Abstract
Type IV pili of Geobacter sulfurreducens are composed of PilA monomers and are essential for long-range extracellular electron transfer to insoluble Fe(III) oxides and graphite anodes. A previous analysis of pilA expression indicated that transcription was initiated at two positions, with two predicted ribosome-binding sites and translation start codons, potentially producing two PilA preprotein isoforms. The present study supports the existence of two functional translation start codons for pilA and identifies two isoforms (short and long) of the PilA preprotein. The short PilA isoform is found predominantly in an intracellular fraction. It seems to stabilize the long isoform and to influence the secretion of several outer-surface c-type cytochromes. The long PilA isoform is required for secretion of PilA to the outer cell surface, a process that requires coexpression of pilA with nine downstream genes. The long isoform was determined to be essential for biofilm formation on certain surfaces, for optimum current production in microbial fuel cells, and for growth on insoluble Fe(III) oxides.
INTRODUCTION
Geobacteraceae are anaerobic bacteria belonging to the Deltaproteobacteria. Geobacter species are Fe(III) reducers that are highly abundant in subsurface environments, where Fe(III) accepts electrons derived from the fermentation of various electron donor substances, e.g., acetate, alcohols, and toxic aromatic pollutants (2, 15, 20, 33, 35, 47, 68). Besides Fe(III), Geobacter species use other insoluble metal oxides as electron acceptors, including Mn(IV), U(VI), and V(V) (9, 36–38, 48), as well as humic substances (34, 67) and graphite anodes (5, 32). Investigations of the mechanism of electron transfer to insoluble electron acceptors have been conducted primarily in Geobacter sulfurreducens due to the availability of a complete genomic sequence (42) and a genetic system (13). Several components of the cell have been identified as important for long-range electron transfer to Fe(III) oxides and/or to graphite anodes. They include MacA, a c-type cytochrome located in the inner membrane (8); several outer membrane c-type cytochromes, OmcB, OmcS, OmcE, and OmcZ (21, 26, 30, 31, 41, 45, 51, 56); and extracellular filamentous material described as “nanowires.” The nanowires of G. sulfurreducens have been reported to be type IV pili that are essential for electron transfer to Fe(III) oxide (53), for optimal current production when a graphite anode is the sole extracellular electron acceptor (39, 46, 54, 56, 63), and for thick biofilm formation on various surfaces (45, 55). The type IV pili of G. sulfurreducens, like those of other Gram-negative bacteria, are presumably polymers of mainly one protein, termed pilin or PilA, encoded by the pilA gene (GSU1496) (53).
Type IV pilins in Gram-negative bacteria are synthesized as prepilins, with a leader sequence that is cleaved after a conserved glycine (defined as position −1) by a specific leader peptidase, PilD, in the inner membrane (3, 16, 61). Mature pilin subunits have a hydrophobic amino-terminal segment with a consensus sequence that includes a conserved phenylalanine (position 1) and glutamic acid (position 5), which forms the core of the pilus fiber (14). Cleaved pilin monomers assemble into pilus filaments in the periplasmic space via an electrostatic attraction among pilin subunits (14, 40, 71), and the growing filaments cross the outer membrane through a hole in a multimeric outer membrane protein called secretin, or PilQ (12, 71).
Several genes are involved in pilus biogenesis, few of which are conserved across the Gram-negative bacteria (1, 19, 29). Type IV pilins are divided into two subclasses according to the lengths of the leader peptide and the mature protein (1, 10, 11, 27, 52). Type IVa pilins have leader peptides less than 10 amino acids in length, whereas type IVb pilins have leader peptides that are up to 30 amino acids. In addition, type IVa pilin biogenesis genes are scattered throughout the genome, whereas the type IVb pilin genes are typically clustered. The PilA protein, encoded by the pilA gene of G. sulfurreducens, has been reported previously to belong to the type IVa subclass (53).
Transcription of the type IV pilin genes is regulated by the PilS-PilR two-component regulatory system (7, 19, 22, 23). PilS autophosphorylates in response to an environmental signal and then transfers the phosphate group to the response regulator PilR. Phosphorylated PilR modulates transcription at a promoter upstream of the pilin gene. In G. sulfurreducens, pilS and pilR are located just upstream of pilA (Fig. 1A). PilR (GSU1495) likely functions as an RpoN-dependent enhancer-binding protein that binds to a specific consensus sequence located in a predicted promoter region upstream of the pilA gene (24). Mapping the 5′ end of the pilA transcript revealed the presence of long and short transcripts of pilA. The long transcript was RpoN dependent, and unlike the short transcript (RpoN independent), it was not detected in RNA isolated from the pilR mutant strain. Juárez and coworkers identified two transcription start sites and predicted two translation start codons with independent ribosomal binding sites (24) but did not investigate whether the two pilA transcripts produced different PilA preprotein isoforms. Characterization of the PilR-deficient mutant strain revealed phenotypes similar to those found in the PilA-deficient strain. Both the pilR and pilA mutant strains were unable to grow on insoluble Fe(III) oxide and exhibited a decreased ability to attach to glass (24, 53, 55). These observations suggested that the PilA isoform resulting from the long transcript, not detected in the pilR mutant strain, is necessary for growth and attachment (24). The purpose of this study is to investigate the hypothesis that the two translation start codons are functional and correspond to two PilA preprotein isoforms and to determine the roles that these isoforms have in growth and attachment.
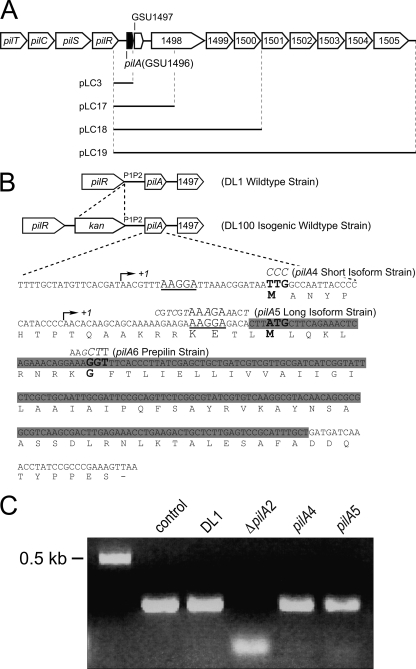
Fig 1.
(A) Genomic organization of the pilus biosynthesis genes and gene cluster downstream of pilA (GSU1496). The black bars indicate DNA cloned into the plasmids constructed for complementation experiments. (B) Sequences of the pilA gene and PilA protein, including the mutations introduced for each strain used in this work. In the isogenic wild-type and all mutant strains, a kanamycin cassette was inserted upstream of the P1 and P2 promoter regions. The arrows indicate the two transcription initiation sites. The two ribosome-binding sites are underlined. The translation start codons (TTG and ATG) and the peptidase cleavage site (GGT) are in boldface. The base changes leading to amino acid replacements are depicted in italics for each mutant strain. The deleted sequence of the pilA gene in the ΔpilA2 strain is highlighted in gray. (C) DNA gel of semiquantitative reverse transcriptase PCRs. (1) proC control; (2) wild-type pilA in DL1; (3) ΔpilA2; (4) short-isoform (pilA4) strain; and (5) long-isoform (pilA5) strain.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The wild-type (Wt) and mutant strains of G. sulfurreducens and the plasmids generated in this work are listed in Tables 1 and 2, respectively. Escherichia coli strain TOP10 was purchased from Invitrogen Co. (Carlsbad, CA) and was used to subclone PCR products and for DNA manipulations.
Table 1.
G. sulfurreducens strains used in this work
| Strain name or genotypea | Descriptionb |
|---|---|
| DL1 (Wt) | Wild type |
| DL100 (isogenic Wt) | Wild-type pilA; Kanr |
| pilA4 (short isoform) | Long isoform start codon mutation; TTG→CCC |
| pilA5 (long isoform) | Short isoform ribosome-binding site mutation |
| pilA6 (prepilin) | Peptidase cleavage site mutation |
| pilA7 (pilA4 and pilA5 double mutant) | Long isoform start codon mutation (TTG→CCC) and short isoform ribosome-binding site mutation |
| ΔpilA2 (in-frame deletion) | In-frame deletion mutation |
All strain were generated in this work and were derived from DL1 (9).
All strains are Kanr, except DL1.
Table 2.
Plasmids used in this work
| Plasmida | Descriptionb | Parent plasmid |
|---|---|---|
| pCD341 | Source of kanamycin resistance cassette | (43) |
| pLC1 | Expression vector with tacUV5 and lac promoters removed | pRG5 (25) |
| pLC3 | pilA (wild type) | pLC1 |
| pLC8 | pilA long isoform start codon mutation (TTG→CCC) | pLC3 |
| pLC13 | pilA short isoform ribosome-binding site mutation | pLC3 |
| pLC14 | pilA long isoform start codon mutation (TTG→CCC) and short isoform ribosome-binding site mutation | pLC3 |
| pLC17 | pilA GSU1497, 1,271 bp of GSU1498 | pLC1 |
| pLC18 | pilA GSU1497, GSU1498, GSU1499, GSU1500 | pLC17 |
| pLC19 | pilA GSU1497, GSU1498, GSU1499, GSU1500, GSU1501, GSU1502, GSU1503, GSU1504, GSU1505 | pLC18 |
| pLC20 | pilA peptidase cleavage site mutation | pLC3 |
All plasmids were generated in this work except as noted.
All plasmids are Specr, except pCD341 (Kanr).
DNA manipulations and plasmid construction.
