Abstract
A nasal carriage survey for methicillin-resistant Staphylococcus aureus (MRSA) in an intensive care unit detected four strains of MRSA with reduced susceptibility to vancomycin. The vanA gene was found in two of these vancomycin-intermediate Staphylococcus aureus (VISA) strains. The absence of selective vancomycin pressure might have resulted in reduced expression of the resistant gene.
TEXT
Gram-positive bacteria, particularly Staphylococcus aureus and Enterococcus spp., are extremely important pathogens, not only in the hospital setting but also in the community. Methicillin-resistant S. aureus (MRSA) is now endemic in health care facilities. Vancomycin-resistant enterococci (VRE) were first reported in 1988 (9), but these organisms quickly became endemic in hospital intensive care units (ICUs). In vitro conjugative transfer of the vanA gene from enterococci to S. aureus was demonstrated in 1992 (13). However, it was not until 1996, when the first case of vancomycin-intermediate S. aureus (VISA; MIC, 8 to 16 μg/ml) was detected, that decreased susceptibility to vancomycin became a clinical reality (3). The isolation of these glycopeptide-intermediate S. aureus (GISA) isolates, a broader term, has raised concern since, after vancomycin and teicoplanin, few therapeutic options exist for treatment of MRSA infections. In June 2002, the first clinical S. aureus isolate with high-level vancomycin resistance (VRSA; MIC, ≥32 μg/ml) was detected in a patient from Michigan who had extensive exposure to vancomycin (4). That organism contained the vanA gene, suggesting transfer of genetic material from a vanA-containing vancomycin-resistant Enterococcus faecalis strain.
None of the VISA strains identified so far contained the vanA gene or any of the other vancomycin-resistant genes found in VRE. We found four MRSA strains, colonizing the anterior nares of hospitalized patients, with reduced susceptibility to vancomycin, of which two were VISA strains harboring vanA. This is believed to be the first report of VISA strains containing vanA isolated from a routine nasal carriage survey in August 2010, from patients in the ICU of a tertiary-care hospital in north India.
As a part of the infection control program, culture specimens were taken weekly from the anterior nares of all the patients admitted to the ICU and examined for the presence of MRSA and VISA/VRSA. During the carriage survey, a total of 135 specimens over a period of 3 months were collected with dry, sterile swabs and inoculated onto mannitol salt agar. After a 48-hour incubation at 35°C, all presumptive S. aureus colonies were isolated on blood agar plates containing 5% sheep blood for further analysis. Identification of S. aureus was confirmed by Gram staining, catalase testing, and tube coagulase testing. MRSA screening was done with cefoxitin disks (30 μg), according to CLSI guidelines (5). Finally, to screen for vancomycin resistance, all MRSA isolates were inoculated onto brain heart infusion (BHI) agar containing 4 μg of vancomycin per milliliter and incubated at 35°C for 24 h.
Broth microdilution susceptibility testing was performed to determine the MICs of vancomycin and teicoplanin (Hi Media, India). Growth of the VISA isolates were also seen in commercial vancomycin screen agar and Hi Comb vancomycin MIC strips (Hi Media, India), as per the criteria of the Centers for Disease Control and Prevention (CDC) (10).
Genomic staphylococcal DNA, which was isolated by the phenol chloroform method, was used as the template for PCR for detection of the presence of vanA and vanB, based on a protocol given elsewhere (1), in the VISA isolates. Furthermore, a study of gene expression for the resistant gene by quantitative real-time reverse transcriptase PCR was done (11).
Analysis of colony size was done by plating the strains on both Mueller-Hinton agar and BHI and reading them after 24 and 48 h.
Morphological changes in these isolates were assessed by thin section electron microscopy (EM) (12).
Genomic DNA from the isolates was digested with SmaI endonuclease, and DNA fragments were separated by pulsed-field gel electrophoresis (PFGE) for molecular typing (12).
In the nasal carriage survey, four MRSA strains were found to grow on BHI agar plates containing vancomycin (4 μg/ml) after 24 h of incubation, as small colonies. No heteroresistant population was found. Vancomycin MICs for these strains ranged from 6 to 8 μg/ml. The teicoplanin MIC ranged from 4 to 32 μg/ml. The MICs for each strain are shown in Table 1. All the VISA isolates had a similar profile of resistance to multiple antimicrobial agents, including aminoglycosides and fluoroquinolones. Moreover, all the isolates were susceptible to vancomycin by the disk diffusion method.
Table 1.
Detailed description of the MRSA isolates
| Isolate | MIC (μg/ml) |
Susceptibility pattern |
PCR Result | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Van | Teico | Pen | Gen | Amk | Cip | Lev | Ery | Tri/Sul | Van | Liz | ||
| 1 | 6 | 2 | R | S | R | R | R | R | S | S | S | − |
| 2 | 8 | 16 | R | R | R | R | R | R | R | S | S | vanA |
| 3 | 8 | 32 | R | R | R | R | R | R | R | S | S | vanA |
| 4 | 6 | 4 | R | R | R | R | R | R | R | S | S | − |
S, susceptible; R, resistant; Liz, linezolid; Van, vancomycin; Teico; teicoplanin; Tri/Sul, trimethoprim-sulfamethaxole; Pen, penicillin; Gen, gentamicin; Amk, amikacin; Cip, ciprofloxacin; Lev, levofloxacin; Ery, erythromycin.
