cytotoxin-producing K. oxytoca (the first isolate from each patient) were highly diversified, with no unique cluster being identified during the 18-month study period (Fig. 4).
Fig 3.
Workup for Klebsiella oxytoca in a regional hospital in Hong Kong (1 November 2009 to 30 April 2011). a, 21/60 (35%) patients were coinfected with other pathogens, including 5/60 (8.3%) patients with other bacteria detected by culture (Salmonella spp. in 4 cases and Campylobacter jejuni subsp. jejuni in 1 case); 8/60 (13.3%) patients with norovirus detected by RT-PCR, among whom were 2 patients from residential care homes; 3/60 (5%) patients with rotavirus detected by a latex agglutination test; 3/60 (5%) patients with toxigenic strains of Clostridium difficile detected by a cell culture assay; and 2/60 (3.3%) patients with both norovirus and toxigenic strains of C. difficile. b, 7/44 (15.9%) patients were coinfected with other pathogens, including 4/44 (9.1%) patients with a toxigenic strain of C. difficile detected by a cell culture assay, 2/44 (4.5%) patients with rotavirus detected by a latex agglutination test, and 1/44 (2.3%) patients with Aeromonas caviae detected by culture.
Fig 4.
Pulsed-field gel electrophoresis patterns of 34 strains of cytotoxin-producing Klebsiella oxytoca (the first isolate from each patient). The first isolate from each patient is listed in the dendrogram. MH43-1 is a standard strain of K. oxytoca, provided by Christoph Högenauer, Department of Internal Medicine, Medical University of Graz, Graz, Austria. *, copathogens were found in the stool samples; #, non-cytotoxin-producing strains of K. oxytoca, shown as a reference.
Epidemiology of K. oxytoca.
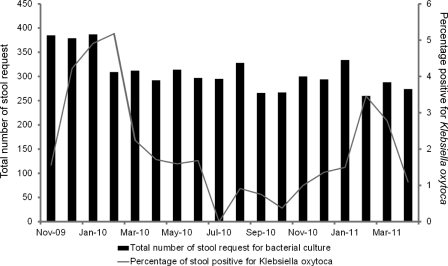
During the study period, 104 patients had K. oxytoca cultured in their diarrheal stool samples. There were 53 male and 51 female patients, with a median age of 63 years (ranging from 1 to 100 years). Twenty-eight (26.9%) of these patients were aged ≤10 years, while 38 (36.5%) were aged ≥60 years. Ninety-seven patients (93.3%) were of Chinese ethnicity. Most of the patients (36 patients; 34.6%) were managed in medical wards, whereas 24 (23.1%) were in pediatric and adolescent wards, 21 (20.2%) were in surgical wards, 14 (13.5%) were in the bone marrow transplant unit, 4 were in the observation ward of the accident and emergency department, 3 were in the obstetrics and gynecology wards, and 1 each was in the adult intensive care unit and in the orthopedic ward. These patients were managed in 43 wards of 2 different hospitals (one acute-care hospital and one chronic-care hospital), without any epidemiological link or clustering. Only 2 patients were referred from residential care homes for the elderly. The detection rate for K. oxytoca was over 2% in the wintertime in Hong Kong (Fig. 5). Community-associated diarrhea was found for 60 (57.7%) of 104 patients, whereas health care-associated diarrhea was noted for 44 (42.3%) patients (Table 2). Patients with community-associated diarrhea were significantly younger and had more copathogens in their stool samples. In contrast, patients with health care-associated diarrhea had significantly lower hemoglobin, lymphocyte, and albumin levels. Among 104 patients, 76 (73.1%) patients had K. oxytoca in their diarrheal stool samples without copathogens, whereas 9 (8.7%) patients were coinfected with norovirus, as determined by RT-PCR; 7 (6.7%) were coinfected with toxigenic strains of C. difficile; 6 (5.8%) were coinfected with other stool pathogens (Salmonella spp. in 4 patients, Campylobacter jejuni subsp. jejuni in 1, and Aeromonas caviae in 1); 4 (3.8%) were coinfected with rotavirus, as detected by a latex agglutination test; and 2 were coinfected with both norovirus and toxigenic C. difficile. No patient was coinfected with vancomycin-resistant enterococci (VRE). The 76 patients with K. oxytoca in diarrheal stool samples without copathogens were further analyzed, with 24 (31.6%) of them carrying cytotoxin-producing strains. Patients with cytotoxin-producing K. oxytoca were significantly associated with antibiotic therapy after hospital admission (Table 3). None of the patients with cytotoxin-producing K. oxytoca developed bloody diarrhea due to hemorrhagic colitis. The epidemiological characteristics of the 24 patients with toxigenic K. oxytoca without copathogens are summarized in Table 4.