Genomic DNA of the G. sulfurreducens wild-type strain DL1 (9) was purified using the MasterPure Complete DNA Purification Kit (Epicentre Technologies, Madison, WI). Plasmid DNA purification, PCR product purification, and gel extraction were performed using the QIAprep Spin Mini Plasmid Purification, QIAquick PCR Purification, and QIAquick Gel Extraction kits, respectively (Qiagen Inc., Valencia, CA). Restriction enzymes, Klenow fragment, and T4 DNA ligase were purchased from New England BioLabs Inc. (Beverly, MA). Primers for PCR DNA amplification and for site-directed mutagenesis were purchased from Operon Biotechnologies Inc. (Huntsville, AL). All PCRs were performed with high-fidelity Phusion polymerase using reaction conditions specified by the manufacturer (Finnzymes Inc., Woburn, MA).
The pLC1 expression vector was generated from the pRG5 expression vector (25) by removing the tacUV5 and lac promoters. pRG5 was digested with NcoI and AseI, followed by treatment with Klenow fragment. The double-digested vector was gel purified and precipitated with ethanol. The precipitate was washed with 70% ethanol and resuspended in 0.5× TE buffer (0.5 mM Tris HCl, pH 8.0, 0.5 mM EDTA). To generate pLC1, the DNA was closed with T4DNA ligase and transformed into E. coli Top10 chemically competent cells, and colonies were screened for growth on spectinomycin plates. The identity of pLC1 was verified by sequencing. The replication of pLC1 in E. coli was tested and found to have a copy number comparable to that of pRG5 under identical conditions. Also, no change in growth rate or cell yield was observed with the wild-type G. sulfurreducens harboring pLC1 compared to the wild type without plasmid (data not shown).
To construct pLC3, a plasmid expressing wild-type pilA under the control of the native promoters, the coding sequence from the last 200 bases of the upstream gene pilR (GSU1495), the P1 and P2 promoter regions, and all of pilA (GSU1496) was amplified using DL1 genomic DNA as a template (Fig. 1A). The PCR primers rLC41F and rLC42R amplified this region and introduced BamHI and FspI restriction sites at the 5′ and 3′ ends, respectively. The PCR product and pLC1 were digested with BamHI and FspI, followed by ligation and transformation into E. coli TOP10 cells. The identity of pLC3, containing the final 200 bp of pilR, the P1 and P2 promoter regions of pilA, and the entire coding sequence of pilA, was verified by DNA sequencing.
pLC3 served as the template for all the pilA mutations investigated in this work, using the QuikChange II XL Site-Directed Mutagenesis Kit (Stratagene Inc., La Jolla, CA). In addition to the specific mutation, the PCR primers either introduced a restriction site or eliminated a naturally occurring site within pilA (Table 3). The introduction of the restriction site was used for screening to verify that the plasmid carried the desired mutation(s). Also, all plasmids were sequenced to confirm that the pilA gene carried only the desired mutation.
Table 3.
Primers used in this work
| Purpose | Name | Sequencea | Description |
|---|---|---|---|
| Strain construction | |||
| pilA4, pilA5, pilA6, and pilA7 strains | rLC43F | AGAGCAGGTGAAGGAAGGGAGTTT | Amplifies the last 500 bp of pilR with the kanamycin cassette |
| rLC46R | GACGATTTTCGTCACTGGCTCCTCTTGGATCCCCCGGGCTGCAGGAATTCG | ||
| rLC47F | CGAATTCCTGCAGCCCGGGGGATCCAAGAGGAGCCAGTGACGAAAATCGTCA | Amplifies pilA | |
| rLC51R | CTATTGCGCACAATGGCTATTCCCTGCATTGCGA | ||
| ΔpilA2 strain | rLC43F | AGAGCAGGTGAAGGAAGGGAGTTT | Amplifies the last 500 bp of pilR |
| rLC44R | TCACTCCTCATCCATGCCAAATTTTGCCAGGC | ||
| rLC45F | GCCTGGCAAAATTTGGCATGGATGAGGAGTGACTGACGGAACAGCGGGAAGTCCAGC | Amplifies the kanamycin cassette | |
| rLC46R | TGACGATTTTCGTCACTGGCTCCTCTTGGATCCCCCGGGCTGCAGGAATTCG | ||
| rLC47F | CGAATTCCTGCAGCCCGGGGGATCCAAGAGGAGCCAGTGACGAAAATCGTCA | Amplifies the P1, P2 promoter region and introduces the pilA deletion | |
| rLC48Rb | TGTCTCCTTTCTTCTTTTTGCTGCTTGTGTTGGGGTATGGGGGTAATTGGCCAATTATCCGTTTAATCCTTAAACGTTATCGTG | ||
| rLC49Fb | GCAGCAAAAAGAAGAAAGGAGACAGATGATCAAACCTATCCGCCCGAAAGTTAATTGATTAAATACATACTGGAGGAAACCATG | Amplifies 400 bp downstream of pilA and to introduces the pilA deletion | |
| rLC50R | CTACTGCGACTTCCACTCGGTACCAGAAAAATCAG | ||
| Plasmids construction | |||
| pLC3 | rLC41F | GATAGGATCCGTCACCGAGTGCGAACTGCC | Amplifies pilA and native promoters |
| rLC42R | CTATTGCGCACAATGGCTATTCCCTGCATT GCGA | ||
| pLC8 | M1_Fwdc,d | GATTAAACGGATAACCCGCCAATTACCCCCATACCCCAACACAAGC | Introduces the long isoform start codon mutation (TTG → CCC) |
| M1_Revd | CTTGTGTTGGGGTATGGGGGTAATTGGCGGGTTATCCGTTTAATCC | ||
| pLC13 | RBS_Fwdc,e | CCCCCATACCCCAACACAAGCAGCAAAACGTCGTAAAGAAACTCTTATGCTTCAGAAACTCAG | Introduces the short isoform ribosome-binding site mutation |
| RBS_Reve | CTGAGTTTCTGAAGCATAAGAGTTTCTTTACGACGTTTTGCTGCTTGTGTTGGGGTATGGGGG | ||
| pLC17 | rLC108 | GATATGCGCATCCACGCGGGTAAACGGCCTGA | Amplifies fragment 1 (2.5 kb) for pLC19 |
| rLC101 | CTATCCCGGGTCTGCCGCATAACCGACGTAACTTC | ||
| pLC18 | rLC102 | TGGTCATGTGTATAATCCCACCAATTATGGC | Amplifies fragment 2 (3.2 kb) for pLC19 |
| rLC103 | CTATCCCGGGGAGCCATTGGAGAGGACGACAG | ||
| pLC19 | rLC104 | CTGTAGATCGTGACGGATTGGGTG | Amplifies the fragment 3 (5.2 kb) for pLC19 |
| rLC107 | CTATCCCGGGACATACCTGGCCACTGGTTC | ||
| pLC20 | G20L_Fwdc | CTCAGAAACAGGAAGCTTTTCACCCTTATCGAGCTGCTGATCG | Introduces the peptidase cleavage site mutation |
| G20L_Rev | GATCAGCAGCTCGATAAGGGTGAAAAGCTTCCTGTTTCTGAG | ||
| RT-PCR | pilAq_Fwd | GGCCAATTACCCCCATAC | Amplifies the long isoform |
| pilAq_Rev | ACTTTCGGGCGGATAGGTTT |
Annealing nucleotides are shaded; restriction sites are italicized and underlined.
The deletion version of pilA introduced by the primers is in boldface.
Base pairs replaced by site-directed mutagenesis are in boldface.
This mutation disrupts the unique, naturally occurring MscI site in pilA.
This mutation disrupts the unique, naturally occurring BsmAI site in pilA.
pLC19 was constructed in three steps, and each step was verified by PCR, restriction endonuclease digestion, and sequencing. A set of primers was designed to amplify a 10.5-kb segment of DL1 genomic DNA containing all the genes from GSU1496 (pilA) to GSU1505 in three smaller fragments: fragment 1 (2.5 kb), fragment 2 (3.2 kb), and fragment 3 (5.2 kb). The primers that amplified fragment 1 (rLC108 and rLC101) introduced FspI and XmaI sites at the 5′ and 3′ ends, respectively (Table 3), for ligation into expression vector pLC1 to create pLC17.
The forward primer that amplified fragment 2 (rLC102) overlapped slightly with fragment 1 and created an NheI site unique to the 10.5-kb segment. The reverse primer (rLC103) introduced an XmaI site at the 3′ end of fragment 2 (Table 3). These two sites were used to insert fragment 2 into pLC17, creating plasmid pLC18. The forward primer for amplification of fragment 3 (rLC104) overlapped slightly with fragment 2 and incorporated an XhoI site unique to the 10.5-kb segment. The reverse primer (rLC107) introduced an XmaI site at the 3′ end of fragment 3 (Table 3). These two sites were used to insert fragment 3 into pLC18 to create pLC19. Plasmid pLC19 contains the entire 10.5-kb segment comprised of the final 200 bp of pilR, the P1 and P2 promoter regions, pilA, GSU1497, and eight genes (GSU1498 to GSU1505) downstream (Fig. 1A). This plasmid was used to express pilA and the associated genes in trans to test the requirements for coordinated gene product activity for full PilA function.
Construction of G. sulfurreducens mutant strains.
A recombinant PCR protocol (13) was used to construct all the mutant strains described in this study (Table 1). Briefly, pLC3-derived plasmids carrying pilA with the desired mutations were used as templates for amplification of pilA to generate linear mutagenic fragments. The point mutations in pilA were introduced into the chromosome of DL1 by homologous recombination using a kanamycin resistance marker to select for recombinants; this marker was inserted 5′ of the pilA P1/P2 promoter region. Mutations were confirmed by PCR using Phusion polymerase and different sets of primers to map the pilA region, by digestion at the introduced restriction sites in the pilA gene, and by genomic-DNA sequencing.
In the case of the pilA in-frame deletion mutant (ΔpilA2) strain, the last 500 bp of pilR was amplified from DL1 chromosomal DNA using rLC43F and rLC44R. The kanamycin cassette was amplified from the pCD341 plasmid (Table 2) using rLC45F and rLC46R. rLC47F and rLC48R amplified the promoter region of the pilA gene and introduced an in-frame deletion of pilA. A 400-bp segment downstream of the pilA gene was amplified from DL1 genomic DNA using rLC49F and rLC50R. rLC48 and rLC49 primers introduced the first 18 and last 10 codons of the pilA gene, respectively. The in-frame deletion sequence of pilA was confirmed by genomic-DNA sequencing.
