Abstract
Zur regulators control zinc homeostasis by repressing target genes under zinc-sufficient conditions in a wide variety of bacteria. This paper describes how part of a survey of duplicated genes led to the identification of the open reading frame all2473 as the gene encoding the Zur regulator of the cyanobacterium Anabaena sp. strain PCC 7120. All2473 binds to DNA in a zinc-dependent manner, and its DNA-binding sequence was characterized, which allowed us to determine the relative contribution of particular nucleotides to Zur binding. A zur mutant was found to be impaired in the regulation of zinc homeostasis, showing sensitivity to elevated concentrations of zinc but not other metals. In an effort to characterize the Zur regulon in Anabaena, 23 genes containing upstream putative Zur-binding sequences were identified and found to be regulated by Zur. These genes are organized in six single transcriptional units and six operons, some of them containing multiple Zur-regulated promoters. The identities of genes of the Zur regulon indicate that Anabaena adapts to conditions of zinc deficiency by replacing zinc metalloproteins with paralogues that fulfill the same function but presumably with a lower zinc demand, and with inducing putative metallochaperones and membrane transport systems likely being involved in the scavenging of extracellular zinc, including plasma membrane ABC transport systems and outer membrane TonB-dependent receptors. Among the Zur-regulated genes, the ones showing the highest induction level encode proteins of the outer membrane, suggesting a primary role for components of this cell compartment in the capture of zinc cations from the extracellular medium.
INTRODUCTION
In a changing environment, bacterial cells are continuously challenged by either insufficient, elevated, or even toxic concentrations of metals. However, both excess and deficiency of a particular metal in the cytoplasm are detrimental for cell growth. The maintenance of the cytoplasmic concentrations of metals within certain levels needs to be tightly controlled to avoid misincorporation of particular metals into noncognate proteins, and this is achieved primarily by regulating their flow into and out of the cell through the control of specific import systems or efflux pumps (61). Adaptation to the availability of metals in the environment may involve complex signaling systems and vast metabolic rearrangements (61).
Zinc is one of the most important divalent metals in biology. It may act as a structural element, helping to maintain the conformation of particular protein domains, or as a catalytic cofactor in the active site of a variety of enzymes (13). Zinc sensing in bacteria is carried out by regulators of different families, including SmtB/ArsR, MerR, TetR, MarR, and the Fur family (10, 26, 32, 37, 54). Zur (zinc uptake regulator) proteins belong to the Fur family. These proteins function as dimeric transcription factors that bind to palindromic DNA sequences in the promoters of regulated genes (33). In general, proteins of the Fur family work as repressors by binding to DNA targets that overlap promoter sequences, thus blocking the access of the RNA polymerase (17, 33). The Fur family includes proteins that, despite showing broad sequence similarity and a similar composition of structural domains, are diverse enough to respond to distinct stimuli. Thus, this family includes members like Zur, Fur, Nur, and Mur, which sense distinct divalent metals (Zn, Fe, Ni, and Mn, respectively), and PerR and Irr, which sense cytoplasmic peroxides and heme, respectively (33). Like other members of the Fur family, Zur proteins have two structural domains connected by a mobile hinge, an N-terminal winged-helix DNA-binding domain and a C-terminal dimerization domain (39, 55). Zur contains several coordination sites for zinc (39, 41, 55) and senses the cytoplasmic concentration of exchangeable zinc by binding to this metal, which in turn allows Zur binding to DNA (33). Effective sensing of zinc is presumed to require the concentration of the metal in the cytoplasm to approach the affinity of the regulatory coordination site(s) (41, 55), and zinc has been estimated to be present in subpicomolar concentrations in Escherichia coli (45). Occupancy of the regulatory site probably induces a conformational reorientation of the two domains so that, in the dimer, the DNA-binding domains adopt an optimal orientation for binding to DNA (39, 41, 55).
Cyanobacteria are oxygenic photosynthetic prokaryotes, distributed throughout a wide variety of environments ranging from oceanic and fresh waters to continental habitats at all latitudes (62). Although some may form symbiotic associations with fungi and plants, most species are free-living organisms. Their capacity to fix atmospheric CO2 and release O2 by photosynthesis together with their abundance on Earth make these organisms quantitatively important in some biogeochemical cycles. For instance, it is estimated that oceanic cyanobacteria account for a significant portion of global primary production (11, 23, 35, 47). Cyanobacteria have a strong dependency on zinc, since one of the most important and abundant enzymes for CO2 fixation in these organisms, carbonic anhydrase, is most commonly a zinc metalloenzyme which generates a substrate for this reaction from bicarbonate (57). Although several aspects of detoxification of excess zinc have been exhaustively analyzed in cyanobacteria (7–9, 12, 30, 36, 49), the response of these organisms to zinc deficiency remains poorly investigated. Indeed, the Zur regulator has scarcely been characterized in this phylum (7, 12, 59).