Fig 5.
Seasonality pattern of detection of Klebsiella oxytoca in stool specimens.
Table 2.
Demographic characteristics of patients with health care-associated and community-associated diarrhea with stool cultures that were positive for Klebsiella oxytocaa
| Parameter | Value for group |
P value | |
|---|---|---|---|
| Community-associated diarrhea (n = 60) | Health care-associated diarrhea (n = 44) | ||
| Mean age (yr) ± SD | 38.0 ± 31.7 | 50.2 ± 24.4 | 0.04 |
| No. (%) of male patients | 31 (51.7) | 22 (50) | 0.87 |
| No. (%) of patients of Chinese ethnicity | 54 (90) | 43 (97.7) | 0.12 |
| No. (%) of patients residing in RCHE | 2 (3.3) | 0 | 0.22 |
| No. (%) of patients with cytotoxin-producing strain | 18 (30) | 16 (36.4) | 0.49 |
| No. (%) of patients with underlying disease | |||
| Cerebrovascular accident | 1 (1.7) | 2 (4.5) | 0.39 |
| Chronic cardiopulmonary conditionsb | 7 (11.7) | 9 (20.5) | 0.22 |
| Chronic renal failure | 4 (6.7) | 3 (6.8) | 0.98 |
| Diabetes mellitus | 5 (8.3) | 2 (4.5) | 0.45 |
| Malignancy | 10 (16.7) | 17 (38.6) | 0.39 |
| Hematological parametersc | |||
| Mean white cell count (109 cells/liter) ± SD | 11.3 ± 8.6 | 9.2 ± 6.6 | 0.19 |
| Mean neutrophil count (109 cells/liter) ± SD | 7.3 ± 6.0 | 7.0 ± 4.8 | 0.78 |
| Mean lymphocyte count (109 cells/liter) ± SD | 2.4 ± 2.7 | 1.2 ± 0.9 | <0.01 |
| Mean hemoglobin count (1012 cells/liter) ± SD | 12.3 ± 2.2 | 10.9 ± 2.0 | <0.01 |
| Mean platelet count (109 cells/liter) ± SD | 255 ± 127 | 224 ± 129 | 0.25 |
| Biochemical parametersc | |||
| Mean urea level (mmol/liter) ± SD | 6.9 ± 5.7 | 8.7 ± 8.7 | 0.22 |
| Mean creatinine level (μmol/liter) ± SD | 96 ± 142 | 144 ± 228 | 0.20 |
| Mean albumin level (g/liter) ± SD | 38 ± 9 | 32 ± 8 | <0.01 |
| Mean globulin level (g/liter) ± SD | 30 ± 7 | 34 ± 8 | 0.04 |
| Mean total bilirubin level (μmol/liter) ± SD | 9.4 ± 9.2 | 21.0 ± 64.6 | 0.19 |
| Mean alkaline phosphatase level (U/liter) ± SD | 162 ± 129 | 153 ± 203 | 0.78 |
| Mean alanine aminotransferase level (U/liter) ± SD | 38 ± 43 | 35 ± 42 | 0.78 |
| Mean aspartate aminotransferase level (U/liter) ± SD | 54 ± 88 | 37 ± 29 | 0.24 |
RCHE, residential care homes for the elderly.
