Abstract
Glutamine synthetase (GS) and glucose-6-phosphate isomerase (GPI) were identified as novel adhesive moonlighting proteins of Lactobacillus crispatus ST1. Both proteins were bound onto the bacterial surface at acidic pHs, whereas a suspension of the cells to pH 8 caused their release into the buffer, a pattern previously observed with surface-bound enolase and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) of L. crispatus. The pH shift was associated with a rapid and transient increase in cell wall permeability, as measured by cell staining with propidium iodide. A gradual increase in the release of the four moonlighting proteins was also observed after the treatment of L. crispatus ST1 cells with increasing concentrations of the antimicrobial cationic peptide LL-37, which kills bacteria by disturbing membrane integrity and was here observed to increase the cell wall permeability of L. crispatus ST1. At pH 4, the fusion proteins His6-GS, His6-GPI, His6-enolase, and His6-GAPDH showed localized binding to cell division septa and poles of L. crispatus ST1 cells, whereas no binding to Lactobacillus rhamnosus GG was detected. Strain ST1 showed a pH-dependent adherence to the basement membrane preparation Matrigel. Purified His6-GS and His6-GPI proteins bound to type I collagen, and His6-GS also bound to laminin, and their level of binding was higher at pH 5.5 than at pH 6.5. His6-GS also expressed a plasminogen receptor function. The results show the strain-dependent surface association of moonlighting proteins in lactobacilli and that these proteins are released from the L. crispatus surface after cell trauma, under conditions of alkaline stress, or in the presence of the antimicrobial peptide LL-37 produced by human cells.
INTRODUCTION
Moonlighting proteins are characterized by their multiple autonomous functions, which are biologically unrelated and often localize to separate cellular compartments. The independent functions are not partitioned into different protein domains, indicating that the moonlighting proteins have not evolved through gene fusions but rather through modification and adaptation within one polypeptide chain. Structure analyses have provided evidence that moonlighting proteins utilize separate protein surfaces for their multiple functions (26, 29).
Moonlighting proteins have been detected in plants, animals, yeast, as well as prokaryotes, and their functions are involved in a range of biologically important processes. Research on bacterial moonlighting proteins has focused on their role in bacterial pathogenesis, and several moonlighting proteins indeed have a role in the virulence of important human pathogens, such as Streptococcus pyogenes, Streptococcus pneumoniae, and Staphylococcus aureus (24). Many of the identified moonlighting proteins localize to the bacterial surface but were originally identified as cytoplasmic enzymes of the glycolytic pathway or as having other metabolic functions, or they are molecular chaperones. The identified moonlighting functions include adhesion to host epithelia, extracellular matrices (ECMs), and/or secreted mucins as well as the engagement of the host proteolytic plasminogen (Plg) system and the modulation of host immune responses (24). Moonlighting proteins appear to be common in bacteria, and they have been identified in commensal bacteria as well. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and enolase of Lactobacillus crispatus were detected in the cytoplasm, on the cell surface, and released into cell-free buffer, and these proteins bind plasminogen and enhance its activation by human physiological plasminogen activators (27). Subsequently, GAPDH and enolase were found on the surface of Lactobacillus plantarum (16, 32, 51) and Lactobacillus jensenii (54) cells, where they have adhesive functions. Other adhesive moonlighting proteins detected in lactobacilli include elongation factor Tu, triosephosphate isomerase, the heat shock protein GroEL, DnaK, and pyruvate kinase (11, 15, 21, 31, 48).
The bacterial moonlighting proteins were originally described as being “anchorless” because their sequences do not contain known sequence motifs for surface anchoring, nor do the protein sequences contain identified secretion signals (46). Lactic acid bacteria are efficient producers of lactate and rapidly acidify their environment down to pH 4. GAPDH, enolase, and most other moonlighting proteins of Gram-positive bacteria have pIs of around 5 (4, 61). Thus, enolase and GAPDH have a positive net charge at an acidic pH that prevails in the natural niches of L. crispatus, e.g., the human vagina and the human and chicken intestines. In L. crispatus strain ST1, enolase and GAPDH are bound to the L. crispatus ST1 cell surface at acidic pH, but at a neutral or slightly alkaline pH or in the presence of high salt concentrations, these proteins are released into buffer (4). The pH-induced release is rapid and not diminished by chloramphenicol, nor are there transcriptional differences in the enolase and GAPDH genes of L. crispatus cells at pH 5 and pH 8 (4), which indicates that the increased release does not require de novo protein synthesis. Enolase and GAPDH of L. crispatus also bind to acidic lipoteichoic acids (LTAs) at low pH but not at neutral pH, which suggests that LTAs play a role in anchoring these proteins to the bacterial cell surface via ionic interactions (4). The pH-dependent expression of GAPDH on the cell surface of Streptococcus gordonii was also reported previously (44).
The mechanism(s) of how bacterial moonlighting proteins translocate to the cell exterior has remained unknown. They can be released from dead or damaged cells and then bind to neighboring cells, or they can be secreted onto the cell surface by an as-yet-undescribed mechanism (24). Indirect evidence for both hypotheses has been described. The spontaneous lysis of Bacillus subtilis cells at the stationary growth phase leads to a leakage of as much as 5% of the activity of isocitrate dehydrogenase, a cytoplasmic enzyme marker (55). On the other hand, several moonlighting proteins have been identified in the growth medium of B. subtilis during the stationary growth phase, and secretion was argued on the basis of the concomitant decrease of intracellular carboxylesterase and increase in the medium (62). The process was not inhibited by chloramphenicol or a proton motive force inhibitor. In L. plantarum, cell-surface-associated GAPDH activity was increased at the stationary growth phase, where plasma membrane permeability was also increased, which suggested a connection between membrane permeability and the efflux of GAPDH (51). Other types of stress, such as iron starvation, also increase the release of GAPDH from S. pyogenes (19) and S. gordonii cell surfaces (44). The growth of Bifidobacterium animalis subsp. lactis in the presence of bile salts led to higher levels of the DnaK protein and plasminogen activation on the bacterial surface (15). The binding of extracellular moonlighting proteins from buffer onto the bacterial surface has been described. Enolase and GAPDH bound at acidic pH to the surface of L. crispatus from buffer (4), and pneumococcal enolase binds to S. pneumoniae cells at a neutral pH (10). Supporting the secretion hypothesis, the genetic modification of GAPDH and enolase has been found to prevent their translocation to the cell surface. The genetic fusion of a C-terminal hydrophobic tail of 12 amino acids to GAPDH prevented its cell surface export in S. pyogenes (12), and the deletion of a central hydrophobic α-helical domain of 19 amino acids abolished the surface translocation of enolase in B. subtilis (62). An accessory secA2 gene in Listeria monocytogenes is involved in the secretion of enolase and several other proteins (38), and a homologous gene is present in pathogenic Gram-positive bacteria, including streptococci, but is lacking in the lactobacilli studied here. Taken together, our knowledge of the translocation and surface anchoring of bacterial moonlighting proteins remains fragmentary, and it is not known whether several translocation and/or anchoring mechanisms are used by different bacterial species and/or for different moonlighting proteins.
Cathelicidins are antimicrobial, highly cationic peptides that are important effector molecules of innate immunity and are produced mainly by phagocytes and epithelial cells (56). Humans have one cathelicidin protein, hCAP-18, which is processed by serine proteases into the antimicrobial peptide LL-37. hCAP-18/LL-37 is an integral part of the human innate immune system and is present in leukocytes; various epithelial cells, including colonic and vaginal cells; as well as body fluids, such as saliva and wound fluids (63). LL-37 binds to negatively charged targets in the bacterial cell wall and the cytoplasmic membrane and is effective against both Gram-positive and Gram-negative bacteria. LL-37 is thought to kill bacteria by disrupting membrane integrity, and a recent study suggested that LL-37 also halts bacterial growth by affecting cell wall biogenesis (53). At the molecular level, both functions are indicative of specific interactions between LL-37 and lipid molecules.