In this study, the response of Anabaena sp. strain PCC 7120 (also known as Nostoc sp. strain PCC 7120) to zinc deficiency was analyzed. The Zur regulator was identified with in vitro and in vivo evidence of the specificity of this factor for zinc. An empirical characterization of the Zur-binding sequence is provided, and components of the Zur regulon are identified.
MATERIALS AND METHODS
Organisms and growth conditions.
Anabaena sp. strain PCC 7120 was routinely grown in BG11 medium (48) at 30°C, illuminated with white fluorescent lamps at 75 μE m−2 s−1, in Erlenmeyer flasks that were shaken or aerated by bubbling with a mixture of air enriched with 1% CO2. Bubbled cultures were buffered with 10 mM NaHCO3. Solid medium was prepared by the addition of 1% agar to BG11. The zur mutant was cultured in medium containing 2 to 5 μg ml−1 streptomycin and 2 to 5 μg ml−1 spectinomycin. When indicated, N,N,N′,N′-tetrakis(2-pyridilmethyl)ethylenediamine (TPEN) was added to the cultures at a final concentration of 20 μM. TPEN was prepared in dimethyl sulfoxide (DMSO) at a concentration of 20 mM. In experiments using TPEN, DMSO was added to control cultures not containing TPEN at the same final concentration as those containing it.
Escherichia coli was grown in Luria-Bertani (1) medium with antibiotics, when needed, at the following concentrations: ampicillin, 50 μg ml−1; kanamycin, 25 μg ml−1; chloramphenicol, 30 μg ml−1; streptomycin, 25 μg ml−1; spectinomycin, 100 μg ml−1. Escherichia coli DH5α was used for routine cloning. XL1-Blue and BL21(DE3) were used for overexpression of genes under the control of the trc promoter and the T7 promoter, respectively.
Generation of an Anabaena zur mutant.
A deletion-insertion mutant of the all2473 gene (furB/zur) was generated as follows. A DNA fragment containing the all2473 gene and flanking sequences was amplified by PCR using Anabaena genomic DNA as a template and primers 2473_DEL-1F and 2473_DEL_1R. The PCR product was cloned in plasmid pBluescript SK+ (Novagen). The resulting plasmid, pCMN34, was PCR amplified with divergent primers 2473_DEL-2F and 2473_DEL_2R and ligated to the C.S3 cassette (15) conferring resistance to streptomycin and spectinomycin. The insert was cloned into the pRL278 vector and transferred by triparental conjugation to Anabaena (14).
Overexpression and purification of proteins.
all1691 (furA), all2473 (furB or zur), and alr0957 (furC) were PCR amplified from Anabaena genomic DNA using the primer pairs furA-1F–furA-1R, 2473-CLN-1F–2473-CLN-1R, and 0957-CLN-1F–0957-CLN-1R, respectively, cloned in the pCMN28b expression vector (M. Napolitano and I. Luque, unpublished data) in frame with the N-terminal Strep-Tag II sequence tag, and introduced into BL21(DE3). Cells were incubated for 16 h at 16°C after the addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Cell extracts were prepared, and proteins were purified through 1-ml Strep-Tactin Superflow columns (IBA) by following the instructions of the manufacturer.
RNA extraction and Northern blotting.
RNA preparation from cyanobacterial cells and Northern assays were carried out as described in reference 40.
5′-RACE.