Including coronary artery disease, congestive heart failure, and chronic obstructive airway disease.
At the time of K. oxytoca isolation.
Table 3.
Risk factors for patients with cytotoxin-producing Klebsiella oxytoca without copathogens
| Parameter | Value for group |
P value | |
|---|---|---|---|
| Cytotoxin-producing K. oxytoca (n = 24) | Non-cytotoxin-producing K. oxytoca (n = 52) | ||
| Mean age (yr) ± SD | 45.6 ± 30.9 | 44.2 ± 26.9 | 0.85 |
| No. (%) of male patients | 15 (62.5) | 26 (50) | 0.31 |
| No. (%) of patients of Chinese ethnicity | 21 (87.5) | 49 (94.2) | 0.31 |
| No. (%) of patients with health care-associated diarrhea | 13 (54.1) | 24 (46.2) | 0.52 |
| No. (%) of patients who received antibiotica 3 mo before isolation of K. oxytoca | 9 (37.5) | 11 (21.2) | 0.13 |
| No. (%) of patients who received antibiotica at admission before isolation of K. oxytoca | 12 (50) | 11 (21.2) | 0.01 |
| No. (%) of patients with underlying disease | |||
| Cerebrovascular accident | 0 | 2 (3.8) | 0.33 |
| Chronic cardiopulmonary conditionsb | 4 (16.7) | 9 (17.3) | 0.95 |
| Chronic renal failure | 0 | 6 (11.5) | 0.83 |
| Diabetes mellitus | 1 (4.2) | 5 (9.6) | 0.41 |
| Malignancy | 8 (33.3) | 12 (23.1) | 0.35 |
Including penicillin group antibiotics, β-lactam/β-lactamase inhibitors, cephalosporin group antibiotics, carbapenem group antibiotics, and fluoroquinolones.
Including coronary artery disease, congestive heart failure, and chronic obstructive airway disease.
Table 4.
Demographic characteristics of 24 patients with cytotoxin-producing Klebsiella oxytoca without copathogens between 1 November 2009 and 31 March 2011 in Hong Konga
| Case (strain) | Sex/age (yr) | Epidemiological statusb | Clinical specialty, ward, date of specimen collection | Underlying disease(s) | Time between admission and isolation of K. oxytoca (days) | Antibiotic(s) used (cumulative no. of days of antibiotics use from admission to isolation of K. oxytoca) |
|---|---|---|---|---|---|---|
| 1 (1691474) | M/1 | Community associated | Pediatric, A, 1 Nov 2009 | None | 1 | None |
| 2 (1693177) | M/19 | Health care associated | BMT, B, 1 Nov 2009 | Lymphoma | 5 | None |
| 3 (1798034) | F/49 | Health care associated | Surgery, C, 13 Nov 2009 | None | 4 | Ceph (4) |
| 4 (1968101) | F/59 | Health care associated | Medicine, D, 12 Dec 2009 | AML | 3 | BLBI (3) |
| 5 (2987095) | F/37 | Community associated | Obstetric, E, 29 Dec 2009 | None | 2 | None |
| 6 (0017901) | M/41 | Community associated | Medicine, F, 4 Jan 2010 | None | 0c | None |
| 7 (0061917) | F/80 | Health care associated | Surgery, G, 12 Jan 2010 | CHF, DM | 46 | BLBI (27), Carbap (15) |
| 8 (0121046) | M/70 | Health care associated | Medicine, H, 22 Jan 2010 | CHF | 8 | BLBI (2) |
| 9 (0163242) | M/69 | Community associated | Medicine, I, 29 Jan 2010 | AML | 2 | Ceph (2) |
| 10 (0190800) | F/67 | Community associated | Surgery, J, 3 Feb 2010 | None | 2 | BLBI (1), Carbap (2) |
| 11 (0199562) | M/5 | Health care associated | Pediatric, A, 4 Feb 2010 | VSD | 6 | None |
| 12 (0309321) | M/74 | Community associated | Medicine, I, 25 Feb 2010 | RA | 0 | None |
| 13 (0310862) | F/53 | Health care associated | Surgery, K, 25 Feb 2010 | None | 8 | Ceph (8) |
| 14 (0519647) | F/1 | Health care associated | Pediatric, A, 1 Apr 2010 | Anoxic brain damage | 4 | None |
| 15 (0724195) | F/53 | Community associated | BMT, B, 7 May 2010 | AML | 1 | None |
| 16 (0872665) | M/28 | Health care associated | Medicine, L, 2 Jun 2010 | Lymphosarcoma | 103d | Ceph (1), FQ (18) |