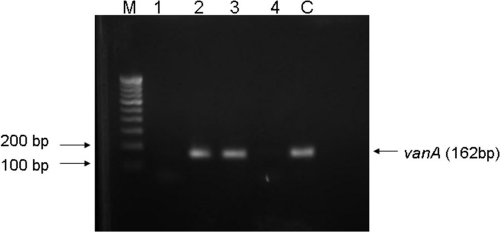
PCR assays for vancomycin resistance loci revealed the presence of vanA in two of the isolates (Fig. 1). The other two strains did not show the presence of either vanA or vanB. Furthermore, quantitative real-time PCR showed a lack of expression of the vanA gene in these isolates.
Fig 1.
Agarose gel electrophoresis of PCR-amplified vancomycin resistance genes. Lanes: M, 100-bp ladder; 1 to 4, S. aureus isolates; C, known VRE (vanA) strain.
PFGE of the isolates revealed four different clones.
Sections of the isolates examined by EM showed that they had a thicker cell wall than MRSA strains.
None of the patients carrying these VISA strains were on vancomycin at the time of survey, and the history of only one of the carriers revealed the use of vancomycin 2 months earlier. The rate of isolation of VRE from the stool samples in the hospitalized patients in the same unit was as high as 55.5% during the same time period (our unpublished data). Moreover, stool samples from both the patients with vanA strains were also positive for VRE.
Strains of vancomycin-intermediate S. aureus (VISA) with vancomycin MICs of 8 μg/ml have been reported from Japan, the United States, France, the United Kingdom, and Germany (17). Most of these isolates appear to have developed from preexisting MRSA infections. Studies from India have also reported reduced susceptibility of MRSA to vancomycin (2, 6). But all the reported VISA isolates were clinical strains, and nasal carriage was not reported, even in studies with the purpose of seeking VRSA in VRE-colonized patients (7). Even though VISA isolates are rare causes of clinical infections, we sought VISA colonization by surveying a high-risk hospitalized population in the ICU, where isolation of VRE was high and vancomycin was extensively used, being administered in nearly 20% of the admitted patients at one point in time.
Vancomycin resistance among MRSA strains might arise in several ways. Other than plasmid-mediated vanA vancomycin resistance gene transfer from enterococcal species to S. aureus, laboratory studies have demonstrated that S. aureus strains resistant to vancomycin can be produced by a step pressure procedure (8). But interspecies transfer of resistant genes was thought not to be responsible for intermediate resistance to vancomycin in S. aureus (10). Instead, VISA strains have been observed to have lower growth rates and thicker cell walls than fully susceptible strains. Increased cell wall thickness helps in resistance by sequestering vancomycin molecules in the cell wall peptidoglycan, thus reducing the susceptibility of S. aureus to vancomycin (16). But the genetic mechanisms by which these changes occur are not fully understood. None of the VISA strains have been shown to have any of the van determinants (vanA, vanB, vanC1, vanC2, or vanC3) that are present in VRE. In this study, vanA was seen in two of the isolates with intermediate resistance. The lower MIC of vancomycin might have been due not to reduced expression of vanA gene in the absence of selective vancomycin pressure, as seen by quantitative PCR, but rather to a thick cell wall. However, an increase in MICs after serial passage of the vanA-positive strains in vancomycin-enriched medium for a short period could not be demonstrated. Low-level resistance of a VRSA strain due to a longer lag phase before the induction of resistance along with loss of the vanA operon has been demonstrated (14). VRSA from clinical isolates have been reported in the absence of vancomycin exposure (18). Similarly, it was found that concurrent or recent vancomycin exposure was not a prerequisite for the development of VISA, just as in the case of VRSA. The factors leading to the reduced expression of vanA gene could not be exactly determined. The other two strains, which did not show the presence of vanA or vanB, however, were not tested further for other van determinants.
A previous study from the same center had reported six VISA and two VRSA strains from clinical samples, but none of these isolates was found to have either vanA or vanB by PCR (17).
These strains are as troublesome as the VRSA strains from the treatment point of view, as suggested by the cases of the few affected patients in the United States (16). The vanA phenotype observed in enterococci confers resistance to both vancomycin and teicoplanin. All VISA strains reported to date have reduced susceptibility to teicoplanin (10), and historically, teicoplanin resistance was acquired prior to vancomycin resistance. Teicoplanin MICs corresponding to intermediate and resistant levels were also noted for these isolates.
The importance of screening of MRSA isolates for VISA has been repeatedly emphasized, but a survey of laboratories indicated that many did not use methods that can detect VISA strains (16). Because these strains appear to have developed from strains of MRSA instead of a single clone, they have often been missed by disk diffusion testing, as in this study. Surveillance of patient populations, especially in units where the probability of MRSA carriage and prolonged glycopeptide therapy is high, should be regularly done using vancomycin agar screening tests.