Here, we describe glutamine synthetase (GS) and glucose-6-phosphate isomerase (GPI) as novel adhesive moonlighting enzymes of L. crispatus that associate on the cell surface at an acidic pH. We also show that LL-37 enhances the release of moonlighting proteins from the L. crispatus cell surface, which indicates that the surface architecture of L. crispatus is modified upon contact with the central innate immunity system of humans.
MATERIALS AND METHODS
Bacterial strains and protein extraction.
Lactobacillus crispatus ST1 isolated from chicken crop (18, 45) and the probiotic strain Lactobacillus rhamnosus GG (30) were cultivated for 16 to 18 h at 37°C in static De Man, Rogosa, and Sharpe (MRS) broth (Difco Laboratories). For analysis of the buffer-released extracellular proteins, the bacteria were collected and suspended without washing in 50 mM Tris-HCl buffer at pH 4 or at pH 8. Cells were removed by centrifugation, and the supernatant was filtered through a 0.2-μm membrane to remove any remaining cells (27).
Cloning, overproduction, and purification of GS and GPI.
The bacteria were treated with lysozyme (20 mg/ml; Sigma-Aldrich) and mutanolysin (50 U/ml; Roche Diagnostics) for 1 h, and chromosomal DNA was then extracted by using the Qiagen Genomic Tip 20/G system (Qiagen) according to the manufacturer's instructions. The DNA primers used to amplify the GS-encoding gene glnA and the GPI-encoding gene pgi of L. crispatus ST1 and L. rhamnosus GG as well as the eno and the gap genes of L. rhamnosus GG were designed on the basis of available genomic sequences (30, 45). The correct nucleotide sequences of the amplicons were verified by DNA sequencing. The eno and gap genes of L. crispatus ST1 were described previously (27). The genes were cloned into the pQE-30 expression vector plasmid (Qiagen) for expression as His6 fusion proteins in Escherichia coli M15, and the His6-tagged proteins were purified under nondenaturing conditions and dialyzed against phosphate-buffered saline (PBS) (pH 7.1), as described previously (27).
Subcellular locations of GS and GPI.
Antisera against purified His6-GS and His6-GPI of L. crispatus ST1 and L. rhamnosus GG as well as His6-enolase and His6-GAPDH of L. rhamnosus GG were raised in rabbits by use of routine immunization procedures (Medprobe). Immunoglobulin G (IgG) from hyperimmune sera was purified by affinity chromatography using protein A-Sepharose CL-4B (Pharmacia LKB Biotechnology). IgGs raised against His6-enolase and His6-GAPDH of L. crispatus ST1 were available from a previous study (27). We stained the L. crispatus ST1 population with immunoglobulins using indirect immunofluorescence as described previously (4). Briefly, L. crispatus ST1 cells (109 cells/ml) were washed once with 50 mM Tris-HCl (pH 4) and fixed with 3.5% (wt/vol) paraformaldehyde in PBS (pH 4.0) prior detection with anti-His6-enolase, anti-His6-GAPDH, anti-His6-GS, and anti-His6-GPI IgGs (0.2 mg/ml) as primary antibodies and Alexa-488 (Invitrogen)-conjugated anti-rabbit IgG (1 μg/ml) as a secondary antibody. The bacteria were then examined with an epifluorescence microscope (Olympus BX50) equipped with a filter for Alexa-488 (excitation at 450 to 490 nm and emission at 515 nm), and the images were digitally recorded by using the Image-Pro Plus program (Media Cybernetics).
For studying the release of GS, GPI, enolase, and GAPDH from the cell surface, 1010 cells/ml were incubated for 1 h in the pH 4 and pH 8 buffers, and the cells and the corresponding supernatants were collected by centrifugation. Surface-attached proteins were extracted by boiling the cell pellet for 1 min in reducing sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer (4). The peptides from the cell surface and in the supernatants were compared by 12% (wt/vol) SDS-PAGE and Western blotting by analyzing samples representing the same culture volume.
PI staining.
To visualize the viability of the bacterial cells, bacteria were treated for 10 min with 0.075 μM propidium iodide (PI; Invitrogen) in a dark room and examined with an epifluorescence microscope with a filter for PI (excitation at 510 to 550 nm and emission at 590 nm) (7). The mean number of PI-positive cells in 10 randomly chosen microscopic fields of 3.6 × 104 μm2 was counted, and the assay was independently repeated three times.
Cell treatment with the cathelicidin LL-37.
To detect the effect of LL-37 on the release of GS, GPI, enolase, and GAPDH from L. crispatus ST1, 2.3 × 109 cells were incubated for 2 h in 50 mM Tris-HCl (pH 8.0) (total volume, 500 μl) containing synthetic LL-37 (Peptide 2.0 Inc.) at concentrations ranging from 0 to 16 μM. The cells and supernatant fractions were separated, and the bacteria were stained with 0.075 μM propidium iodide (7). The numbers of stained and unstained cells in 10 randomly selected microscopic fields of 3.6 × 104 μm2 were calculated, and the number of viable cells was also assessed by plating. The amounts of GS, GPI, enolase, and GAPDH in the filtered supernatant fractions from 108 LL-37-treated cells and, for comparison, from lysed nontreated cells (5 × 107 cells) were analyzed by Western blotting using specific antibodies. To lyse L. crispatus cells, 5 × 109 cells were treated with lysozyme (20 mg/ml; Sigma-Aldrich) and mutanolysin (50 U/ml; Roche Diagnostics) in 50 mM Tris-HCl buffer (pH 8.0) in a volume of 1 ml for 2 h at 37°C. The cells were then lysed by boiling for 10 min.
GPI enzyme assay.
The GPI enzymatic activity in the buffer extract as well as that of His6-GPI of L. crispatus ST1 were measured as described previously by Mathur and Garg (40). Briefly, the assay was performed at room temperature with 50 mM Tris-HCl buffer (pH 8.0) containing 0.5 mM NADP+, 1 mM fructose-6-phosphate, and 0.5 U glucose-6-phosphate dehydrogenase in a final volume of 50 μl. The reaction was initiated by addition of the buffer extract from 108 cells, and the increase in the NADPH concentration was measured spectrophotometrically at 340 nm.
Protein chemistry.
The GS/GPI-containing fraction extracted from L. crispatus ST1 cells was electrophoresed on SDS-PAGE gels and transferred by blotting onto a polyvinylidene difluoride (PVDF) membrane. The peptides were excised, and the N-terminal sequencing of the peptides was performed at the Microchemical Facility, The Babraham Institute, Cambridge, United Kingdom. The obtained sequences were blasted against the genome sequence of L. crispatus ST1 (45).
Adherence and binding assays.
The adherence of L. crispatus ST1 cells to the basement membrane (BM) preparation Matrigel (BD Biosciences) was tested as described previously (60). Briefly, Matrigel (diluted 1:10 in PBS) and bovine serum albumin (BSA) (25 μg/ml) were coated onto diagnostic slides overnight at 22°C in a moist chamber. L. crispatus ST1 cells were washed with 50 mM Tris-HCl (pH 4 or 8), and the same buffer was used for the subsequent adhesion, washing, and staining procedures. The bacteria were tested at a concentration of 109 cells/ml, and incubation was performed for 2 h at 20°C. After washing, adherent bacteria were stained with methylene blue, and the slides were examined by light microscopy. Adherent bacteria on 20 randomly chosen microscopic fields of 1.6 × 104 μm2 were counted.