| 17 (1369771) | M/82 | Health care associated | Medicine, I, 25 Aug 2010 | CA of pancreas | 6 | Ceph (6) |
| 18 (1977932) | M/69 | Health care associated | Surgery, M, 10 Dec 2010 | IHD | 60 | BLBI (18), Ceph (4), FQ (8) |
| 19 (0307532) | M/87 | Community associated | Surgery N, 24 Feb 2011 | CA of rectum | 2 | None |
| 20 (0336147) | M/2 | Community associated | Pediatric, O, 2 Mar 2011 | Lymphoma | 1 | None |
| 21 (0365099) | M/1 | Community associated | Surgery, P, 7 Mar 2011 | None | 1 | None |
| 22 (0390772) | M/64 | Health care associated | Medicine, Q, 11 Mar 2011 | Diverticulosis | 4 | Ceph (2) |
| 23 (0404130) | M/83 | Health care associated | Surgery, R, 14 Mar 2011 | CA of stomach | 30 | BLBI (20) |
| 24 (0464840) | F/11 | Community associated | Pediatric, A, 24 Mar 2011 | None | 1 | None |
AML, acute myeloid leukemia; BLBI, β-lactam/β-lactamase inhibitors; BMT, bone marrow transplantation; CA, carcinoma; Carbap, carbapenem group; Ceph, cephalosporin group; CHF, congestive heart failure; DM, diabetes mellitus; F, female; FQ, fluoroquinolones; IHD, ischemic heart disease; M, male; Pen, penicillin group; RA, rheumatoid arthritis; VSD, ventricular septal defect.
None of the patients were referred from a residential care home for elderly.
Day 0 indicates that K. oxytoca was isolated upon the date of admission.
The patient had an intermittent home leave arrangement for 19 days (1 March, 5 March, 8 March, 10 March, 13 March, 17 March, 25 March, 31 March, 6 April, 8 April, 17 April, 27 April, 3 May, 7 May, 15 May, 17 May, 24 May, and 26 to 27 May 2011) during hospitalization.
DISCUSSION
We evaluated the use of SCITB agar as a differential and selective growth medium for studying the epidemiology of K. oxytoca in patients with diarrhea because there is presently no standard growth medium for the detection of the organism. SCITB agar contains the dual carbon sources of citrate and inositol, which can facilitate the detection of Klebsiella spp. In addition, citrate was utilized by 97 to 99% of Klebsiella sp. strains but not by E. coli, while inositol was fermented by 97 to 99% of Klebsiella sp. strains but by less than 1% of E. coli strains (37). When supplemented by the spot indole test, SCITB agar was shown to be a simple screening medium with good sensitivity (93.8%) and specificity (99.9%) in our study. Compared with the use of MacConkey agar as a screening test, SCITB agar plus the spot indole test reduced the workload associated with the performance of standard biochemical identification by 23-fold, which markedly saved time and the cost of microbial identification by the automicrobic microbiological identification system. A recent study used a modified MacConkey agar with adonitol replacing lactose as a selective and differential growth medium for the identification of K. oxytoca in stool samples (35). However, the adonitol-fermenting pink or red colonies had to be subcultured onto blood agar for further testing, including a motility-indole-ornithine test, a triple-sugar-iron test, and a phenylalanine deaminase test (35), which was labor-intensive. In contrast, the direct inoculation of stool samples onto MacConkey agar, followed by identification using an API 20E instrument (bioMérieux), has been used by a reference center (20, 40). Although this method reduces the likelihood of missing inositol-negative K. oxytoca, which may occur in up to 7% of cases (37), it requires the meticulous detection of all mucoid and lactose-fermenting colonies, and K. oxytoca may be overgrown by E. coli in MacConkey agar.