Promoter mapping by 5′ rapid amplification of cDNA ends (5′-RACE) was carried out as described in reference 4, with some modifications. Briefly, 50 μg of RNA from Anabaena cells was incubated with 20 U of tobacco alkaline phosphatase (TAP) (1) for 2 h at 37°C. A control reaction mixture was incubated in parallel without enzyme. Both reaction mixtures were ligated to 15 pmol of an RNA oligonucleotide named Bensing RNA (see Table S2 in the supplemental material) with T4 RNA ligase. Aliquots (5 to 7 μg of RNA) of both reaction mixtures were annealed to specific primers for each of the genes in the all4729-all4721 cluster and subjected to retrotranscription with 100 U of Superscript II reverse transcriptase (Invitrogen) at 47°C. cDNAs were amplified by using a forward primer named Bensing DNA (see Table S2) partially overlapping the sequence of the RNA oligonucleotide mentioned above and reverse primers all4727 4R, all4727 5R, all4726 1R, all4726 3R, all4725 3R, all4725 4R, all4724_1R, all4724 2R, all4724 3R, all4724 4R, thrS2 5R, thrS2 6R, thrS2 7R, thrS2 8R, all4722_2R, all4722 3R, all4722 4R, all4722 5R, all4722 6R, all4722 7R, and all4721 1R annealing with the 5′ region of the corresponding open reading frames (ORFs) (see Table S2). The PCR products were resolved on agarose gels, and bands found exclusively in the lanes corresponding to TAP-treated samples were sequenced.
Electrophoresis mobility shift assay (EMSA).
Unless otherwise stated, radioactive DNA fragments to be used in EMSAs were generated by annealing partially overlapping oligonucleotides (all4725-GS-1F and all4725-GS-1R for all4725, all4723-GS-1F and all4723-GS-1R for all4723, all4722-GS-1F and all4722-GS-1R for all4722, all4721-GS-1F and all4721-GS-1R for all4721; see Table S2 in the supplemental material) and filling in with Klenow DNA polymerase in the presence of [32P]dCTP. Forward primer all4725-GS-1F and reverse primers all4725-GS-2R to all4725-GS-14R (see Table S2) were used to generate DNA fragments with mutated versions of the Zur-binding site at the all4725 promoter. DNA was used at 0.1 to 0.5 fmol per reaction, incubated in a buffer containing 20 mM Tris-HCl (pH 8), 50 mM KCl, 1 mM dithiothreitol (DTT), and 20% glycerol for 15 min at room temperature in the absence or presence of 0.01 to 20 pmol of protein, and resolved on 4 to 5% acrylamide gels. For the determination of the Kd, retarded and nonretarded bands were quantified in a Cyclone Plus storage phosphor system (Perkin Elmer), and the percentage of retarded bands was plotted against the concentration of protein in the assay and fitted to a simplified version of the Hill equation (53).
Real-time PCR.
RNAs from wild-type (WT) Anabaena and the zur mutant were treated with DNase and inspected for DNA contamination by conventional PCR. DNA-free RNA (5 μg) was used for cDNA synthesis with a Superscript First Strand cDNA synthesis kit (Invitrogen) using random hexamers as primers. cDNAs were used as templates to set up PCRs with the SensiMix SYBR kit (Bioline) in an iQ5 multicolor real-time PCR detection system (Bio-Rad). The rnpB gene (60) was used as a standard gene for normalization.
RESULTS
Description of a gene cluster encoding several metalloproteins in Anabaena.
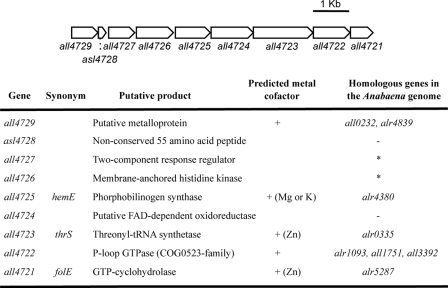
In a survey of duplicated genes in the genome of the cyanobacterium Anabaena sp. strain PCC 7120 (referred to here as Anabaena), a cluster of 9 genes (all4729 to all4721), of which 7 had homologues at different locations in the genome, was observed (Fig. 1). Features of the genes in the cluster are as follows: (i) they appear to be not functionally related; (ii) 6 of their putative products are predicted to contain metal binding sites; (iii) their expression is very low or silent under standard laboratory growth conditions; and (iv) some genes (all4725, all4723, and all4721) encode proteins with putative essential housekeeping functions. In contrast, the respective paralogues (alr4380, alr0335, and alr5287) of the latter genes were highly expressed under standard growth conditions (data not shown).
Fig 1.
Structure of a divalent metal-responsive gene cluster of Anabaena. A diagram of the gene cluster is at the top. +, presence of a putative metal cofactor (specified in parentheses in cases where it is predictable). Anabaena genes homologous to those in the cluster are indicated with their locus tags or with an asterisk when many paralogues are present in the genome.