For binding assays with His6 fusion proteins (20 μg/ml), the ECM proteins laminin (10 μg/ml for GS binding and 200 μg/ml for GPI binding; Sigma-Aldrich), collagen I (1 μg/ml; Sigma-Aldrich), collagen IV (200 μg/ml; Sigma-Aldrich), and fibronectin (200 μg/ml; BD Biosciences) were immobilized on a CM5 sensor chip surface by amine coupling according to the manufacturer's instructions. The binding of recombinant His6-GS and His6-GPI to the coated surface was determined by use of citrate buffer (10 mM citrate-NaOH, 150 mM NaCl, 0.005% P-20) with BiacoreX at a flow rate of 20 μl/min. To explore the binding of His6-GS and His6-GPI at different pHs, the purified adhesins were diluted in citrate buffer at the indicated pH, and a buffer at the same pH was used as the running buffer for surface plasmon resonance (Biacore). The protein concentration giving a clear signal at pH 6.0 was selected to be tested at all pHs (5.5, 6.0, and 6.5). The highest concentration tested was 200 μg/ml. The binding to an activated-deactivated flow cell without any immobilized protein was used as a control and was subtracted from the binding to the coated flow cell. After each run, the chip was regenerated with 5 mM NaOH (5 to 10 μl).
To analyze the binding of His6-GS, His6-GPI, His6-enolase, and His6-GAPDH to cell surfaces of L. crispatus ST1 and L. rhamnosus GG cells, 5 × 108 cells from a culture grown overnight were washed twice with 50 mM Tris-HCl (pH 8.0), incubated with 50 μg/ml His6 fusion proteins in 50 mM Tris-HCl buffer at pH 4 for 30 min, and then fixed with 3.5% (wt/vol) paraformaldehyde and washed with 50 mM Tris-HCl (pH 4.0). The surface localization of His6 fusion proteins was visualized by immunofluorescence with a monoclonal His6 antibody (diluted 1:200 in PBS; Santa Cruz Biotechnology, Inc.) and Alexa-488-conjugated secondary antibodies.
The binding of plasminogen to His6-GS and His6-GPI was measured by time-resolved fluorometry as described previously (27). Briefly, polystyrene microtiter plates were coated with His6-tagged proteins (180 nM). A laminin (Sigma-Aldrich)-coated surface was used as a positive control and a BSA (Sigma-Aldrich)-coated surface was used as a negative control for plasminogen binding. One microgram of Glu-plasminogen (American Diagnostica) was added to 100 μl PBS–0.1% Tween 20 in the presence or absence of 4 mM ε-aminocaproic acid (EACA; Sigma-Aldrich). Anti-plasminogen antibody (0.72 μg per well; American Diagnostica) and Eu3+-labeled anti-rabbit antibody (80 ng per well; Wallac Oy) were used for the detection of binding. The enhancement of tissue-type plasminogen activator (tPA)-catalyzed plasminogen activation was analyzed as previously detailed (27). Recombinant His6-GS and His6-GPI (both at 137 nM) were incubated with 4 μg human or bovine Glu-Plg, 2 ng tPA (American Diagnostica), and 0.45 mM chromogenic substrate of plasmin S-2251 (Kabivitrum) in a final volume of 200 μl, and the increase in plasmin activity was assessed at intervals by measuring the absorbance at 405 nm.
RESULTS
Identification of GS and GPI released into buffer.
To extract loosely bound proteins, “the buffersome,” from the cell surface of L. crispatus ST1 cells, the bacteria were incubated for 1 h in pH 8 buffer (27). After incubation, the pH of the cell suspension slightly varied in different experiments, between 7.5 and 7.8. The filtered, cell-free supernatant was analyzed by SDS-PAGE (Fig. 1A). The proteins were fractioned by gel filtration and anion-exchange chromatography, and we previously described enolase and GAPDH in this extract (27). A fraction containing two peptides with an apparent molecular mass of ca. 50 kDa that could not be separated by the procedures (Fig. 1A) showed adhesiveness to basement membrane material (see below) and was therefore chosen for further studies. To identify the peptides, their N-terminal amino acid sequences were determined to be RCQYTAEEIKQEV(G/N)D(R/D)KV(T/V)RF and SLIKFDSSKLTPFVHENLS. The sequences were blasted against the genomic sequence of L. crispatus ST1 (45). The first sequence variants gave by far the highest score (65% identity) with glutamine synthetase (GS), which is encoded by the glnA gene and has the predicted N-terminal sequence SKQYTAEEIKQEVEDKDVRF and a predicted mass of 50,428 Da. The next best matches in a BLAST search were GMP synthase (score, 9%; predicted mass, 57,672 Da) and NAD synthetase (score, 14%; predicted mass, 31,032 Da). The other peptide sequence perfectly matched the N terminus of glucose-6-phosphate isomerase (GPI), which has a predicted mass of 49,537 Da.
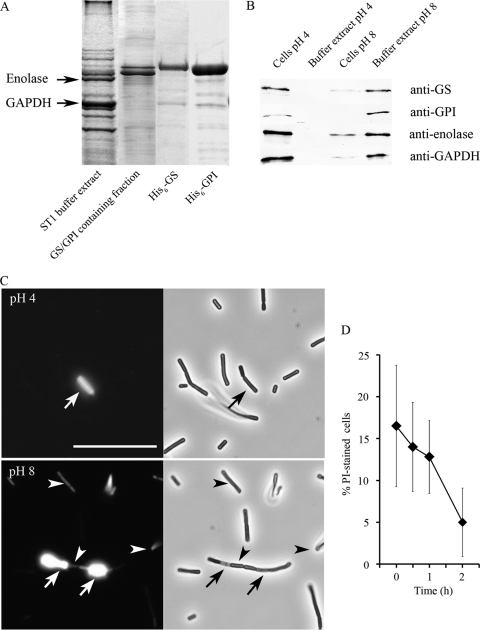
Fig 1.
Release of surface proteins from L. crispatus ST1 and changes in cell wall permeability after a pH upshift. (A) SDS-PAGE image showing the proteins released from L. crispatus ST1 cells upon their suspension in pH 8 buffer, i.e., the buffer extract, as well as the His6-tagged GS and GPI peptides of L. crispatus ST1. The GS- and GPI-containing fraction from the L. crispatus ST1 extract is also shown, and the migrations of enolase and GAPDH are indicated. (B) L. crispatus ST1 cells from cultures grown overnight were incubated at pH 4.0 and 8.0, and the proteins were detected by Western blotting using IgGs raised against purified recombinant proteins. The presence of GS and GPI on the cell surface and in the buffer extracts is shown; data for the assay for enolase and GAPDH proteins are shown for comparison. (C) Staining of L. crispatus ST1 cells with propidium iodide (on the left) and their visualization by light microscopy (on the right) after incubation at pH 4 (top) or at pH 8 (bottom). Arrows indicate brightly stained translucent cells, and arrowheads indicate less-stained permeabilized cells in the pH 8 buffer. Size bar, 20 μm. (D) Proportion of PI-stained cells in the L. crispatus ST1 population over time after the pH upshift. Means and standard deviations from three independent assays are shown.
The glnA and pgi genes were cloned from L. crispatus ST1, and the proteins were expressed as His6 fusions in E. coli and purified under nondenaturing conditions. A comparison of the extracted GS and GPI peptides and the fusion proteins by SDS-PAGE showed the expected apparent masses for the His6-GS and His6-GPI peptides (Fig. 1A). We next isolated IgG molecules from hyperimmune sera raised against His6-GS and His6-GPI, and in Western blotting experiments, these IgGs recognized the GS and the GPI peptides in the pH 8 buffer extract from L. crispatus ST1 cells, whereas at pH 4, the two proteins were detected to be mainly bound on the L. crispatus ST1 cell surface (Fig. 1B). This pH-dependent distribution is very similar to that shown by enolase and GAPDH of L. crispatus ST1 (Fig. 1B), which we described previously (4). The predicted pIs of GS and GPI of L. crispatus ST1 are 5.3 and 4.9, which are close to those of L. crispatus ST1 enolase (pI 4.7) and GAPDH (pI 5.7). We also concluded that GS and GPI remain bound on L. crispatus ST1 cells at pH 4, i.e., below their isoelectric points, and are released from the cell surface at a neutral or basic pH.
Suspension in pH 8 buffer causes a change in permeability of L. crispatus ST1 cells.