Reverse transcriptase PCR (RT-PCR).
G. sulfurreducens strains were cultured in NB medium (13) with 15 mM acetate and 40 mM fumarate (NBAF) at the pilus-inducing temperature, 25°C (53). Cells were harvested in mid-log phase, and the total RNA was purified with an RNeasy Midi kit (Qiagen Inc., Valencia, CA). The RNA was treated to remove contaminating DNA with a DNA-free kit (Ambion, Austin, TX). The absence of DNA was verified by a PCR using the RNA sample as a template. The purity of the RNA was assessed on a 1% agarose gel. RNA concentrations were measured using a NanoDrop ND-1000 spectrophotometer (Wilmington, DE), and samples were stored in small aliquots at −80°C until use. cDNA samples were synthesized from equivalent amounts of total RNA as starting material, using random nonamer primers with an Enhanced Avian RT First Strand Synthesis kit (Sigma-Aldrich, St. Louis, MO). PCRs were performed using the cDNAs from the different strains as templates with primers that amplified the long transcript of the pilA gene (pilAq_Fwd and pilAq_Rev [Table 3]).
Culture conditions and growth media.
G. sulfurreducens wild-type and mutant strains were cultured in NBAF medium under strict anaerobic conditions (80/20 N2-CO2) at 25°C. Plating and incubation on solid agar medium were performed as previously described (13). Liquid cultures were streaked or spread on NBAF plates containing 1.5% agar, 0.1% yeast extract, and 1 mM cysteine and incubated in an anaerobic chamber at 30°C.
For microbial fuel cell analyses, bacteria were first grown in 200-ml batch cultures of freshwater medium (38) with 10 mM acetate as the sole electron donor and the graphite anode poised at +300 mV (versus an Ag/AgCl reference electrode) as the sole electron acceptor (6). The system was switched to flowthrough mode with 10 mM acetate at 30 ml/h once current production was observed (54).
Western blots and heme-staining analyses.
PilA protein fractions were prepared from G. sulfurreducens cultures grown at 25°C under strict anaerobic conditions in NBAF. Cells were fractionated based on a published protocol (72) to separate secreted from nonsecreted PilA with the following modifications: late-log-phase cells were harvested by centrifugation for 15 min at 5,000 × g and 4°C (Sorvall RC5B Plus; F14S-6*250 rotor), without first subjecting the cells to shear. Supernatants were collected and concentrated 100-fold in a Microcon Centrifugal Filter Device (Millipore Corp., Billerica, MA) with a 5,000-molecular-weight-cutoff (MWCO) membrane. Further filtration was applied through 3,000-MWCO Ultrafiltration tubes (Millipore Corp., Billerica, MA) to a final volume of 500 μl. The retentate is fraction 1 and is also referred to as the “secreted protein” fraction. The cell pellet was resuspended in 50 mM Tris-HCl buffer (pH 7.5) and sonicated for 4 min with a 15-s on, 20-s off duty cycle (Sonic dismembrator F550; Fisher Scientific, PA). The resuspended and sonicated pellet was then centrifuged for 15 min at 5,000 × g and 4°C. The supernatant is fraction 2 and is also referred to as the “soluble nonsecreted protein” fraction. The pellet is fraction 3 and is also referred to as the “membrane-associated protein” fraction. This fractionation procedure was adopted upon observing a substantial amount of the total expressed PilA protein in fraction 1, the secreted protein fraction. The protein fractions were separated by electrophoresis on 15% acrylamide tris-tricine SDS gels, and the protein bands were transferred to polyvinylidene difluoride (PVDF) membranes using a semidry transfer unit (Trans-Blot; Bio-Rad Laboratories, Hercules, CA). The membranes were treated with PilA-specific polyclonal antibody (39, 74), and the PilA bands were visualized using a One-Step Western kit (Genscript Corp., Piscataway, NJ).
For detection of the outer-surface heme-bound c-type cytochromes, samples were prepared as described previously (41) and separated by electrophoresis using 12% Next gels (Amresco Inc., Solon OH). N,N,N′,N′-tetramethylbenzidine was used for heme staining according to the published protocols (17, 64).
SeeBlue Plus 2 (Invitrogen Corp., Carlsbad, CA) prestained MW standards were used for all electrophoresis gels.
Biofilm characterization/analysis.
All bacterial biofilms, regardless of the surface material, were stained with Syto 9 (L7012 Component A DNA stain; Invitrogen Corp., Carlsbad, CA). The stained biofilms were imaged using a Leica TCS SP5 confocal laser scanning microscope (CLSM) with an HCX PL APO 100× objective (numerical aperture [NA], 1.4). CLSM images were processed with LAS AF software (Leica Microsystems GmbH, Wetzlar, Germany) to produce three-dimensional projection images and cross sections of the biofilms. Statistical analyses of thickness, coverage, and roughness were conducted using the biofilm analysis software Phobia laser scanning microscopy imaging processor (PHLIP) (39, 44) from a minimum of 15 image stacks per condition.
Cell attachment assays.
Cells were exposed to graphite or glass surfaces in the presence of 10 mM acetate and 40 mM fumarate at 25°C under anaerobic conditions for 4 days and then prepared for confocal imaging as described above. For Fe(III) oxide attachment assays, cells were grown in freshwater medium with 10 mM acetate in the presence of soluble electron acceptor and 40 mM fumarate and incubated with Fe(III) oxide-coated borosilicate coverslips prepared as described previously (65). Six slides were prepared for each strain, and cells were incubated in the presence of soluble electron donor and acceptor for 24 h, at which time three slides per strain were imaged by CLSM. The remaining three slides were incubated for 4 days more with Fe(III) oxide coating as the sole electron acceptor (with fumarate removed) and then examined by CLSM.
RESULTS
Semiquantitative transcription analysis of pilA in the short (pilA4)- and long (pilA5)-isoform strains.
To test the hypothesis that two PilA preprotein isoforms exist and to determine the function of each preprotein variant, two mutant strains were generated. The strain expressing only the short isoform of the PilA preprotein (pilA4) had a mutation that changed the predicted translation start codon (TTG) of the long isoform into a non-start codon (CCC) (Fig. 1B). These mutations were upstream of the start site for the short-isoform transcript of pilA and therefore were not expected to affect the amino acid sequence of the short-isoform PilA preprotein (Fig. 1B). In the long-isoform (pilA5) strain, a mutation was introduced into the ribosome-binding site of the short pilA transcript (Fig. 1B). This strategy was chosen in preference to a point mutation in the translation start codon (ATG) because the latter approach would have changed this methionine—a residue within the long PilA isoform—which would influence PilA function in an unknown manner. Instead, site-directed mutagenesis was used to disrupt the ribosome-binding site while maintaining the wild-type PilA preprotein sequence, using codons that occur with similar frequencies in the genome of G. sulfurreducens (CGT for Arg, AAA for Lys, GAA for Glu, and ACT for Thr). Both the short- and long-isoform strains were selected by means of a kanamycin resistance cassette located before the P1 and P2 promoters of pilA (Fig. 1B). The isogenic control strain, DL100, with a kanamycin resistance marker in the same location (Fig. 1B), expressed PilA like the wild-type strain, DL1 (data not shown).
To determine whether the mutations engineered into pilA had any effect on transcription, semiquantitative reverse transcriptase PCRs were performed on RNA samples extracted from the wild-type strain (DL1), from the short (pilA4)- and long (pilA5)-isoform strains, and from the deletion (ΔpilA2) strain. PCRs were conducted with cDNA as a template and primers designed to amplify the long transcript of pilA; the forward primer annealed to a region upstream of the transcription start site for the short pilA transcript (Table 3 lists primer sequences). Long-transcript RNA was detected in all mutant strains at levels comparable to those in the wild-type strain (Fig. 1C).
Analysis of the strains expressing only the short (pilA4) or long (pilA5) isoform provides evidence of two PilA preprotein isoforms and cooperation among isoforms for function.
To investigate the effects of the mutations on pilA expression and cellular distribution, the presence and mobility of PilA in three cell fractions were determined with Western blots. These blots (Fig. 2A) provided evidence of significant differences in the subcellular distribution of PilA between the wild-type and engineered strains. Unfortunately, it was not feasible to analyze the cellular location of PilA by electron microscopy (EM). Direct comparisons of EM images from the wild-type and ΔpilA strains are uninformative, because G. sulfurreducens possesses several filament-like structures that have morphologies, lengths, and diameters similar to those of type IV pili, which prevents the unambiguous identification of type IV pili (28). Also, the anti-PilA antibody used in our Western blot experiments does not bind to native PilA, so immunoelectron microscopy could not be used to determine the cellular location of PilA. Consequently, we used the Western blot data, together with the known properties of PilA, to assess the presence and form of PilA in the cell fractions.
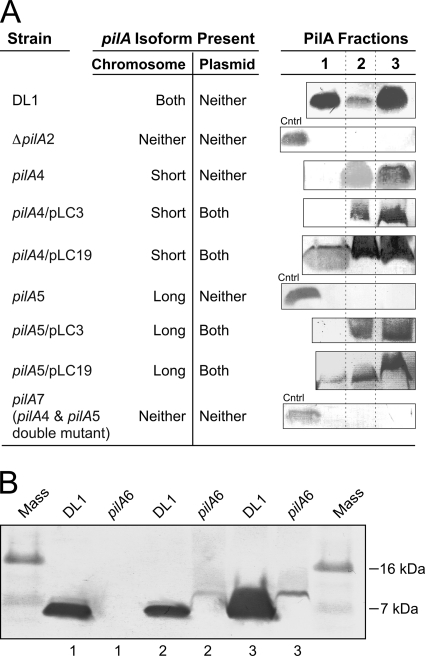
Fig 2.