In our prospective analysis, K. oxytoca was found in about 3% of patients who presented with diarrhea during hospitalization, a rate which was lower than that reported in a recent study, in which the organism was detected in almost 10% of patients who had stool specimens submitted for a C. difficile cytotoxin assay (35). The difference in our study was that the “added test” included all stool samples from diarrheal patients submitted for bacterial culturing for stool pathogens, a C. difficile cytotoxin assay, norovirus RT-PCR, and rotavirus antigen detection, as previously described (11). The incidence of K. oxytoca in patients with diarrhea was therefore representative of our population. In particular, cytotoxin-producing strains were found in 33% of the patients, which was comparable with previously reported figures of 28% to 40% (35, 40).
Although the causative role of cytotoxin-producing K. oxytoca in patients with antibiotic-associated hemorrhagic colitis has been well established (20), the relationship between K. oxytoca and nonhemorrhagic antibiotic-associated diarrhea requires further investigation. In one study of 371 consecutive patients, 107 of whom received antibiotics and experienced diarrhea, all 89 patients with nonbloody diarrhea had a culture that was negative for K. oxytoca (40). In contrast, based on a different culture method, our study showed that 76 patients with nonbloody diarrhea had a culture that was positive for K. oxytoca without copathogens in their stool, and 23 (30%) of them received antibiotics at some time between admission and the isolation of K. oxytoca. Twelve (52%) of these 23 patients had cytotoxin-producing strains. The prior use of antibiotics, including penicillins, cephalosporins, carbapenems, and fluoroquinolones, during hospitalization was significantly associated with the detection of cytotoxin-producing K. oxytoca strains compared with non-cytotoxin-producing strains. Since none of the patients in our cohort had bloody diarrhea, it appeared that K. oxytoca may present as mild and uncomplicated diarrhea in our population. It is notable that non-cytotoxin-producing K. oxytoca was found in many diarrheal specimens. This is not surprising, as there have been many reports of nontoxigenic Vibrio cholerae (4, 7, 26, 31), E. coli (3, 33, 34), and C. difficile (23, 24) strains causing diarrhea. Other yet-unidentified virulence factors may contribute to its pathogenesis.
Health care-associated diarrhea was found for 44 (42%) of 104 patients with K. oxytoca, which should alert infection control professionals to the possibility of outbreaks. In fact, nosocomial outbreaks due to K. oxytoca were reported previously (32, 39), including a report of the detection of a multidrug-resistant strain (13), which warranted proactive infection control measures. However, unlike the situation with C. difficile and methicillin-resistant Staphylococcus aureus, where the major reservoir is patients from nursing homes, who act as silent carriers in nosocomial outbreaks (9, 12), none of our patients with health care-associated diarrhea due to K. oxytoca were admitted from nursing homes. In fact, there was no clustering or epidemiological link among the patients with K. oxytoca in our health care setting that would be suggestive of a nosocomial outbreak. This was supported by the multiclonality of our K. oxytoca isolates, as demonstrated by PFGE, as nosocomial outbreaks are usually caused by a single strain (32, 39). The lack of evidence for the nosocomial transmission of K. oxytoca might be related to the mild disease in our population. As shown in a previous study, patients with antibiotic-associated hemorrhagic colitis due to cytotoxin-producing K. oxytoca had a higher microbial density, up to 106 CFU per ml, in their diarrheal stool samples (40), while patients with mild disease might have a lower bacterial load. Therefore, single-room isolation with contact precautions should be practiced in cases of patients with antibiotic-associated hemorrhagic colitis due to cytotoxin-producing K. oxytoca, while standard precautions can be practiced in cases of patients with mild disease. It is interesting to note that advanced age and lower lymphocyte counts and hemoglobin and albumin levels were shown to be significant risk factors for health care-associated diarrhea due to K. oxytoca after adjusting for differences in comorbidities.