In this study, though we found VISA isolates with the vanA genotype, this was probably not the mechanism of intermediate resistance to vancomycin. Instead, as in other VISA isolates, a thick cell wall resulted in resistance. An unstable vancomycin resistance phenotype in such isolates (10) and heterologous expression of the enterococcal vanA operon in MRSA (14) have already been reported. The VISA strains with the vanA genotype colonizing the anterior nares may be a potent source of VRSA with reduced expression of the vancomycin resistance gene. When exposed to the appropriate selective step-up pressure, these isolates may eventually take a resistant form. In addition, they may act as carriers, promoting easy transfer of drug resistance determinants. The potential for emergence of VRSA isolates from these strains during asymptomatic colonization, rather than during infection, may contribute to delays in detection (15). Therefore, systematic surveillance for these strains is essential to prevent infection, colonization, and dissemination in the hospital environment.
Footnotes
Published ahead of print 15 February 2012
REFERENCES
- 1. Arbour N, et al. 2008. Real-time PCR detection of VRE. Spartan Bioscience Inc. AN0019, version 1.1 [Google Scholar]
- 2. Assadullah S, et al. 2003. Emergence of low level vancomycin resistance in MRSA. Ind. J. Med. Microbiol. 21:196–198 [PubMed] [Google Scholar]
- 3. Centers for Disease Control and Prevention 1997. Reduced susceptibility of Staphylococcus aureus to vancomycin—Japan, 1996. MMWR Morb. Mortal. Wkly. Rep. 46:624–626 [PubMed] [Google Scholar]
- 4. Chang S, et al. 2003. Infection with vancomycin resistant Staphylococcus aureus containing the vanA resistance gene. N. Engl. J. Med. 348:1342–1347 [DOI] [PubMed] [Google Scholar]
- 5. Clinical Laboratory and Standards Institute 2011. Performance standards for antimicrobial susceptibility testing; 21st informational supplement. M100–S21-31(1) Clinical Laboratory and Standards Institute, Wayne, [PubMed] [Google Scholar]
- 6. Dhawan B, Gadepalli R, Rao C, Kapil A, Sreenivas V. 2010. Decreased susceptibility to vancomycin in methicillin-resistant Staphylococcus aureus: a 5 year study in an Indian tertiary hospital. J. Med. Microbiol. 59:375–376 [DOI] [PubMed] [Google Scholar]
- 7. Franchi D, Climo MW, Wong AHM, Michael EB, Wenzel RP. 1999. Seeking vancomycin resistant Staphylococcus aureus among patients with vancomycin-resistant enterococci. Clin. Infect. Dis. 29:1556–1558 [DOI] [PubMed] [Google Scholar]
- 8. Hiramatsu K. 1998. The emergence of Staphylococcus aureus with reduced susceptibility to vancomycin in Japan. Am. J. Med. 104(5A):7S–10S [DOI] [PubMed] [Google Scholar]
- 9. Leclercq R, Derlot E, Duval J, Courvalin P. 1988. Plasmid-mediated resistance to vancomycin and teicoplanin in Enterococcus faecium. N. Engl. J. Med. 319:157–161 [DOI] [PubMed] [Google Scholar]
- 10. Liu C, Chambers HF. 2003. Staphylococcus aureus with heterogeneous resistance to vancomycin: epidemiology, clinical significance, and critical assessment of diagnostic methods. Antimicrob. Agents Chemother. 47:3040–3045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Livak KL, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 12. Marchese A, Balistreri G, Tonoli E, Debbia A, Schitto GC. 2000. Heterogeneous vancomycin resistance in methicillin-resistant Staphylococcus aureus strains isolated in a large Italian hospital. J. Clin. Microbiol. 38:866–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Noble WC, Virani Z, Cree RG. 1992. Co-transfer of vancomycin and other resistance genes from Enterococcus faecalis NCTC 12201 to Staphylococcus aureus. FEMS Microbiol. Lett. 72:195–198 [DOI] [PubMed] [Google Scholar]
- 14. Perichon B, Courvalin P. 2004. Heterologous expression of the enterococcal vanA operon in methicillin resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 48:4281–4285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ray AJ, Pultz NJ, Bhalla A, Aron DC, Donskey CJ. 2003. Coexistence of vancomycin-resistant enterococci and Staphylococcus aureus in the intestinal tracts of hospitalized patients. Clin. Infect. Dis. 37:875–881 [DOI] [PubMed] [Google Scholar]
- 16. Tenover FC, Biddle JW, Lancaster MV. 2001. Increasing resistance to vancomycin and other glycopeptides in Staphylococcus aureus. Emerg. Infect. Dis. 7:327–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tiwari HK, Sen MR. 2006. Emergence of vancomycin resistant Staphylococcus aureus (VRSA) from a tertiary care hospital from northern part of India. BMC Infect. Dis. 6:156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Whitener CJ, et al. 2004. Vancomycin-resistant Staphylococcus aureus in the absence of vancomycin exposure. Clin. Infect. Dis. 38:1049–1055 [DOI] [PubMed] [Google Scholar]