Localization of wild-type and engineered PilA. (A) Protein bands were detected with the PilA-specific antibody. The PilA fractions are fraction 1 (secreted), fraction 2 (nonsecreted soluble), and fraction 3 (membrane associated), as defined in Materials and Methods. Control (Cntrl) samples (the secreted protein fraction of DL1) were included in null strains to verify the possibility of detecting PilA if it was present. All PilA protein fractions migrated at 7 kDa. (B) Western blot of PilA isolated from the prepilin strain (pilA6) using the PilA-specific antibody. Shown are PilA fraction 1 (secreted), fraction 2 (nonsecreted soluble), and fraction 3 (membrane associated) containing PilA protein of the wild-type DL1 and the prepilin (pilA6) strains. Each lane contains 10 μg total cell protein (A and B).
In the wild-type strain (DL1), as well as the wild-type isogenic control (DL100), PilA was detected in three fractions: the secreted fraction (fraction 1), a soluble cellular fraction (fraction 2), and a membrane-associated (sedimentable) fraction (fraction 3) (Fig. 2A). It migrated at 7 kDa, indicating that preprotein had been cleaved. The PilA in fraction 1 was collected from culture supernatants after the cells were removed by sedimentation, but without first subjecting the cells to shear (e.g., in a blender or by extrusion). Since the mature (7-kDa) protein contains all the molecular elements required for filament assembly (14), we expect that PilA recovered in fraction 1 is present as filaments. To our knowledge, except for the soluble pilin of Neisseria gonorrhoeae (58), an N-terminally truncated form of pilin protein, all the mature type IV pilin proteins that have been studied are assembled into filaments (50, 66, 69). Nevertheless, we cannot rule out the possibility that fraction 1 contains PilA monomer. Also, some of the PilA observed in fraction 2 could arise from filamentous PilA that was sheared from the cell surface during the sonication process (see Materials and Methods).
In the strain expressing only the short isoform (pilA4), where the long-isoform translation start codon was eliminated by replacement with a non-start codon, no secreted protein was detected by Western blotting, but the cell-associated (nonsecreted soluble and membrane-associated) forms were detected (Fig. 2A). This result suggested that the TTG start codon was functional and the long isoform was necessary for PilA secretion and assembly into filaments. The comparable intensities of PilA staining in the strain expressing only the short isoform and the wild type (DL1) suggest that the PilA protein in the cell is generated predominantly from the short isoform with the putative ATG start codon.
If the two preprotein isoforms functioned independently, only the secreted PilA protein would be expected upon elimination of the short isoform. However, no PilA, either the secreted or the cellular form, was detected in the strain expressing only the long isoform (pilA5), in which the short-isoform ribosome-binding site was mutated (Fig. 2A). The absence of PilA derived from the long preprotein isoform was not due to a defect in transcription (Fig. 1C) or a change in protein structure, because the mutagenesis strategy maintained the wild-type PilA amino acid sequence using codons with similar frequencies. Therefore, the result provided evidence that PilA derived from the long prepilin isoform was neither secreted nor accumulated in any of the three fractions without the short isoform present. Also, the pilA7 strain, a double mutant of pilA4 and pilA5 in which both translational initiation regions were mutated, did not produce any PilA in any of the three fractions, as expected (Fig. 2A).
The long and short PilA preproteins are processed into a single mature form.
Despite the fact that the long isoform of the PilA preprotein, with a predicted mass of 9 kDa, contains 20 more amino acids than the short preprotein isoform (7 kDa), only one band was detected on Western blots. This finding was attributed to rapid cleavage of the preprotein by the peptidase PilD at the inner membrane (62), where both isoforms were cleaved after the glycine at position −1 (3, 16), resulting in the same mature protein. Similar observations have been reported for type IV PilA of Myxococcus xanthus (73), where rapid cleavage by PilD prevented the detection of uncleaved PilA in wild-type strains on Western blots. The slower migration of the prepilin protein was detected only when the cleavage site was mutated. Immunofluorescence microscopy suggested that the cleaved PilA was located in or near the cell membrane, while the mutant PilA (uncleaved) was cytoplasmic (73). We observed a slower-migrating form of PilA, corresponding to the 9-kDa preprotein, only after the cleavage site was disrupted by mutagenesis (Fig. 2B), in which the glycine at position −1 was replaced with leucine (GGT to CTT) (Fig. 1B). The 9-kDa PilA was detected only in the cellular fractions of the mutant (Fig. 2B), indicating the necessity for cleavage in PilA secretion. For reasons we do not entirely understand, only one band was observed in the cleavage site mutant (pilA6), which could have been due to an inability to resolve the short preprotein from the long preprotein, to the fact that one preprotein isoform was translated at a substantially lower level than the other (so that only one of the two was detected on the Western blot), and/or to the fact that the uncleaved preproteins were less stable than mature PilA and were degraded. Nonetheless, the observation that the short-form- and long-form-expressing strains (pilA4 and pilA5, respectively) both show impaired and distinct subcellular localization patterns (Fig. 2A) provide evidence that both preprotein forms are required for the formation of type IV pili.
The strain expressing only the short-isoform PilA modulates the distribution of outer membrane cytochrome OmcZ.
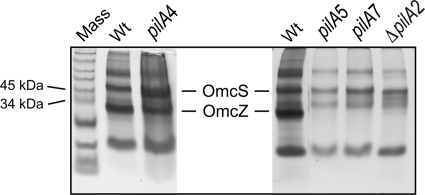
In addition to PilA secretion, the secretion of an outer membrane c-type cytochrome, OmcZ, was also affected by the absence of the short PilA isoform. OmcZ, which is important for extracellular electron transfer (21, 45, 56), was notably absent on a heme-stained gel of the outer cell surface proteins isolated from the strain expressing only the long isoform (pilA5); this profile was also observed with the ΔpilA2 and the pilA7 strains (Fig. 3). On the other hand, OmcZ was observed among the outer membrane proteins of the strain expressing only the short isoform (pilA4) at levels comparable to those in the wild type (DL1) (Fig. 3). Taken together, the data in Fig. 3 demonstrate that the mislocalization of OmcZ in the long-isoform (pilA5), pilA7, and ΔpilA2 strains might be due to the absence of the short isoform.
Fig 3.
Heme-stained SDS-PAGE of loosely bound outer surface c-type cytochromes prepared from the wild-type and pilA mutant strains. Shown are wild-type DL1 (Wt), short-isoform (pilA4), long-isoform (pilA5), null (pilA7), and deletion (ΔpilA2) strains. The Omc distribution in samples from the ΔpilA2 and ΔpilA1 pilA deletion strains (48) were found to be comparable (unpublished observation). The protein concentrations in the loosely bound outer membrane preparations were determined with a bicinchoninic acid (BCA) assay, and 3 μg of protein was loaded into each lane.
Complementation of the short (pilA4)- and long (pilA5)-isoform phenotypes by trans expression of pilA.
To investigate the components required for full restoration of PilA protein expression and secretion, the plasmid pLC3 was first used to express the pilA gene in trans in the strains expressing either the short or long isoform. pLC3 contains the entire coding sequence of the pilA gene under the control of the native P1 and P2 promoters (Fig. 1A). Expression of the pilA gene from pLC3 restored the intracellular soluble and membrane-associated fractions of the PilA protein in the long-isoform strain, but no secreted protein was detected in either a strain expressing only the long isoform or only the short isoform complemented with pLC3 (Fig. 2A). These data provided evidence that plasmid-borne pilA was sufficient to restore the cellular expression of PilA, but not secretion.
Published microarray data collected from cells grown under various conditions, including biofilms harvested from current-producing versus non-current-producing electrodes (42) and a pilR mutant strain versus the wild type (22), had similar expression profiles for pilA and the genes downstream. The downstream gene GSU1497, encoding a hypothetical protein, and a gene cluster from GSU1498 to GSU1505, encoding ATP-dependent transporters, membrane proteins, a cell wall glycosylation enzyme, and other proteins of unknown function (Fig. 1A), were observed to have levels of expression similar to those of pilA. Biofilm cells harvested from the non-current-producing electrodes expressed pilA and the downstream genes at lower levels than cells harvested from current-producing electrodes, and the pilR mutant strain expressed pilA and the downstream genes at lower levels than the wild type grown under the same conditions (22, 42). Genes GSU1500 to GSU1505 were reported to function as an ATP-dependent exporter operon, and deletion of one gene, GSU1501, required the coexpression of the entire operon in trans to restore function (57). Furthermore, PilA was not detected on a Western blot of protein fractions prepared from a GSU1497 deletion strain (data not shown), which suggested that PilA expression, maturation, and/or stability required at least that one additional gene. Therefore, we tested the requirement for coexpressing pilA with nine genes downstream (GSU1497 to GSU1505) to fully complement the defect in pilA. pLC19, in which pilA expression was regulated by the native promoters (like pLC3), was constructed for this purpose, but also to carry the downstream genes (Fig. 1A). Secreted PilA was detected by Western blotting in protein samples prepared from the strain expressing only the short isoform (pilA4) but harboring pLC19 (pilA4 strain/pLC19) and also in samples isolated from the complemented long-isoform strain (pilA5 strain/pLC19) (Fig. 2A). Full complementation (expression and secretion) was not achieved for the strains expressing only the short or the long isoform harboring either of the intermediate plasmids, pLC17 or pLC18 (data not shown). These data were evidence of the requirement for the coexpression of pilA with GSU1497 and eight other genes downstream, in trans, to achieve full complementation of the chromosomal mutations in pilA.
The strain expressing only the short isoform displays diminished current production in microbial fuel cells.