We next sought to visualize the presence of GS, GPI, enolase, and GAPDH on the L. crispatus ST1 cell surface by an indirect immunofluorescence procedure (4), where the pH 4 cells were first washed at pH 4, fixed with 3.5% (wt/vol) paraformaldehyde at pH 4, and then stained with IgGs produced against L. crispatus ST1 His6-GS and His6-GPI and Alexa-488-conjugated IgGs at a neutral pH. Our initial observation was that the cells that were strongly stained with the antibodies shared the appearance of damaged or dying cells (see below). We therefore stained the L. crispatus ST1 cells at pH 4 and pH 8 with the viability stain PI, which colors cells red by binding to DNA in cells with increased cell wall permeability (7). After PI staining, most cells from the pH 4 buffer were nonreactive and appeared healthy under light microscopy, whereas ca. 1 to 2% of cells were stained very strongly by PI and appeared translucent and probably dead by light microscopy. Upon the staining of the cells that were suspended in the pH 8 buffer, we observed that the proportion of PI-stained cells was higher, although the positive cells were stained less brightly by PI (Fig. 1C). We next determined the proportion of PI-stainable cells over time after the transfer of ST1 cells to pH 8 buffer. The analysis showed that immediately after the transfer, 17% of cells were permeabilized with PI, and their frequency in the cell population was reduced to 5% after a 2-h incubation. These results suggest that the suspension of stationary-growth-phase cells of L. crispatus ST1 in pH 8 is an alkaline stress situation and leads to a transient increase in cell wall permeability.
Visualization of GS, GPI, enolase, and GAPDH in the cell population of L. crispatus ST1.
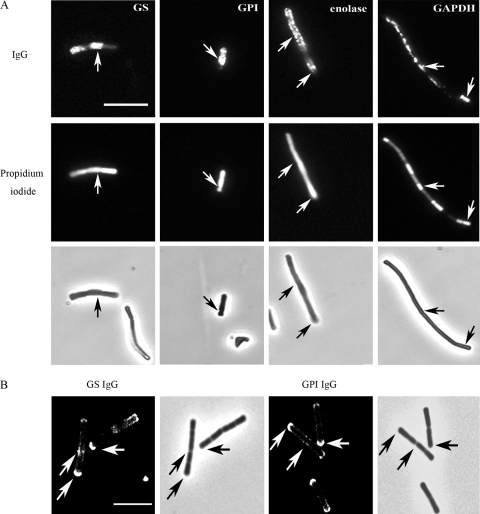
In order to identify cells that carry moonlighting proteins on their surface, we next double stained L. crispatus ST1 cells grown overnight first with IgGs and then with PI at pH 4 (Fig. 2A). The general view was that those bacterial cells which were strongly stained by PI were also positive for the IgGs; i.e., the IgG molecules stained a cell subpopulation that was permeabilized. This was observed for all four moonlighting proteins of L. crispatus ST1. Most reactive pH 4 cells were stained uniformly by IgGs, indicating that the binding was not exclusively at the cell surface. Rarely, in less than 1% of the cells at pH 4, we observed a localized binding of anti-GS and anti-GPI IgGs to the cell surface, to cell poles, and to the cell division area (Fig. 2B).
Fig 2.
Presence of moonlighting proteins on the bacterial surface. (A) Double staining of L. crispatus ST1 cells was done first by indirect immunofluorescence and then with propidium iodide. The anti-His6 fusion protein IgGs are indicated at the top. The middle panels show propidium iodide staining of the same cells, and the bottom panels show the corresponding light microscopic view. Arrows indicate costaining by immunoglobulins and propidium iodide. (B) Immunofluorescence staining of L. crispatus ST1 cells with anti-His6-GS IgGs (on the left) and anti-His6-GPI IgGs (on the right). Arrows indicate IgG binding to cell poles and to division septa. Size bar, 10 μm.
Binding of His6 fusion proteins to the bacterial cell surface is localized and strain dependent.
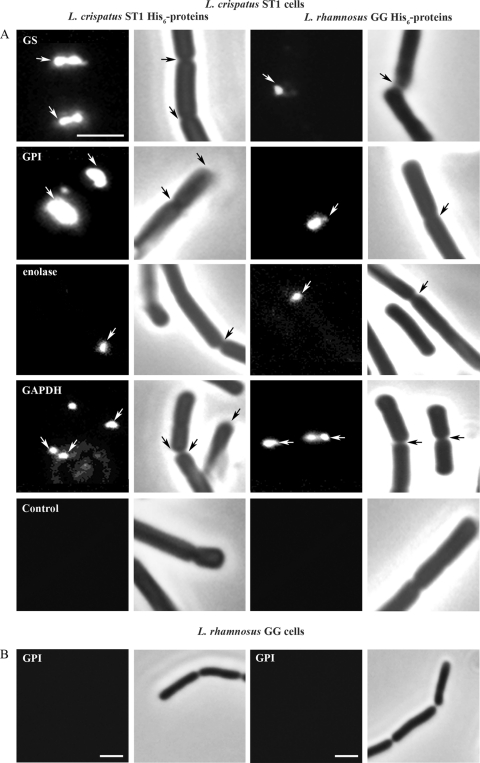
The results described above suggested that alkaline shock increases the release of moonlighting proteins from the L. crispatus surface and that these proteins might reassociate back onto the cell surface under suitable conditions. To assess whether these proteins bind in vitro to the bacterial surface, we tested the binding of His6 fusion proteins to L. crispatus ST1 cells. For comparison, we included the human probiotic strain L. rhamnosus GG in the assays. Bacteria were first washed at pH 8 to remove moonlighting proteins, and the His6-GS, His6-GPI, His6-enolase, and His6-GAPDH proteins were then allowed to bind to the cell surface at pH 4 and at pH 8. After fixing and washing, the surface localization of His6 fusion proteins was assessed by immunofluorescence microscopy using a monoclonal anti-His6 IgG. Binding of the His6 fusion proteins to division septa and poles of L. crispatus ST1 cells was observed (Fig. 3), whereas we observed no binding of the His6 fusion proteins in assays performed with the pH 8 buffer. No binding of the His6 proteins to the L. rhamnosus GG cells was detected at either pH (Fig. 3). We also expressed the glnA, pgi, eno, and gap gene products of L. rhamnosus GG as His6 fusion proteins and tested their binding to L. crispatus ST1 and L. rhamnosus GG cells. These proteins showed localized binding similar to that of the ST1-derived fusion proteins on L. crispatus ST1 cells but did not bind to L. rhamnosus GG cells (Fig. 3). Thus, the four moonlighting proteins reassociate with L. crispatus cells at a low pH but have a poor affinity for the cell surface of L. rhamnosus GG.
Fig 3.
Binding of purified His6-GS, His6-GPI, His6-enolase, and His6-GAPDH to L. crispatus ST1 and L. rhamnosus GG cells. The bacteria were washed at pH 8 and then incubated with the His6 fusion proteins from L. crispatus ST1 and L. rhamnosus GG at pH 4. Detection was done by indirect immunofluorescence with monoclonal anti-His6 IgG and Alexa-488-conjugated secondary IgGs. (A) Binding of the proteins to L. crispatus ST1 cells. The His6 fusion proteins are indicated on the left, and a control staining of cells without added His6 fusion proteins is also shown. Both the immunostained and phase-contrast images of the microscopic fields are shown; the arrows indicate protein binding to bacterial cell division areas and to cell poles. (B) Binding of L. crispatus ST1 His6-GPI (left) and L. rhamnosus GG His6-GPI (right) to L. rhamnosus GG cells at pH 4. Size bar, 3 μm.
Cathelicidin LL-37 increases the release of enolase, GAPDH, GS, and GPI from L. crispatus.