There are several limitations of this study. First, the study duration of 18 months and the relatively small sample size did not allow for a more detailed epidemiological analysis of a disease with such a low prevalence. However, it appears that K. oxytoca is more frequently found in diarrheal stool samples in the winter in Hong Kong, as our study period included two consecutive winters. Second, we did not measure the microbial load in the stool samples. Hence, the differential microbial density of K. oxytoca with respect to antibiotic therapy could not be ascertained in our study. However, the selective SCITB agar, which is more sensitive than conventional MacConkey agar for the detection of K. oxytoca, can facilitate further investigations of this new clinical entity.
ACKNOWLEDGMENTS
We thank Christoph Högenauer, Department of Internal Medicine, Medical University of Graz, Graz, Austria, for providing us standard strains of K. oxytoca (MH43-1) as positive controls. We thank Grace Kwan and Antonio Ngan for their assistance in taking photographs of the culture plate.
The work was supported by the Research Fund for the Control of Infectious Diseases of the Food and Health Bureau of the Hong Kong Special Administrative Region Government.
Footnotes
Published ahead of print 22 February 2012
REFERENCES
- 1. Abbott SL. 2007. Klebsiella, Enterobacter, Citrobacter, Serratia, Plesiomonas, and other Enterobacteriaceae, p 698–715 In Murray PR, Jr, et al. (ed), Manual of clinical microbiology, 9th ed ASM Press, Washington, DC [Google Scholar]
- 2. Akerlund T, et al. 2008. Increased sporulation rate of epidemic Clostridium difficile type 027/NAP1. J. Clin. Microbiol. 46:1530–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Allerberger F, et al. 2000. Nontoxigenic sorbitol-fermenting Escherichia coli O157:H− associated with a family outbreak of diarrhoea. Wien. Klin. Wochenschr. 112:846–850 [PubMed] [Google Scholar]
- 4. Batchelor RA, Wignall FS. 1988. Nontoxigenic 01 Vibrio cholerae in Peru: a report of two cases associated with diarrhea. Diagn. Microbiol. Infect. Dis. 10:135–138 [DOI] [PubMed] [Google Scholar]
- 5. Beaugerie L, et al. 2003. Klebsiella oxytoca as an agent of antibiotic-associated hemorrhagic colitis. Clin. Gastroenterol. Hepatol. 1:370–376 [DOI] [PubMed] [Google Scholar]
- 6. Birgand G, et al. 2010. Investigation of a large outbreak of Clostridium difficile PCR-ribotype 027 infections in northern France, 2006–2007 and associated clusters in 2008–2009. Euro Surveill. 15(25):19597. [DOI] [PubMed] [Google Scholar]
- 7. Chatterjee S, et al. 2009. Incidence, virulence factors, and clonality among clinical strains of non-O1, non-O139 Vibrio cholerae isolates from hospitalized diarrheal patients in Kolkata, India. J. Clin. Microbiol. 47:1087–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen J, Cachay ER, Hunt GC. 2004. Klebsiella oxytoca: a rare cause of severe infectious colitis: first North American case report. Gastrointest. Endosc. 60:142–145 [DOI] [PubMed] [Google Scholar]
- 9. Cheng VC, et al. 2011. Studying the transmission dynamics of meticillin-resistant Staphylococcus aureus in Hong Kong using spa typing. J. Hosp. Infect. 79:206–210 [DOI] [PubMed] [Google Scholar]
- 10. Cheng VC, et al. 2009. Antimicrobial stewardship program directed at broad-spectrum intravenous antibiotics prescription in a tertiary hospital. Eur. J. Clin. Microbiol. Infect. Dis. 28:1447–1456 [DOI] [PubMed] [Google Scholar]