The PilA-deficient strain was reported to have severely impaired current production in microbial fuel cells (54), but investigation of the distribution of the outer-surface cytochromes in PilA-deficient cells revealed an unexpected mislocalization of OmcZ (Fig. 3), an outer membrane cytochrome important for current production (21, 45). When considered with the observation that PilA modulated the distribution of outer membrane cytochromes (see above), the previous findings led us to question whether the reduction in current was due to the missing type IV pili, the mislocalization of OmcZ, or both (45, 54).
The strain expressing only the short isoform lacks secreted PilA and thus presumably the filamentous form of PilA, yet the strain has a wild-type-like distribution of the outer-surface cytochromes, including OmcZ, and at levels comparable to those in the wild-type strain (DL1) (Fig. 3). Therefore, the current produced by biofilms of the short-isoform strain was measured in a microbial fuel cell to assess the requirement for secreted PilA alone for current production. The results plotted in Fig. 4A show that current production by the short-isoform strain followed a different pattern than the wild type. Current production by the isogenic control strain (DL100) increased sharply and reached a maximum current of 15.0 ± 0.5 mA (Fig. 4A), which is typical of the wild-type strain (DL1) under the same experimental conditions (45). Thus, the kanamycin cassette did not interfere with secreted PilA function. In comparison, the maximum current produced by the strain expressing only the short isoform (10.4 ± 0.6 mA) was lower than that produced by DL100 (Fig. 4A) or DL1 (45), and it took significantly longer to achieve the maximum current (∼200 h) than for either DL100 (∼100 h) (Fig. 4A) or DL1 (45).
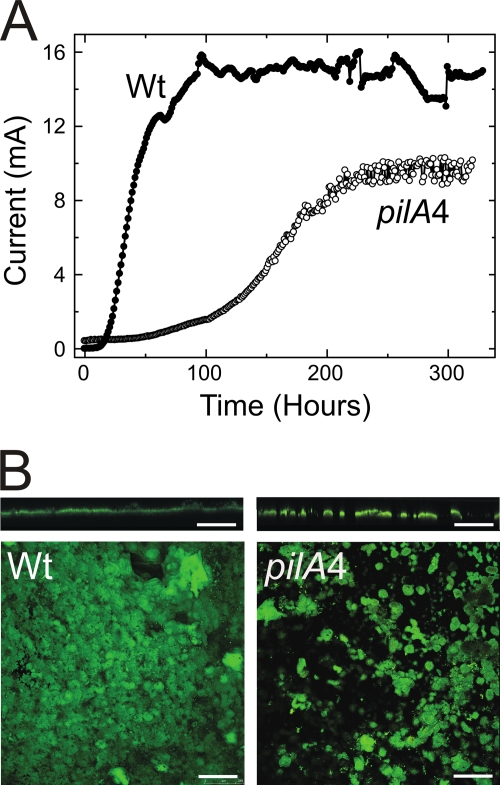
Fig 4.
Current production by the DL100 (Wt) and the short-isoform (pilA4) strains in microbial fuel cells. (A) Plot representative of data obtained with three biological replicates. The ΔpilA strain produces a maximum current of 1.2 mA under the same experimental conditions (45). (B) Confocal scanning micrographs of biofilms formed with the isogenic wild-type DL100 (Wt) and short-isoform (pilA4) strains on graphite anodes producing maximum current for 3 days in microbial fuel cells. Cross-sectional (top) and top-down (bottom) views are shown. Scale bars, 250 μm.
Confocal microscopy was used to assess the structures of biofilms formed by the wild type and mutants, which could account for the differences in current production. The biofilm characterization protocol has been previously described (39); briefly, after 3 days at maximum current, confocal microscope images were taken of biofilms from five random locations in each of three independent experimental samples for each strain (15 images per strain). The biofilm thickness and substrate coverage of the short-isoform and isogenic-control strains were determined. The results are summarized in Table 4. The anode biofilm formed with the short-isoform strain had a maximum thickness (72 μm) greater than that (55 μm) of the isogenic wild-type biofilm, but the short-isoform biofilm had less surface coverage than the wild type (38% versus 67%). Considered together, the average biofilm thicknesses were comparable, but the short-isoform biofilm had greater roughness than the wild type. These differences are reflected in the representative confocal images in Fig. 4B. The projection images show that the short-isoform biofilm covers a smaller fraction of the anode surface, and the cross-section images show that the short-isoform biofilm contains taller pillar structures with deeper channels between the pillars. These differences in biofilm morphology, especially the smaller percent coverage displayed by the strain expressing only the short isoform, provide an explanation for the difference in current.
Table 4.
Analysis of mature biofilms in microbial fuel cellsa
| Strain | Substrate coverage (%) | Thickness (μm) | Roughness | Maximum thickness (μm) |
|---|---|---|---|---|
| Wt (DL100) | 67 ± 7 | 44 ± 4 | 0.26 ± 0.07 | 55 |
| Short isoform (pilA4) | 38 ± 12 | 47 ± 13 | 0.49 ± 0.08 | 72 |
Biofilms were grown on graphite anodes poised at +300 mV (vs. Ag/AgCl) after 3 days at maximum current. The values are averages of triplicate samples ± standard deviations.
Type IV pili of G. sulfurreducens have been previously shown to be electrically conductive (53). Our observation that the maximum current at the anode is reduced when no PilA is secreted (Fig. 4A) is consistent with this property (45, 54, 56). However, a significant difference between the previous studies and the present work lies in the fact that the previous studies were conducted with mutants that lack type IV pili and have an aberrant distribution of outer membrane cytochromes (Fig. 3) while the strain expressing only the short isoform lacks type IV pili yet has a wild-type-like Omc distribution, so that OmcZ—a critical component of electron transfer within the biofilm bulk (21, 45, 56)—is still present (Fig. 3). Thus, the current production by the short-isoform strain—lower than the wild type but greater than the PilA-deficient strains studied previously (45, 54)—is plausible and can be explained by the absence of type IV pili. From investigations conducted with other Gram-negative species, it is known that type IV pili mediate cell attachment to surfaces (49, 70). The differences between the biofilms (Fig. 4B) generated with the wild type and strains expressing the short isoform are consistent with the different cell attachment phenotypes, and we suspect that the longer delay in current production in the absence of secreted PilA is a consequence of compromised attachment (see below). However, as biofilms do form with the strain expressing only the short isoform, it seems that secreted PilA, while important, is not an absolute requirement for cell attachment, biofilm growth, and current production.
Evaluation of the structural role of secreted PilA in biofilm formation.
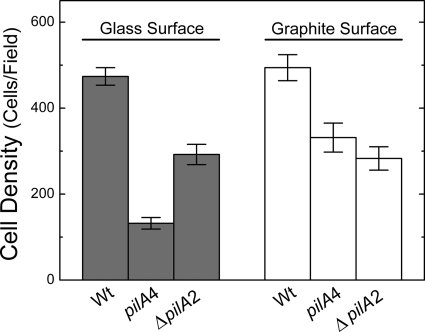
Type IV pili are known to be an important structural component for surface adhesion and colony formation (49, 59, 70). In G. sulfurreducens, type IV pili have been demonstrated to be essential for attachment to glass surfaces when a soluble electron acceptor was provided (55). We examined the strain expressing only the short isoform (pilA4) for the ability to attach to graphite and glass, because this strain is different from the PilA-deficient strain used previously (55): the outer surface of the short-isoform strain created for this study lacks only secreted PilA but seems to maintain the wild-type-like cytochrome composition (Fig. 3). The short-isoform and isogenic wild-type (DL100) strains were incubated under strict anaerobic conditions in freshwater medium, with acetate as the electron donor and fumarate as the electron acceptor, at the pilus-inducing temperature, 25°C, and in the presence of graphite or glass coverslips for 4 days. CLSM image stacks were collected from a minimum of five random locations on graphite or glass for each of three biological replicates within each experiment. Surface coverage was evaluated by calculating the cell density per field. The absence of secreted PilA had a negative impact on cell attachment regardless of the surface material. Cells expressing only the short isoform exhibited significantly less attachment to graphite by an average of 160 fewer cells per field than the wild-type (DL100) cells (unpaired t test; P < 0.01), which exhibited on average 490 ± 180 cells/field (Fig. 5). Also, the attachment of the short-isoform strain to glass was substantially less, by 340 cells/field, than that of DL100 (unpaired t test; P < 0.01), which exhibited on average 470 ± 130 cells/field (Fig. 5). These data are consistent with the published observations made with PilA- and PilR-deficient strains (24, 55). Taken together, these data provide further support for an active role for secreted PilA in surface colonization, even when the surface is not an electron acceptor.
Fig 5.
Average biomass of the mature biofilms formed on glass and graphite surfaces with the isogenic wild-type DL100 (Wt), short-isoform (pilA4), and ΔpilA2 strains. The results are the averages and standard errors of the mean from six biological replicates in two independent experiments.
Secreted PilA is important for growth on insoluble Fe(III) oxide.