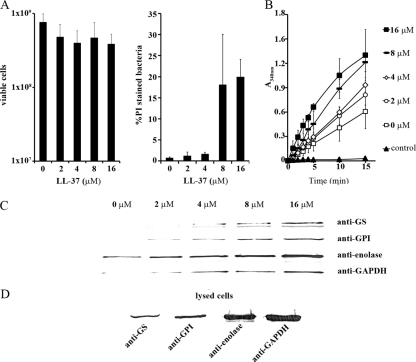
LL-37 is a cationic antimicrobial peptide and an important defense system against bacteria in epithelial cells. It disturbs the cytoplasmic membrane of the bacteria, thereby increasing the permeability of the cells and eventually halting their growth (53). Our hypothesis was that LL-37 potentiates the release of the moonlighting proteins from lactobacilli due to its cell-wall-permeabilizing effect. We treated L. crispatus ST1 cells with LL-37 at a concentration range of 0 μM to 16 μM in 50 mM Tris-HCl (pH 8) (Fig. 4). The pH 8 buffer was used to minimize the reassociation of the moonlighting proteins and to enable measurements of GPI enzyme activity. The highest concentration of LL-37 killed 49% of L. crispatus ST1 cells (Fig. 4A). To analyze the effect of LL-37 on cell wall integrity, L. crispatus ST1 cells were also stained with PI. The proportion of PI-stained cells increased from 0.7% to 20% when the LL-37 concentration was raised (Fig. 4A).
Fig 4.
Effect of LL-37 on cell viability, cell wall permeability, and release of GS, GPI, enolase, and GAPDH from L. crispatus ST1. (A) Effect of increasing LL-37 concentrations on the viability (left) and on the proportion of propidium iodide (PI)-stainable, permeabilized cells (right). Means with standard deviations from three independent assays are shown. (B) GPI enzymatic activity in buffer extracts from L. crispatus ST1 cells treated with LL-37 at the indicated concentrations. The control is an assay performed in the absence of the bacterial buffer extract. The means and standard deviations from three independent assays are shown. (C) Release of GS, GPI, enolase, and GAPDH into buffer from LL-37-treated cells (108 cells). The enzymes were detected by Western blotting using immunoglobulins raised against the His6 fusion proteins. (D) Western blotting of the proteins from 5 × 107 lysed cells is shown for comparison.
To detect possible increases in the release of moonlighting proteins, we first assessed the effect of increasing LL-37 concentrations on the release of GPI activity from 108 L. crispatus ST1 (Fig. 4B). The pH 8 buffer extract obtained from L. crispatus ST1 cells exhibited GPI activity, which was increased stepwise along with the increased LL-37 concentrations. Assays with purified GPI showed that the enzyme activity was not affected by the LL-37 concentrations used in this study (not shown). We next analyzed by Western blotting the presence of GS, GPI, enolase, and GAPDH in buffer extracts from LL-37-treated L. crispatus ST1 cells (Fig. 4C). A gradual increase in the amounts of the proteins in the buffer upon increasing LL-37 concentrations was evident. To compare total amounts of the enzymes in L. crispatus ST1 cells, 5 × 107 cells were lysed first with lysozyme and mutanolysin and then by boiling for 10 min; note that the number of cells analyzed in Fig. 4D is half the amount analyzed in Fig. 4C.
Adhesive and plasminogen-binding properties of GS and GPI.
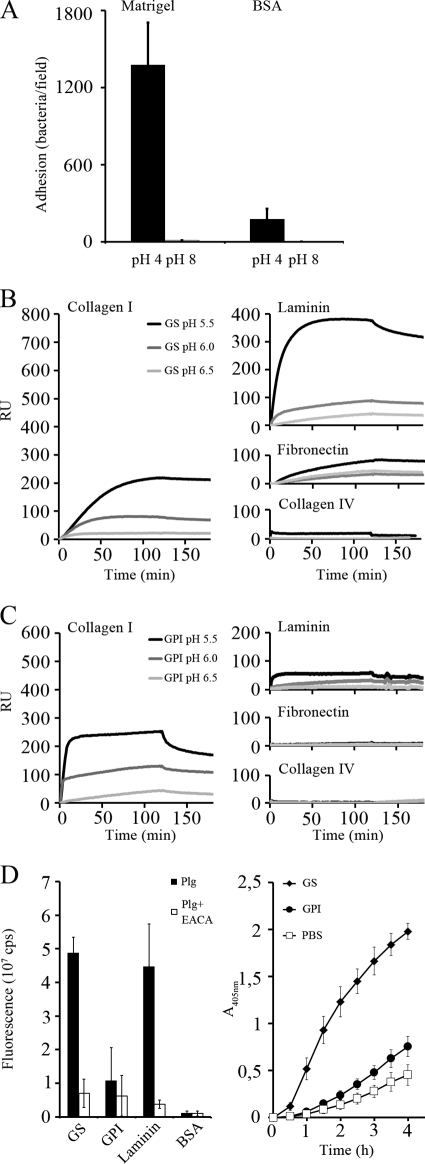
A high number of lactobacillar isolates have been found to be adhesive to proteins of the ECM (2, 23, 41), and on the other hand, moonlighting proteins have been associated with such adhesiveness (5, 16). We assessed the adherence of L. crispatus ST1 cells to the commonly used reconstituted basement membrane (BM) preparation Matrigel to see whether the bacterial adhesiveness was altered by pH, due to the detachment of surface-associated moonlighting proteins. L. crispatus ST1 cells were collected into buffers of pH 4 or pH 8 and washed once with the same buffer, and adherence was tested at the same pH (Fig. 5A). The adherence of L. crispatus ST1 cells to Matrigel was higher at the acidic pH.
Fig 5.
Moonlighting functions of GS and GPI proteins from L. crispatus ST1. (A) Adherence of L. crispatus ST1 cells to immobilized Matrigel at pH 4 and at pH 8. L. crispatus ST1 cells were washed with 50 mM Tris-HCl buffer (pH 4 or 8), and the adhesion assay was performed at the same pH. The mean numbers and standard deviations for bacteria from 20 randomly chosen microscopic fields of 1.6 × 104 μm2 are shown. The adherence of L. crispatus ST1 cells to BSA at both pHs is also shown. (B and C) Binding of His6-GS (B) and His6-GPI (C) to immobilized laminin, collagen I, fibronectin, and collagen IV was tested by surface plasmon resonance (Biacore) at pH 5.5, 6.0, and 6.5. RU, relative units. (D) Binding of human plasminogen to His6-GS and His6-GPI proteins (left) and their effect on tPA-mediated plasminogen activation (right). Binding was tested in the presence and absence of EACA, a lysine analog that inhibits physiological plasminogen binding. For comparison, binding to laminin, a target for plasminogen binding, and to BSA is also shown. PBS in the activation test indicates an assay without added bacterial proteins. Means and standard deviations from three independent assays are shown.
Another feature affecting adhesiveness is that the surface proteins of L. crispatus ST1 could function more effectively at low pHs. We therefore assessed the binding of His6-GS and His6-GPI of L. crispatus ST1 to the ECM proteins laminin, fibronectin, collagen I, and collagen IV at pH 5.5, 6.0, and 6.5 by surface plasmon resonance. The binding profiles of the two His6 fusion proteins differed: His6-GS but not His6-GPI bound to fibronectin (Fig. 5B and C). Both His6 fusion proteins bound to collagen I, and the level of binding was clearly higher at acidic pHs, but neither of them bound to collagen IV. Both proteins also bound to laminin, and, especially at pH 5.5, the level of binding of His6-GS was high.
Another function associated with several bacterial moonlighting proteins is their ability to immobilize plasminogen on the bacterial surface and thus advance its activation by tPA (9, 27). We assessed the binding of human plasminogen to His6-GS and His6-GPI from L. crispatus ST1. The His6-GS protein showed a high level of binding, which was effectively inhibited by EACA, an inhibitor of plasminogen binding (Fig. 5D). His6-GS, but not His6-GPI, also enhanced plasminogen activation by tPA.
DISCUSSION
We describe here two novel lactobacillar moonlighting proteins, GS and GPI, which were released from the bacterial surface into pH 8 extraction buffer. This behavior is very similar to that shown by enolase and GAPDH, previously identified as moonlighting proteins of L. crispatus ST1 (27). Moonlighting proteins were released from the ST1 surface in stress situations, in particular in the presence of the antimicrobial peptide LL-37, and our results give an example of how lactobacilli can modify their surface architecture upon contact with the innate immunity system of humans.