- 11. Cheng VC, et al. 2011. Prevention of nosocomial transmission of norovirus by strategic infection control measures. Infect. Control Hosp. Epidemiol. 32:229–237 [DOI] [PubMed] [Google Scholar]
- 12. Cheng VC, et al. 2011. Clostridium difficile isolates with increased sporulation: emergence of PCR ribotype 002 in Hong Kong. Eur. J. Clin. Microbiol. Infect. Dis. 30:1371–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Decre D, Burghoffer B, Gautier V, Petit JC, Arlet G. 2004. Outbreak of multi-resistant Klebsiella oxytoca involving strains with extended-spectrum beta-lactamases and strains with extended-spectrum activity of the chromosomal beta-lactamase. J. Antimicrob. Chemother. 54:881–888 [DOI] [PubMed] [Google Scholar]
- 14. Georgiadou SP, Loukeris D, Smilakou S, Daikos GL, Sipsas NV. 2011. Effective control of an acute gastroenteritis outbreak due to norovirus infection in a hospital ward in Athens, Greece, April 2011. Euro Surveill. 16(28):19915. [PubMed] [Google Scholar]
- 15. Goorhuis A, et al. 2008. Emergence of Clostridium difficile infection due to a new hypervirulent strain, polymerase chain reaction ribotype 078. Clin. Infect. Dis. 47:1162–1170 [DOI] [PubMed] [Google Scholar]
- 16. Graf K, et al. 2009. An outbreak of Clostridium difficile-associated disease (CDAD) in a German university hospital. Eur. J. Clin. Microbiol. Infect. Dis. 28:543–545 [DOI] [PubMed] [Google Scholar]
- 17. Ho PL, Chow KH, Lai EL, Lau EH, Cheng VC. 2011. Extended-spectrum-beta-lactamase-positive Escherichia coli mainly adds to, rather than replaces, extended-spectrum-beta-lactamase-negative E. coli in causing bacteraemia in Hong Kong, 2000–10. J. Antimicrob. Chemother. 67:778–780 [DOI] [PubMed] [Google Scholar]
- 18. Ho PL, Ho AY, Chow KH, Cheng VC. 2010. Surveillance for multidrug-resistant Acinetobacter baumannii: a lesson on definitions. Int. J. Antimicrob. Agents 36:469–471 [DOI] [PubMed] [Google Scholar]
- 19. Hoffmann KM, et al. 2010. Antibiotic-associated hemorrhagic colitis caused by cytotoxin-producing Klebsiella oxytoca. Pediatrics 125:e960–e963 [DOI] [PubMed] [Google Scholar]
- 20. Hogenauer C, et al. 2006. Klebsiella oxytoca as a causative organism of antibiotic-associated hemorrhagic colitis. N. Engl. J. Med. 355:2418–2426 [DOI] [PubMed] [Google Scholar]
- 21. Joainig MM, et al. 2010. Cytotoxic effects of Klebsiella oxytoca strains isolated from patients with antibiotic-associated hemorrhagic colitis or other diseases caused by infections and from healthy subjects. J. Clin. Microbiol. 48:817–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Koga H, et al. 1999. Can quinolones cause hemorrhagic colitis of late onset? Report of three cases. Dis. Colon Rectum 42:1502–1504 [DOI] [PubMed] [Google Scholar]
- 23. Martirosian G, Szczesny A, Cohen SH, Silva J., Jr 2005. Analysis of Clostridium difficile-associated diarrhea among patients hospitalized in tertiary care academic hospital. Diagn. Microbiol. Infect. Dis. 52:153–155 [DOI] [PubMed] [Google Scholar]
- 24. Martirosian G, Szczesny A, Cohen SH, Silva J., Jr 2004. Isolation of non-toxigenic strains of Clostridium difficile from cases of diarrhea among patients hospitalized in hematology/oncology ward. Pol. J. Microbiol. 53:197–200 [PubMed] [Google Scholar]