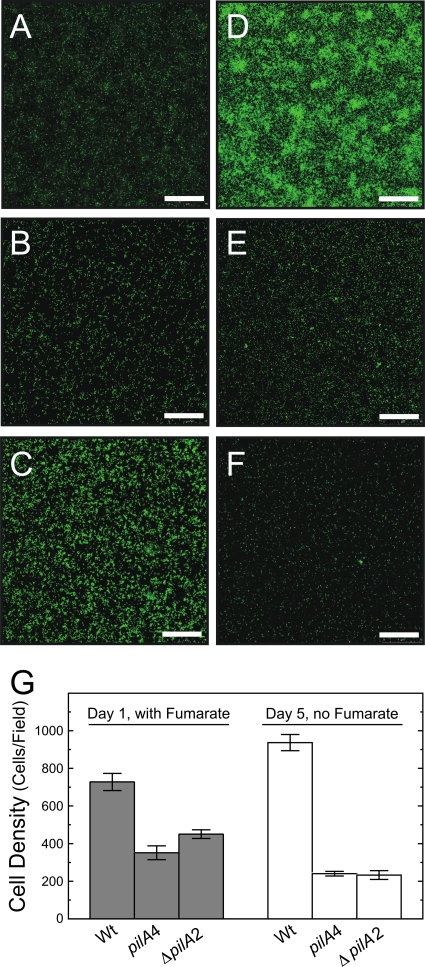
The strain expressing only the short-isoform preprotein (pilA4) was examined for the ability to grow on an insoluble electron acceptor. Cell growth was initiated on Fe(III) oxide-coated glass slides in the presence of a soluble electron donor and a soluble electron acceptor. After 24 h, the soluble acceptor was removed so that insoluble Fe(III) oxide was the only electron acceptor. The representative CLSM images in Fig. 6A to F and the average cell densities presented in Fig. 6G of the isogenic wild type (DL100), the strain expressing only the short isoform (pilA4), and the deletion strain (ΔpilA2) illustrate the similarities and differences in the attachment and growth of these strains on Fe(III) oxide-coated glass. Figure 6 shows that wild-type and mutant cells were capable of attachment to the Fe(III) oxide-coated glass when both a soluble electron donor and acceptor were provided (Fig. 6A to C). However, attachment was less by an average of 370 cells/field (unpaired t test; P < 0.01) than for DL100, which exhibited an average of 690 ± 200 cells/field (Fig. 6G, gray bars). This observation was consistent with earlier reports of a PilA-deficient strain that, while still able to access Fe(III) oxide particles (53), generated a biofilm on Fe(III) oxide-coated glass in the presence of soluble electron acceptor with a thickness ca. 50% smaller than those of the biofilms of wild-type cells (55). Significant growth was noted with the isogenic wild-type cells after removing fumarate from the medium and incubating the cells with Fe(III) oxide as the sole electron acceptor (compare Fig. 6A and D). This was quantified as an average increase of 230 cells per field (unpaired t test; P < 0.01) (Fig. 6G). In contrast, the short-isoform strain was not able to use Fe(III) oxide particles as an electron acceptor. No growth was detected in any of the six biological replicates; instead, the average number of cells/field decreased by 80 (unpaired t test; P < 0.05) (Fig. 6B, E, and G). The ΔpilA2 strain was similarly defective in growth in the absence of a soluble electron acceptor (Fig. 6C, F, and G). These data are consistent with the previous report for the PilA-deficient strain (55): the short-isoform strain, which does not secrete PilA, and the PilA-deficient strain are both incapable of using Fe(III) oxide as an electron acceptor.
Fig 6.
Confocal microscope images of biofilms formed with wild-type DL100 (A and D) and the short-isoform (pilA4) (B and E) and ΔpilA2 (C and F) strains on Fe(III) oxide-coated glass. The images were taken after 24 h in medium with 40 mM fumarate present (A, B, and C) and 4 days later after the fumarate was removed (D, E, and F). The scale bars represent 75 μm. (G) Average numbers of cells in biofilms of the isogenic DL100 (Wt) and short-isoform (pilA4) and ΔpilA2 strains formed on Fe(III) oxide-coated glass. Cell densities were measured after 24 h of incubation in the presence of 40 mM fumarate and 4 days later after the fumarate was removed. The results are the averages of six biological replicates from two independent experiments. The error bars are standard errors of the mean.
DISCUSSION
Type IV (PilA) pili of G. sulfurreducens are an essential component of the conductive anode biofilm, promoting cell-to-cell electron transfer through the 50-μm-thick biomass layer (18, 39, 45, 56, 63). The biofilms are electrically conductive, and the evidence is consistent with type IV pili functioning as the final electron conduit onto insoluble Fe(III) oxide (53). The 273-bp pilA gene encodes the pilus structural subunit protein PilA. pilA transcription is regulated by a two-component regulatory system composed of a histidine kinase, PilS, and the response regulator PilR, an RpoN-dependent enhancer-binding protein (24). The molecular analysis of pilA transcription revealed two start sites, a long RpoN-dependent transcript and a short RpoN-independent transcript (24). Each transcript was predicted to have different ribosome-binding sites and start codons, which could give rise to two PilA preprotein isoforms.
We tested the hypothesis that two PilA preprotein isoforms are generated and investigated what roles these two isoforms may have by generating mutant strains that eliminated one isoform at a time. When the translation start codon of the long isoform was mutated (TTG into CCC), no secreted PilA protein was detected by Western blotting developed with an anti-PilA antiserum. However, PilA protein was present in the intracellular and membrane fractions. These data demonstrate that the TTG start codon is functional and produces a preprotein isoform essential for PilA secretion and/or export to the surface of the cell. Heme-stained gels of the outer membrane cytochromes of the strain expressing only the short isoform (pilA4) revealed the wild-type-like composition of OmcS and OmcZ, which is necessary for efficient Fe(III) oxide reduction and electricity generation in microbial fuel cells (41, 45, 56). From these data, we can conclude that the short isoform, but not the long isoform, plays a significant role in generating this Omc distribution.
On the other hand, by disrupting expression of the short isoform to create a strain expressing only the long isoform (pilA5), the processing of both isoforms was eliminated, according to Western blot data. Despite the fact that the long isoform was still transcribed, no PilA protein was detected in any of the three cell fractions collected, indicating that expression of the short isoform preprotein either is necessary for translation of the long-isoform preprotein or contributes to the stability of the long-isoform preprotein. Consequently, the long-isoform strain behaves like a pilA deletion strain; specifically, OmcZ did not properly localize to the cell surface, which implies that this phenotype is due to the absence of the short isoform. OmcZ is a loosely bound outer membrane cytochrome reported to be missing in the extracellular matrix of G. sulfurreducens strains deficient in extracellular polysaccharide (EPS) production (57). In M. xanthus, the expression of extracellular polysaccharide was correlated with the expression of type IV pili. M. xanthus strains in which pilA was deleted exhibited severe reduction in EPS production (4, 73). Taking the data together, it is plausible to speculate that deletion of the pilA gene in G. sulfurreducens, in particular the short isoform, might play an indirect role in proper localization of OmcZ by regulating the expression or assembly of EPS.
Although expression of pilA from a plasmid (in trans) in the long-isoform strain complemented the defect in the nonsecreted soluble and membrane-bound fractions of PilA, coexpression of pilA with nine downstream genes is needed to restore PilA secretion. This requirement for full complementation suggests that a highly coordinated process involving several gene products is necessary for all the functions carried out by PilA in its various forms.
A PilA-deficient strain lacked proper localization of some c-type cytochromes on the outer cell surface—cytochromes that are important for extracellular electron transfer (45, 56). These observations prompted us to use the strain expressing only the short isoform to test for defects in cell attachment and growth on an insoluble electron acceptor. The strain expressing the short isoform alone lacks only type IV pili (it does not secrete PilA); the distribution of outer membrane cytochromes resembles that of the wild type. In contrast, ΔpilA strains are defective in pilus production and have an aberrant distribution of cytochromes. Therefore, we were able to study the role of type IV pili in cell attachment, biofilm formation, and extracellular electron transfer to insoluble electron acceptors without the additional influence of mislocalized cytochromes. The absence of type IV pili affected the cells' ability to attach to surfaces. The short-isoform strain exhibited a moderate decrease in attachment to graphite with the soluble electron donor and acceptor both present. Attachment to glass was substantially diminished under the same conditions. These data are consistent with the previous findings of Reguera et al. (55), who used a ΔpilA strain (with both isoforms deleted), and Juárez et al. (24), who used a ΔpilR strain (the long pilA transcript was eliminated). Both strains were deficient in attachment to glass (24, 55). Our data also indicate that the long pilA isoform, initiated at the TTG translation start codon, is necessary for robust attachment.
In microbial fuel cells, the strain expressing only the short isoform (pilA4) was able to form a conductive biofilm on the graphite anode. These biofilms cover a smaller portion of the anode surface than the isogenic wild type (DL100) and form taller cell pillars with deeper channels between them. Due to the altered structure of the biofilm, short-isoform cells are more limited in accessing the graphite anode. The maximum current produced by a mature biofilm of the strain expressing only the short isoform is 10.4 ± 0.6 mA (versus 15.0 ± 0.5 mA in a DL100 biofilm), which is attributed to the absence of type IV pili, as reported previously (18, 45, 56). However, the current observed in the biofilm of the short-isoform strain is greater than the currents observed with the ΔpilA strain (45, 54). This difference is plausibly attributed to aberrant distribution of OmcZ in the ΔpilA strain. Deletion of omcZ reduces the current production of cells in microbial fuel cells substantially, despite the fact that these cells possess pili in transmission electron microscope images (45). Also, CLSM images of the ΔomcZ current-producing biofilm reveal a reduction in biofilm thickness with changes in its morphology (no pillar structures were observed) (45). As OmcZ and PilA both contribute to biofilm structure and current production in biofilms of wild-type cells, it seems plausible that cells defective only in the production of pili—short-isoform strain cells—would generate more biofilm current than cells that are defective in the production of pili and in the distribution of OmcZ (ΔpilA strain cells).
G. sulfurreducens is incapable of growth on insoluble Fe(III) oxide when both pilA transcripts are disrupted, as in a ΔpilA strain, or when only the long transcript is absent, as in a pilR strain (24, 53, 55). Here, we have shown that cells expressing only the short isoform can attach to Fe(III)-coated glass, albeit to a lesser extent than the wild-type cells, but the short-isoform strain cannot grow when Fe(III) oxide is the sole electron acceptor. Taken together, these observations all serve to confirm the importance of type IV pili and the distribution of outer membrane cytochromes in extracellular electron transfer.
A molecular model for PilA function and type IV pilus formation.