The release and reassociation of moonlighting proteins on the L. crispatus cell surface were sensitive to the pH of the buffer. At a pH value below the pI of the proteins, GS, GPI, enolase, and GAPDH bound to the ST1 cell surface, whereas no binding in the pH 8 buffer was observed. The results suggest that in L. crispatus ST1, these moonlighting proteins are anchored at the cell surface through ionic interactions, and we have previously identified LTAs as a class of negatively charged anchoring molecules that bind enolase and GAPDH (4). The failure of the recombinant moonlighting proteins from L. crispatus or L. rhamnosus GG to bind to the L. rhamnosus GG surface indicates that the mechanisms of the surface association of moonlighting proteins vary in bacterial species. The biochemical explanation for the difference remains open. It was reported previously that the capsule of L. rhamnosus GG protects the bacterium against LL-37 (35), and by analogy, the capsule might also prevent the reassociation of the His6 fusion protein with the cell wall of L. rhamnosus GG. We observed previously that alkaline stress releases fewer proteins from the GG cell surface than from the ST1 cell surface (27), which might reflect smaller amounts of moonlighting proteins on the L. rhamnosus GG cell surface.
Our results are in agreement with those described previously by Saad and coworkers (51), who found that stationary-growth-phase cells of L. plantarum had a high level of surface GAPDH activity, which was correlated with increased cell membrane permeability. They did not detect any release of GAPDH into the culture medium, which however, remained highly acidic throughout their assays. We found that the suspension of L. crispatus ST1 cells in the pH 8 buffer as well as their treatment with LL-37 were stress situations, where both the permeability of the cells and the release of moonlighting proteins were increased. We found that the proportion of PI-stainable cells in the ST1 population increased upon suspension in the pH 8 buffer and then decreased back to the original level in 2 h. This is very similar to the recovery of L. plantarum from acid stress, where the bacterial population displays phenotypic and morphological heterogeneity after a rapid pH downshift and then recovers over a period of ca. 2 h (28). Our hypothesis is that alkaline stress increases the release of moonlighting proteins through a temporary change in the cell wall organization. Our previous work showed that in L. crispatus ST1 cells, the pH-induced release of enolase and GAPDH is not associated with de novo protein synthesis (4). Proteomic studies have shown that growth under alkaline stress conditions increases the cellular content of enolase but reduces the amount of GS of L. plantarum (37), whereas here, the two proteins were released from L. crispatus ST1 cells by LL-37 treatment and in the pH 8 buffer in the same apparent ratio as they occurred in lysed cell samples.
The LL-37 molecule displays both antimicrobial activity as well as a variety of functions related to inflammation (56). LL-37 is present throughout the human body in concentrations that vary according to the body site and the physiological situation; in particular, the concentration of hCAP-18/LL-37 is increased during inflammation by bacterial products and in wound repair and cellular differentiation (13, 56). LL-37 is thought to kill bacteria only in settings of epithelial surfaces or neutrophils and phagocytes (13). LL-37 concentrations in secretions vary from 1 to 2 μg/ml in saliva, up to 20 μg/ml in tracheal aspirates, and 85 μg/ml (19.7 μM) in seminal plasma (13). We detected an increased permeability of L. crispatus ST1 cells at concentrations of 4 to 16 μM, which was associated with a stepwise increase in the release of moonlighting proteins into buffer. These effects can thus take place in vivo as well. Antimicrobial peptides, such as LL-37, studied here, are the first line of defense of the human mucosa which lactobacilli will also face during colonization, and our hypothesis is that such an encounter will lead to changes in the cell wall architecture and properties of lactobacilli.
The binding of His6-GS and His6-GPI to L. crispatus ST1 cells was localized to cell division sites and cell poles. The cell division septa are sites of cell wall synthesis, and several proteins functioning in cell division are complexed at the division site into a ringlike structure, whose remnants may be detected at the poles of newborn cells (59). Septation also involves changes in the synthesis of peptidoglucan of E. coli and the capsule of S. pneumoniae (25, 59), and the division septum of S. aureus is a preferred target for binding by telavancin, a bactericidal peptide that inhibits cell wall synthesis and disrupts membrane barrier function by binding to peptidoglycan precursors (39). By immunofluorescence, we occasionally observed L. crispatus ST1 cells in pH 4 buffer, which were stained by anti-GS and anti-GPI IgGs at the division septa and at cell poles. However, we cannot deduce whether they represent moonlighting proteins bound from the culture medium or proteins secreted by an as-yet-unknown transport system that would localize them to cell poles and septa (24).
Adherence to host tissues is a common function in bacterial moonlighting proteins (24). We found that the adhesiveness of L. crispatus ST1 to the reconstituted basement membrane preparation Matrigel was efficient at pH 4 and almost nonexistent at pH 8, which is in accordance with the hypothesis that adhesive moonlighting proteins are released from the cell surface. Matrigel has been widely used in studies of the adherence and invasiveness of bacterial pathogens as well as metastatic tumor cells (34, 43), and it contains type IV collagen and laminin as major components (20). We found that the GS of L. crispatus ST1 indeed shows an affinity for laminin. It remains open whether L. crispatus ST1 has other, as-yet-unidentified BM/ECM-binding moonlighting proteins. GS and GPI also bound type I collagen, which is the most abundant type of collagen in the human body and is present in, e.g., scar tissue resulting from tissue repair and healing (36). Several isolates of lactobacilli have been found to adhere to proteins of mammalian ECMs and BMs (2, 3, 6, 23, 41). BMs in most tissues are remarkably stable structures with a long half-life and a short turnover of their components, and they provide scaffolds upon which epithelial cells migrate (47, 57). In E. coli, the adherence of renal BMs to collagen is critical for the persistent colonization of the kidneys (42, 52), and it might be that the frequently observed in vitro adhesiveness of lactobacillar isolates to BM/ECM proteins has a biological function in enhancing the long-term colonization of the host.
The immobilization of plasminogen is another function commonly attributed to bacterial moonlighting proteins, and we found that His6-GS, but not His6-GPI, bound human plasminogen and increased its activation by tPA. The binding was inhibited by EACA, indicating that it is mediated by the lysine-binding kringle domains of plasminogen (58). GS is a major plasminogen-binding protein of Mycobacterium tuberculosis (61) and was also identified as a plasminogen-binding protein of B. animalis subsp. lactis (14). The activation of the human plasminogen system is an established virulence function in several invasive bacterial infections (33). At a neutral pH, plasminogen/plasmin remains bound on the pathogen surface and, thus, inaccessible to the circulating plasmin inhibitor α2-antiplasmin (34). In contrast, plasminogen activation by tPA is enhanced by cell-free buffer extracts from L. crispatus ST1 cells, and the plasmin formed is not protected from α2-antiplasmin (27) and thus will be rapidly inactivated in vivo. This indicates that the plasmin created in the presence of L. crispatus ST1 cells remains active only when complexed onto a lysine-containing surface. For tPA-catalyzed activation, this surface is most often the fibrin clot, which is a major physiological target for plasmin proteolysis (49, 50). This suggests that the enhancement of plasmin activity by lactobacilli remains local and could function, e.g., to contribute to the dissolution of fibrin clots or thrombi over a wound site. Along these lines, it was reported previously that the treatment of mice suffering from pneumococcal pneumonia with Lactobacillus casei lowers the amount of fibrin(ogen) deposits in the lung (1, 22), and the potential of synbiotics to reduce procoagulatory factors has been recognized (8). The mechanisms of these in vivo effects obviously are complex, but our results encourage further studies of the role and the mechanisms of the modulation of the fibrinolysis/coagulation cascades by commensal bacteria.