- 25. McDonald LC, et al. 2007. Recommendations for surveillance of Clostridium difficile-associated disease. Infect. Control Hosp. Epidemiol. 28:140–145 [DOI] [PubMed] [Google Scholar]
- 26. Morris JG, Jr, et al. 1984. Isolation of nontoxigenic Vibrio cholerae O group 1 from a patient with severe gastrointestinal disease. J. Clin. Microbiol. 19:296–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Muto CA, et al. 2007. Control of an outbreak of infection with the hypervirulent Clostridium difficile BI strain in a university hospital using a comprehensive “bundle” approach. Clin. Infect. Dis. 45:1266–1273 [DOI] [PubMed] [Google Scholar]
- 28. Niemi RM, Mentu J, Siitonen A, Niemela SI. 2003. Confirmation of Escherichia coli and its distinction from Klebsiella species by gas and indole formation at 44 and 44.5 degrees C. J. Appl. Microbiol. 95:1242–1249 [DOI] [PubMed] [Google Scholar]
- 29. Pepin J, et al. 2005. Emergence of fluoroquinolones as the predominant risk factor for Clostridium difficile-associated diarrhea: a cohort study during an epidemic in Quebec. Clin. Infect. Dis. 41:1254–1260 [DOI] [PubMed] [Google Scholar]
- 30. Philbrick AM, Ernst ME. 2007. Amoxicillin-associated hemorrhagic colitis in the presence of Klebsiella oxytoca. Pharmacotherapy 27:1603–1607 [DOI] [PubMed] [Google Scholar]
- 31. Saha PK, et al. 1996. Nontoxigenic Vibrio cholerae 01 serotype Inaba biotype El Tor associated with a cluster of cases of cholera in southern India. J. Clin. Microbiol. 34:1114–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sardan YC, et al. 2004. A cluster of nosocomial Klebsiella oxytoca bloodstream infections in a university hospital. Infect. Control Hosp. Epidemiol. 25:878–882 [DOI] [PubMed] [Google Scholar]
- 33. Schlager TA, Wanke CA, Guerrant RL. 1990. Net fluid secretion and impaired villous function induced by colonization of the small intestine by nontoxigenic colonizing Escherichia coli. Infect. Immun. 58:1337–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schmidt H, Scheef J, Huppertz HI, Frosch M, Karch H. 1999. Escherichia coli O157:H7 and O157:H(−) strains that do not produce Shiga toxin: phenotypic and genetic characterization of isolates associated with diarrhea and hemolytic-uremic syndrome. J. Clin. Microbiol. 37:3491–3496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Smith SA, et al. 2009. A study of the prevalence of cytotoxic and non-cytotoxic Klebsiella oxytoca fecal colonization in two patient populations. Can. J. Infect. Dis. Med. Microbiol. 20:e169–e172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tenover FC, et al. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Van Kregten E, Westerdaal NA, Willers JM. 1984. New, simple medium for selective recovery of Klebsiella pneumoniae and Klebsiella oxytoca from human feces. J. Clin. Microbiol. 20:936–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Widdowson MA, et al. 2002. An outbreak of diarrhea in a neonatal medium care unit caused by a novel strain of rotavirus: investigation using both epidemiologic and microbiological methods. Infect. Control Hosp. Epidemiol. 23:665–670 [DOI] [PubMed] [Google Scholar]
- 39. Zarate MS, et al. 2008. Outbreak of OXY-2-producing Klebsiella oxytoca in a renal transplant unit. J. Clin. Microbiol. 46:2099–2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zollner-Schwetz I, et al. 2008. Role of Klebsiella oxytoca in antibiotic-associated diarrhea. Clin. Infect. Dis. 47:e74–e78 [DOI] [PubMed] [Google Scholar]