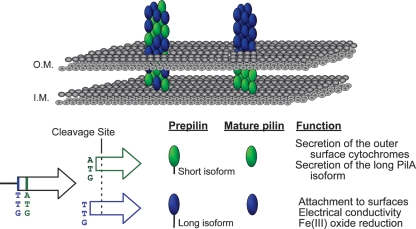
Our data support the presence of two functional translation start codons of the pilA gene that lead to two preprotein isoforms, the long isoform and the short isoform (Fig. 7). Both isoforms are cleaved at the same site (after glycine −1) once they reach the inner membrane, resulting in mature proteins with identical primary sequences. Production of the short-isoform preprotein seems to be required to secrete and/or stabilize mature PilA, as well as to influence the localization of the outer-surface c-type cytochromes. The long isoform seems to be required only for PilA secretion, which assembles into filaments. Coexpression of the pilA gene and the nine genes downstream is essential for proper secretion. This suggests that pilA expression, maturation, and assembly are coordinated processes involving not only the long- and short-isoform preproteins, but also several other gene products. A process of similar complexity has been documented in the formation of type IV pilus filament bundles in enteropathogenic E. coli (EPEC) (60). The long preprotein isoform is required for cell attachment to various surfaces, for optimal current production, and for extracellular electron transfer to insoluble Fe(III) oxides. These are all processes in which pilus filaments may participate directly and for which the coordinated expression, maturation, and assembly process is required.
Fig 7.
Two models of pilus assembly and functions of the PilA isoforms. The short and long isoforms are depicted in green and blue, respectively. Both isoforms are shown cleaved after glycine −1 once they reach the inner membrane (I.M.), generating identical mature proteins. Model I (right) suggests that the mature PilA derived from the short PilA isoform remains intracellular and forms the base of the pilus fiber anchoring the long-isoform-derived PilA in the cell membrane. Model II (left) suggests that secreted PilA is derived from both preprotein isoforms, and long- and short-isoform-derived PilA proteins are assembled into the intra- and extracellular regions of the pilus fiber. O.M., outer membrane.
An operational model that is consistent with our observations is shown in Fig. 7. Our data demonstrate that the PilA originating from the short-isoform preprotein is not secreted in the absence of the long-isoform preprotein, suggesting that it may be directed exclusively to intracellular and membrane-associated locations, such as the base of the pilus at the cell surface (Fig. 7, right). This scenario implies that only the long-isoform preprotein is targeted for secretion and/or pilus formation. On the other hand, we must also consider the possibility that the pili are comprised of mature PilA protein derived from both the short and long preprotein isoforms (Fig. 7, left). The long preprotein isoform is still a requirement for pilus formation (and secretion), so in its absence no secreted PilA is observed. However, pilus formation permits the incorporation of both long and the short isoforms. Additional studies that can discriminate between the fates of the short- and long-isoform preproteins are warranted.
ACKNOWLEDGMENTS
We thank Ashley Franks for assistance with confocal microscopy and Hanno Richter and Muktak Aklujkar for critical reading of the manuscript.
The U.S. Department of Energy Office of Science (BER) supported this research under Cooperative Agreement no. DEFC02-02ER63446.
Footnotes
Published ahead of print 9 March 2012
REFERENCES
- 1. Alm RA, Mattick JS. 1997. Genes involved in the biogenesis and function of type-4 fimbriae in Pseudomonas aeruginosa. Gene 192:89–98 [DOI] [PubMed] [Google Scholar]
- 2. Anderson RT, et al. 2003. Stimulating the in situ activity of Geobacter species to remove uranium from the groundwater of a uranium-contaminated aquifer. Appl. Environ. Microbiol. 69:5884–5891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Arts J, van Boxtel R, Filloux A, Tommassen J, Koster M. 2007. Export of the pseudopilin XcpT of the Pseudomonas aeruginosa type II secretion system via the signal recognition particle-Sec pathway. J. Bacteriol. 189:2069–2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Black WP, Xu Q, Yang Z. 2006. Type IV pili function upstream of the Dif chemotaxis pathway in Myxococcus xanthus EPS regulation. Mol. Microbiol. 61:447–456 [DOI] [PubMed] [Google Scholar]
- 5. Bond DR, Holmes DE, Tender LM, Lovley DR. 2002. Electrode-reducing microorganisms that harvest energy from marine sediments. Science 295:483–485 [DOI] [PubMed] [Google Scholar]
- 6. Bond DR, Lovley DR. 2003. Electricity production by Geobacter sulfurreducens attached to electrodes. Appl. Environ. Microbiol. 69:1548–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Boyd JM, Lory S. 1996. Dual function of PilS during transcriptional activation of the Pseudomonas aeruginosa pilin subunit gene. J. Bacteriol. 178:831–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Butler JE, Kaufmann F, Coppi MV, Nunez C, Lovley DR. 2004. MacA, a diheme c-type cytochrome involved in Fe(III) reduction by Geobacter sulfurreducens. J. Bacteriol. 186:4042–4045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Caccavo F, Jr, et al. 1994. Geobacter sulfurreducens sp. nov., a hydrogen- and acetate-oxidizing dissimilatory metal-reducing microorganism. Appl. Environ. Microbiol. 60:3752–3759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Carbonnelle E, Helaine S, Nassif X, Pelicic V. 2006. A systematic genetic analysis in Neisseria meningitidis defines the Pil proteins required for assembly, functionality, stabilization and export of type IV pili. Mol. Microbiol. 61:1510–1522 [DOI] [PubMed] [Google Scholar]
- 11. Carbonnelle E, Helaine S, Prouvensier L, Nassif X, Pelicic V. 2005. Type IV pilus biogenesis in Neisseria meningitidis: PilW is involved in a step occurring after pilus assembly, essential for fibre stability and function. Mol. Microbiol. 55:54–64 [DOI] [PubMed] [Google Scholar]
- 12. Collins RF, et al. 2005. Interaction with type IV pili induces structural changes in the bacterial outer membrane secretin PilQ. J. Biol. Chem. 280:18923–18930 [DOI] [PubMed] [Google Scholar]
- 13. Coppi MV, Leang C, Sandler SJ, Lovley DR. 2001. Development of a genetic system for Geobacter sulfurreducens. Appl. Environ. Microbiol. 67:3180–3187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Craig L, et al. 2006. Type IV pilus structure by cryo-electron microscopy and crystallography: implications for pilus assembly and functions. Mol. Cell 23:651–662 [DOI] [PubMed] [Google Scholar]
- 15. Finneran KT, Anderson RT, Nevin KP, Lovley DR. 2002. Potential for bioremediation of uranium-contaminated aquifers with microbial U(VI) reduction. Soil Sediment Contam. 11:339–357 [Google Scholar]
- 16. Francetic O, Buddelmeijer N, Lewenza S, Kumamoto CA, Pugsley AP. 2007. Signal recognition particle-dependent inner membrane targeting of the PulG pseudopilin component of a type II secretion system. J. Bacteriol. 189:1783–1793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Francis RT, Jr, Becker RR. 1984. Specific indication of hemoproteins in polyacrylamide gels using a double-staining process. Anal. Biochem. 136:509–514 [DOI] [PubMed] [Google Scholar]
- 18. Franks AE, Nevin KP, Glaven RH, Lovley DR. 2010. Microtoming coupled to microarray analysis to evaluate the spatial metabolic status of Geobacter sulfurreducens biofilms. ISME J. 4:509–519 [DOI] [PubMed] [Google Scholar]
- 19. Hobbs M, Mattick JS. 1993. Common components in the assembly of type 4 fimbriae, DNA transfer systems, filamentous phage and protein-secretion apparatus: a general system for the formation of surface-associated protein complexes. Mol. Microbiol. 10:233–243 [DOI] [PubMed] [Google Scholar]
- 20. Holmes DE, Finneran KT, O'Neil RA, Lovley DR. 2002. Enrichment of members of the family Geobacteraceae associated with stimulation of dissimilatory metal reduction in uranium-contaminated aquifer sediments. Appl. Environ. Microbiol. 68:2300–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Inoue K, et al. 2010. Purification and characterization of OmcZ, an outer-surface, octaheme c-type cytochrome essential for optimal current production by Geobacter sulfurreducens. Appl. Environ. Microbiol. 76:3999–4007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ishimoto KS, Lory S. 1992. Identification of pilR, which encodes a transcriptional activator of the Pseudomonas aeruginosa pilin gene. J. Bacteriol. 174:3514–3521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jin S, Ishimoto KS, Lory S. 1994. PilR, a transcriptional regulator of piliation in Pseudomonas aeruginosa, binds to a cis-acting sequence upstream of the pilin gene promoter. Mol. Microbiol. 14:1049–1057 [DOI] [PubMed] [Google Scholar]
- 24. Juarez K, et al. 2009. PilR, a transcriptional regulator for pilin and other genes required for Fe(III) reduction in Geobacter sulfurreducens. J. Mol. Microbiol. Biotechnol. 16:146–158 [DOI] [PubMed] [Google Scholar]
- 25. Kim BC, et al. 2005. OmcF, a putative c-Type monoheme outer membrane cytochrome required for the expression of other outer membrane cytochromes in Geobacter sulfurreducens. J. Bacteriol. 187:4505–4513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kim BC, Qian X, Leang C, Coppi MV, Lovley DR. 2006. Two putative c-type multiheme cytochromes required for the expression of OmcB, an outer membrane protein essential for optimal Fe(III) reduction in Geobacter sulfurreducens. J. Bacteriol. 188:3138–3142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kirn TJ, Bose N, Taylor RK. 2003. Secretion of a soluble colonization factor by the TCP type 4 pilus biogenesis pathway in Vibrio cholerae. Mol. Microbiol. 49:81–92 [DOI] [PubMed] [Google Scholar]