Lactobacilli are exposed to various environmental stresses, such as extremes in temperature, pH, oxygen, starvation, and antimicrobials, which may affect the physiological status and properties of the cells (17). Our results indicate that lactobacillar moonlighting proteins are more efficiently released into their surroundings after cell wall trauma. The findings are compatible with the observation that stress induced by bacterial growth under conditions of iron starvation or in the presence of bile acids increases the amount, or the release, of moonlighting proteins in streptococci and bifidobacteria (15, 19, 44). Antimicrobial peptides, bile acids, and iron limitation represent in vivo challenges for bacteria colonizing a mammalian host, and the modification of the cell surface may have importance in host interactions and adaptation to various conditions by lactic acid bacteria. An intriguing conclusion from our in vitro observations is that released moonlighting proteins are able to reassociate with different bacterial species, thus providing a novel mechanism of bacterium-bacterium interactions.
ACKNOWLEDGMENTS
This study has been supported by the Foundation for Nutritional Research, Valio Ltd., and the University of Helsinki.
We thank Raili Lameranta, Annukka Jaatinen, Hanna Pauloff, Laura Kalin, and Hanna Järvinen for technical assistance.
Footnotes
Published ahead of print 2 March 2012
REFERENCES
- 1. Aguero G, Villena J, Racedo S, Haro C, Alvarez S. 2006. Beneficial immunomodulatory activity of Lactobacillus casei in malnourished mice pneumonia: effect on inflammation and coagulation. Nutrition 22:810–819 [DOI] [PubMed] [Google Scholar]
- 2. Aleljung P, et al. 1991. Collagen binding by lactobacilli. Curr. Microbiol. 23:33–38 [Google Scholar]
- 3. Antikainen J, Korhonen TK, Kuparinen V, Toba T, Roos S. 2009. Surface proteins of Lactobacillus involved in host interactions, p 95–114 In Ljungh Å, Wadström T. (ed), Lactobacillus molecular biology, from genomics to probiotics. Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
- 4. Antikainen J, Kuparinen V, Lähteenmäki K, Korhonen TK. 2007. pH-dependent association of enolase and glyceraldehyde-3-phosphate dehydrogenase of Lactobacillus crispatus with the cell wall and lipoteichoic acids. J. Bacteriol. 189:4539–4543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Antikainen J, Kuparinen V, Lähteenmäki K, Korhonen TK. 2007. Enolases from Gram-positive bacterial pathogens and commensal lactobacilli share functional similarity in virulence-associated traits. FEMS Immunol. Med. Microbiol. 51:526–534 [DOI] [PubMed] [Google Scholar]
- 6. Antikainen J, Anton L, Sillanpää J, Korhonen TK. 2002. Domains in the S-layer protein CbsA of Lactobacillus crispatus involved in adherence to collagens, laminin and lipoteichoic acids and in self-assembly. Mol. Microbiol. 46:381–394 [DOI] [PubMed] [Google Scholar]
- 7. Banning N, Toze S, Mee BJ. 2002. Escherichia coli survival in groundwater and effluent measured using a combination of propidium iodide and the green fluorescent protein. J. Appl. Microbiol. 93:69–76 [DOI] [PubMed] [Google Scholar]
- 8. Bengmark S. 2003. Use of some pre-, pro- and synbiotics in critically ill patients. Best Pract. Res. Clin. Gastroenterol. 17:833–848 [DOI] [PubMed] [Google Scholar]
- 9. Bergmann S, Hammerschmidt S. 2007. Fibrinolysis and host response in bacterial infections. Thromb. Haemost. 98:512–520 [PubMed] [Google Scholar]
- 10. Bergmann S, Rohde M, Chhatwal GS, Hammerschmidt S. 2001. Alpha-enolase of Streptococcus pneumoniae is a plasmin(ogen)-binding protein displayed on the bacterial cell surface. Mol. Microbiol. 40:1273–1287 [DOI] [PubMed] [Google Scholar]
- 11. Bergonzelli GE, et al. 2006. GroEL of Lactobacillus johnsonii La1 (NCC 533) is cell surface associated: potential role in interactions with the host and the gastric pathogen Helicobacter pylori. Infect. Immun. 74:425–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Boël G, Jin H, Pancholi V. 2005. Inhibition of cell surface export of group A streptococcal anchorless surface dehydrogenase affects bacterial adherence and antiphagocytic properties. Infect. Immun. 73:6237–6248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bucki R, Leszczynska K, Namiot A, Sokolowski W. 2010. Cathelicidin LL-37: a multitask antimicrobial peptide. Arch. Immunol. Ther. Exp. (Warsz.) 58:15–25 [DOI] [PubMed] [Google Scholar]
- 14. Candela M, et al. 2007. Binding of human plasminogen to Bifidobacterium. J. Bacteriol. 189:5929–5936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Candela M, et al. 2010. DnaK from Bifidobacterium animalis subsp. lactis is a surface-exposed human plasminogen receptor upregulated in response to bile salts. Microbiology 156:1609–1618 [DOI] [PubMed] [Google Scholar]
- 16. Castaldo C, et al. 2009. Surface displaced alfa-enolase of Lactobacillus plantarum is a fibronectin binding protein. Microb. Cell Fact. 8:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. De Angelis M, Gobbetti M. 2004. Environmental stress responses in Lactobacillus: a review. Proteomics 4:106–122 [DOI] [PubMed] [Google Scholar]
- 18. Edelman S, et al. 2002. In vitro adhesion specificity of indigenous lactobacilli within the avian intestinal tract. Appl. Environ. Microbiol. 68:5155–5159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Eichenbaum Z, Green BD, Scott JR. 1996. Iron starvation causes release from the group A Streptococcus of the ADP-ribosylating protein called plasmin receptor or surface glyceraldehyde-3-phosphate-dehydrogenase. Infect. Immun. 64:1956–1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Farina AR, et al. 1996. Identification of plasminogen in Matrigel and its activation by reconstitution of this basement membrane extract. Biotechniques 21:904–909 [DOI] [PubMed] [Google Scholar]
- 21. Granato D, et al. 2004. Cell surface-associated elongation factor Tu mediates the attachment of Lactobacillus johnsonii NCC533 (La1) to human intestinal cells and mucins. Infect. Immun. 72:2160–2169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Haro C, Villena J, Zelaya H, Alvarez S, Aguero G. 2009. Lactobacillus casei modulates the inflammation-coagulation interaction in a pneumococcal pneumonia experimental model. J. Inflamm. (Lond.) 6:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Harty DW, Oakey HJ, Patrikakis M, Hume EB, Knox KW. 1994. Pathogenic potential of lactobacilli. Int. J. Food Microbiol. 24:179–189 [DOI] [PubMed] [Google Scholar]
- 24. Henderson B, Martin A. 2011. Bacterial virulence in the moonlight: multitasking bacterial moonlighting proteins are virulence determinants in infectious diseases. Infect. Immun. 79:3476–3491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Henriques MX, Rodrigues T, Carido M, Ferreira L, Filipe SR. 2011. Synthesis of capsular polysaccharide at the division septum of Streptococcus pneumoniae is dependent on a bacterial tyrosine kinase. Mol. Microbiol. 82:515–534 [DOI] [PubMed] [Google Scholar]
- 26. Huberts DH, van der Klei IJ. 2010. Moonlighting proteins: an intriguing mode of multitasking. Biochim. Biophys. Acta 1803:520–525 [DOI] [PubMed] [Google Scholar]
- 27. Hurmalainen V, et al. 2007. Extracellular proteins of Lactobacillus crispatus enhance activation of human plasminogen. Microbiology 153:1112–1122 [DOI] [PubMed] [Google Scholar]
- 28. Ingham CJ, Beerthuyzen M, van Hylckama Vlieg J. 2008. Population heterogeneity of Lactobacillus plantarum WCFS1 microcolonies in response to and recovery from acid stress. Appl. Environ. Microbiol. 74:7750–7758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jeffery CJ. 2004. Molecular mechanisms for multitasking: recent crystal structures of moonlighting proteins. Curr. Opin. Struct. Biol. 14:663–668 [DOI] [PubMed] [Google Scholar]
- 30. Kankainen M, et al. 2009. Comparative genomic analysis of Lactobacillus rhamnosus GG reveals pili containing a human-mucus binding protein. Proc. Natl. Acad. Sci. U. S. A. 106:17193–17198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Katakura Y, Sano R, Hashimoto T, Ninomiya K, Shioya S. 2010. Lactic acid bacteria display on the cell surface cytosolic proteins that recognize yeast mannan. Appl. Microbiol. Biotechnol. 86:319–326 [DOI] [PubMed] [Google Scholar]
- 32. Kinoshita H, et al. 2008. Cell surface Lactobacillus plantarum LA 318 glyceraldehyde-3-phosphate dehydrogenase (GAPDH) adheres to human colonic mucin. J. Appl. Microbiol. 104:1667–1674 [DOI] [PubMed] [Google Scholar]
- 33. Lähteenmäki K, Edelman S, Korhonen TK. 2005. Bacterial metastasis: the host plasminogen system in bacterial invasion. Trends Microbiol. 13:79–85 [DOI] [PubMed] [Google Scholar]
- 34. Lähteenmäki K, Kuusela P, Korhonen TK. 2001. Bacterial plasminogen activators and receptors. FEMS Microbiol. Rev. 25:531–552 [DOI] [PubMed] [Google Scholar]
- 35. Lebeer S, Claes IJ, Verhoeven TL, Vanderleyden J, De Keersmaecker SC. 2011. Exopolysaccharides of Lactobacillus rhamnosus GG form a protective shield against innate immune factors in the intestine. Microb. Biotechnol. 4:368–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. LeBleu VS, Macdonald B, Kalluri R. 2007. Structure and function of basement membranes. Exp. Biol. Med. (Maywood) 232:1121–1129 [DOI] [PubMed] [Google Scholar]
- 37. Lee K, Rho BS, Pi K, Kim HJ, Choi YJ. 2011. Proteomic analysis of protein expression in Lactobacillus plantarum in response to alkaline stress. J. Biotechnol. 153:1–7 [DOI] [PubMed] [Google Scholar]
- 38. Lenz LL, Mohammadi S, Geissler A, Portnoy DA. 2003. SecA2-dependent secretion of autolytic enzymes promotes Listeria monocytogenes pathogenesis. Proc. Natl. Acad. Sci. U. S. A. 100:12432–12437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lunde CS, Rexer CH, Hartouni SR, Axt S, Benton BM. 2010. Fluorescence microscopy demonstrates enhanced targeting of telavancin to the division septum of Staphylococcus aureus. Antimicrob. Agents Chemother. 54:2198–2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mathur D, Garg LC. 2007. Functional phosphoglucose isomerase from Mycobacterium tuberculosis H37Rv: rapid purification with high yield and purity. Protein Expr. Purif. 52:373–378 [DOI] [PubMed] [Google Scholar]
- 41. McGrady JA, Butcher WG, Beighton D, Switalski LM. 1995. Specific and charge interactions mediate collagen recognition by oral lactobacilli. J. Dent. Res. 74:649–657 [DOI] [PubMed] [Google Scholar]
- 42. Miettinen A, et al. 1993. Binding of bacterial adhesins to rat glomerular mesangium in vivo. Kidney Int. 43:592–600 [DOI] [PubMed] [Google Scholar]
- 43. Mignatti P, Rifkin DB. 1993. Biology and biochemistry of proteinases in tumor invasion. Physiol. Rev. 73:161–195 [DOI] [PubMed] [Google Scholar]
- 44. Nelson D, et al. 2001. pH-regulated secretion of a glyceraldehyde-3-phosphate dehydrogenase from Streptococcus gordonii FSS2: purification, characterization, and cloning of the gene encoding this enzyme. J. Dent. Res. 80:371–377 [DOI] [PubMed] [Google Scholar]
- 45. Ojala T, et al. 2010. Genome sequence of Lactobacillus crispatus ST1. J. Bacteriol. 192:3547–3548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pancholi V, Chhatwal GS. 2003. Housekeeping enzymes as virulence factors for pathogens. Int. J. Med. Microbiol. 293:391–401 [DOI] [PubMed] [Google Scholar]
- 47. Price RG, Spiro RG. 1977. Studies on the metabolism of the renal glomerular basement membrane. Turnover measurements in the rat with the use of radiolabeled amino acids. J. Biol. Chem. 252:8597–8602 [PubMed] [Google Scholar]
- 48. Ramiah K, van Reenen CA, Dicks LM. 2008. Surface-bound proteins of Lactobacillus plantarum 423 that contribute to adhesion of Caco-2 cells and their role in competitive exclusion and displacement of Clostridium sporogenes and Enterococcus faecalis. Res. Microbiol. 159:470–475 [DOI] [PubMed] [Google Scholar]
- 49. Redlitz A, Plow EF. 1995. Receptors for plasminogen and t-PA: an update. Baillieres Clin. Haematol. 8:313–327 [DOI] [PubMed] [Google Scholar]
- 50. Rijken DC, Lijnen HR. 2009. New insights into the molecular mechanisms of the fibrinolytic system. J. Thromb. Haemost. 7:4–13 [DOI] [PubMed] [Google Scholar]
- 51. Saad N, et al. 2009. Lactobacillus plantarum 299v surface-bound GAPDH: a new insight into enzyme cell walls location. J. Microbiol. Biotechnol. 19:1635–1643 [DOI] [PubMed] [Google Scholar]
- 52. Selvarangan R, et al. 2004. Interaction of Dr adhesin with collagen type IV is a critical step in Escherichia coli renal persistence. Infect. Immun. 72:4827–4835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sochacki KA, Barns KJ, Bucki R, Weisshaar JC. 2011. Real-time attack on single Escherichia coli cells by the human antimicrobial peptide LL-37. Proc. Natl. Acad. Sci. U. S. A. 108:E77–E81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Spurbeck RR, Arvidson CG. 2010. Lactobacillus jensenii surface-associated proteins inhibit Neisseria gonorrhoeae adherence to epithelial cells. Infect. Immun. 78:3103–3111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Stephenson K, Bron S, Harwood CR. 1999. Cellular lysis in Bacillus subtilis; the affect of multiple extracellular protease deficiencies. Lett. Appl. Microbiol. 29:141–145 [Google Scholar]
- 56. Tjabringa GS, et al. 2003. The antimicrobial peptide LL-37 activates innate immunity at the airway epithelial surface by transactivation of the epidermal growth factor receptor. J. Immunol. 171:6690–6696 [DOI] [PubMed] [Google Scholar]
- 57. Trier JS, Allan CH, Abrahamson DR, Hagen SJ. 1990. Epithelial basement membrane of mouse jejunum. Evidence for laminin turnover along the entire crypt-villus axis. J. Clin. Invest. 86:87–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Verstraete M. 1985. Clinical application of inhibitors of fibrinolysis. Drugs 29:236–261 [DOI] [PubMed] [Google Scholar]
- 59. Vicente M, Rico AI, Martinez-Arteaga R, Mingorance J. 2006. Septum enlightenment: assembly of bacterial division proteins. J. Bacteriol. 188:19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Virkola R, et al. 1996. Interaction of Haemophilus influenzae with the mammalian extracellular matrix. J. Infect. Dis. 173:1137–1147 [DOI] [PubMed] [Google Scholar]
- 61. Xolalpa W, et al. 2007. Identification of novel bacterial plasminogen-binding proteins in the human pathogen Mycobacterium tuberculosis. Proteomics 7:3332–3341 [DOI] [PubMed] [Google Scholar]
- 62. Yang CK, et al. 2011. Nonclassical protein secretion by Bacillus subtilis in the stationary phase is not due to cell lysis. J. Bacteriol. 193:5607–5615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zanetti M. 2005. The role of cathelicidins in the innate host defenses of mammals. Curr. Issues Mol. Biol. 7:179–196 [PubMed] [Google Scholar]