- 28. Klimes A, et al. 2010. Production of pilus-like filaments in Geobacter sulfurreducens in the absence of the type IV pilin protein PilA. FEMS Microbiol. Lett. 310:62–68 [DOI] [PubMed] [Google Scholar]
- 29. Lauer P, Albertson NH, Koomey M. 1993. Conservation of genes encoding components of a type IV pilus assembly/two-step protein export pathway in Neisseria gonorrhoeae. Mol. Microbiol. 8:357–368 [DOI] [PubMed] [Google Scholar]
- 30. Leang C, Coppi MV, Lovley DR. 2003. OmcB, a c-type polyheme cytochrome, involved in Fe(III) reduction in Geobacter sulfurreducens. J. Bacteriol. 185:2096–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Leang C, Lovley DR. 2005. Regulation of two highly similar genes, omcB and omcC, in a 10 kb chromosomal duplication in Geobacter sulfurreducens. Microbiology 151:1761–1767 [DOI] [PubMed] [Google Scholar]
- 32. Lovley DR. 2006. Bug juice: harvesting electricity with microorganisms. Nat. Rev. Microbiol. 4:497–508 [DOI] [PubMed] [Google Scholar]
- 33. Lovley DR, et al. 1989. Oxidation of aromatic contaminants coupled to microbial iron reduction. Nature 339:297–300 [Google Scholar]
- 34. Lovley DR, Coates JD, Blunt-Harris EL, Phillips EJP, Woodward JC. 1996. Humic substances as electron acceptors for microbial respiration. Nature 382:445–447 [Google Scholar]
- 35. Lovley DR, et al. 1993. Geobacter metallireducens gen. nov. sp. nov., a microorganism capable of coupling the complete oxidation of organic compounds to the reduction of iron and other metals. Arch. Microbiol. 159:336–344 [DOI] [PubMed] [Google Scholar]
- 36. Lovley DR, Holmes DE, Nevin KP. 2004. Dissimilatory Fe(III) and Mn(IV) reduction. Adv. Microb. Physiol. 49:219–286 [DOI] [PubMed] [Google Scholar]
- 37. Lovley DR, Phillips EJP, Gorby YA, Landa ER. 1991. Microbial reduction of uranium. Nature 350:413–416 [Google Scholar]
- 38. Lovley DR, Phillips EJ. 1988. Novel mode of microbial energy metabolism: organic carbon oxidation coupled to dissimilatory reduction of iron or manganese. Appl. Environ. Microbiol. 54:1472–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Malvankar NS, et al. 2011. Tunable metallic-like conductivity in microbial nanowire networks. Nat. Nanotechnol. 6:573–579 [DOI] [PubMed] [Google Scholar]
- 40. Mattick JS. 2002. Type IV pili and twitching motility. Annu. Rev. Microbiol. 56:289–314 [DOI] [PubMed] [Google Scholar]
- 41. Mehta T, Coppi MV, Childers SE, Lovley DR. 2005. Outer membrane c-type cytochromes required for Fe(III) and Mn(IV) oxide reduction in Geobacter sulfurreducens. Appl. Environ. Microbiol. 71:8634–8641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Methe BA, et al. 2003. Genome of Geobacter sulfurreducens: metal reduction in subsurface environments. Science 302:1967–1969 [DOI] [PubMed] [Google Scholar]
- 43. Morales VM, Backman A, Bagdasarian M. 1991. A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene 97:39–47 [DOI] [PubMed] [Google Scholar]
- 44. Mueller LN, de Brouwer JF, Almeida JS, Stal LJ, Xavier JB. 2006. Analysis of a marine phototrophic biofilm by confocal laser scanning microscopy using the new image quantification software PHLIP. BMC Ecol. 6:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nevin KP, et al. 2009. Anode biofilm transcriptomics reveals outer surface components essential for high density current production in Geobacter sulfurreducens fuel cells. PLoS One 4:e5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nevin KP, et al. 2008. Power output and columbic efficiencies from biofilms of Geobacter sulfurreducens comparable to mixed community microbial fuel cells. Environ. Microbiol. 10:2505–2514 [DOI] [PubMed] [Google Scholar]
- 47. North NN, et al. 2004. Change in bacterial community structure during in situ biostimulation of subsurface sediment cocontaminated with uranium and nitrate. Appl. Environ. Microbiol. 70:4911–4920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ortiz-Bernad I, Anderson RT, Vrionis HA, Lovley DR. 2004. Vanadium respiration by Geobacter metallireducens: novel strategy for in situ removal of vanadium from groundwater. Appl. Environ. Microbiol. 70:3091–3095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. O'Toole GA, Kolter R. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295–304 [DOI] [PubMed] [Google Scholar]
- 50. Paranjpye RN, Strom MS. 2005. A Vibrio vulnificus type IV pilin contributes to biofilm formation, adherence to epithelial cells, and virulence. Infect. Immun. 73:1411–1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Qian X, et al. 2011. Biochemical characterization of purified OmcS, a c-type cytochrome required for insoluble Fe(III) reduction in Geobacter sulfurreducens. Biochim. Biophys. Acta 1807:404–412 [DOI] [PubMed] [Google Scholar]
- 52. Ramer SW, et al. 2002. The type IV pilus assembly complex: biogenic interactions among the bundle-forming pilus proteins of enteropathogenic Escherichia coli. J. Bacteriol. 184:3457–3465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Reguera G, et al. 2005. Extracellular electron transfer via microbial nanowires. Nature 435:1098–1101 [DOI] [PubMed] [Google Scholar]
- 54. Reguera G, et al. 2006. Biofilm and nanowire production leads to increased current in Geobacter sulfurreducens fuel cells. Appl. Environ. Microbiol. 72:7345–7348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Reguera G, Pollina RB, Nicoll JS, Lovley DR. 2007. Possible nonconductive role of Geobacter sulfurreducens pilus nanowires in biofilm formation. J. Bacteriol. 189:2125–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Richter H, et al. 2009. Cyclic voltammetry of biofilms of wild type and mutant Geobacter sulfurreducens on fuel cell anodes indicates possible roles of OmcB, OmcZ, type IV pili, and protons in extracellular electron transfer. Energy Environ. Sci. 2:506–516 [Google Scholar]
- 57. Rollefson JB, Stephen CS, Tien M, Bond DR. 2011. Identification of an extracellular polysaccharide network essential for cytochrome anchoring and biofilm formation in Geobacter sulfurreducens. J. Bacteriol. 193:1023–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Rytkonen A, Johansson L, Asp V, Albiger B, Jonsson AB. 2001. Soluble pilin of Neisseria gonorrhoeae interacts with human target cells and tissue. Infect. Immun. 69:6419–6426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Shi W, Ngok FK, Zusman DR. 1996. Cell density regulates cellular reversal frequency in Myxococcus xanthus. Proc. Natl. Acad. Sci. U. S. A. 93:4142–4146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Stone KD, Zhang HZ, Carlson LK, Donnenberg MS. 1996. A cluster of fourteen genes from enteropathogenic Escherichia coli is sufficient for the biogenesis of a type IV pilus. Mol. Microbiol. 20:325–337 [DOI] [PubMed] [Google Scholar]
- 61. Strom MS, Lory S. 1991. Amino acid substitutions in pilin of Pseudomonas aeruginosa. Effect on leader peptide cleavage, amino-terminal methylation, and pilus assembly. J. Biol. Chem. 266:1656–1664 [PubMed] [Google Scholar]
- 62. Strom MS, Lory S. 1992. Kinetics and sequence specificity of processing of prepilin by PilD, the type IV leader peptidase of Pseudomonas aeruginosa. J. Bacteriol. 174:7345–7351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Strycharz-Glaven SM, Snider RM, Guiseppi-Elie A, Tender LM. 2011. On the electrical conductivity of microbial nanowires and biofilms. Energy Environ. Sci. doi:10.1039/c1ee01753e [Google Scholar]
- 64. Thomas PE, Ryan D, Levin W. 1976. An improved staining procedure for the detection of the peroxidase activity of cytochrome P-450 on sodium dodecyl sulfate polyacrylamide gels. Anal. Biochem. 75:168–176 [DOI] [PubMed] [Google Scholar]
- 65. van Schie PM, Fletcher M. 1999. Adhesion of biodegradative anaerobic bacteria to solid surfaces. Appl. Environ. Microbiol. 65:5082–5088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Voisin S, et al. 2007. Glycosylation of Pseudomonas aeruginosa strain Pa5196 type IV pilins with mycobacterium-like alpha-1,5-linked d-Araf oligosaccharides. J. Bacteriol. 189:151–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Voordeckers JW, Kim BC, Izallalen M, Lovley DR. 2010. Role of Geobacter sulfurreducens outer-surface c-type cytochromes in the reduction of soil humic acid and anthraquinone-2,6-disulfonate. Appl. Environ. Microbiol. 76:2371–2375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Vrionis HA, et al. 2005. Microbiological and geochemical heterogeneity in an in situ uranium bioremediation field site. Appl. Environ. Microbiol. 71:6308–6318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wall D, Wu SS, Kaiser D. 1998. Contact stimulation of Tgl and type IV pili in Myxococcus xanthus. J. Bacteriol. 180:759–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ward MJ, Zusman DR. 1999. Motility in Myxococcus xanthus and its role in developmental aggregation. Curr. Opin. Microbiol. 2:624–629 [DOI] [PubMed] [Google Scholar]
- 71. Wolfgang M, van Putten JP, Hayes SF, Dorward D, Koomey M. 2000. Components and dynamics of fiber formation define a ubiquitous biogenesis pathway for bacterial pili. EMBO J. 19:6408–6418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wu SS, Wu J, Cheng YL, Kaiser D. 1998. The pilH gene encodes an ABC transporter homologue required for type IV pilus biogenesis and social gliding motility in Myxococcus xanthus. Mol. Microbiol. 29:1249–1261 [DOI] [PubMed] [Google Scholar]
- 73. Yang Z, Lux R, Hu W, Hu C, Shi W. 2010. PilA localization affects extracellular polysaccharide production and fruiting body formation in Myxococcus xanthus. Mol. Microbiol. 76:1500–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Yi H, et al. 2009. Selection of a variant of Geobacter sulfurreducens with enhanced capacity for current production in microbial fuel cells. Biosens. Bioelectron. 24:3498–3503 [DOI] [PubMed] [Google Scholar]