Characterization of the expression profile of the cluster.
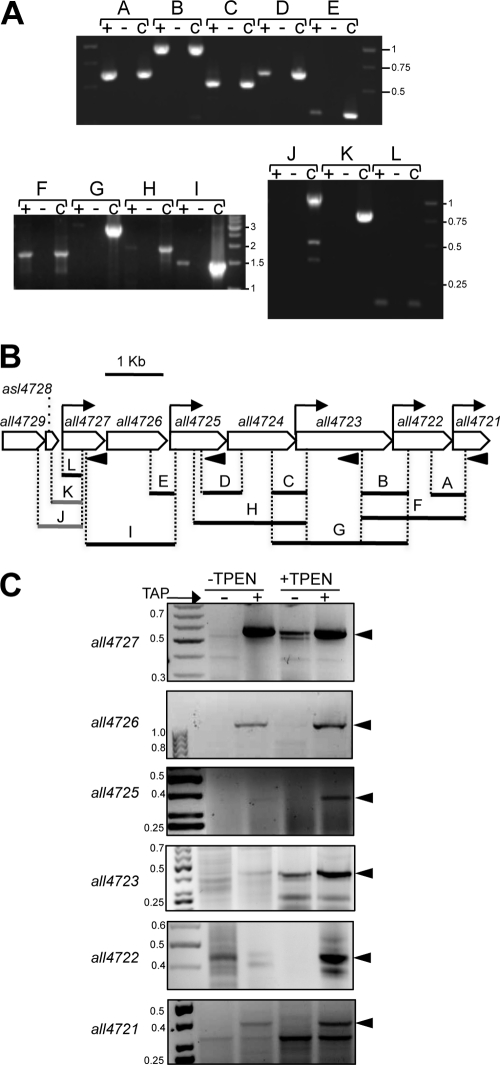
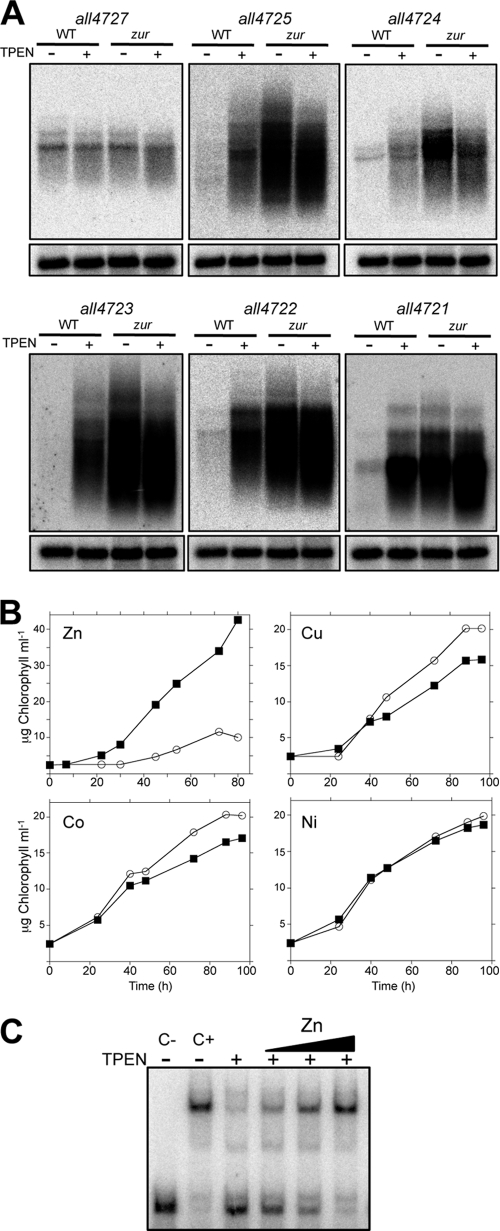
The RNA level of some genes in the cluster was analyzed under several conditions, including diazotrophy, anaerobiosis, nitrogen stress, and high light, and very low mRNA levels were observed under all conditions. Given the presence of putative metal-binding sites in several proteins encoded in the cluster, its expression was analyzed in cells grown for a 24-hour period in the absence or presence of TPEN, a divalent metal chelator. In time course experiments two different profiles were observed; on one side, all4729, all4727, and all4726, in the 5′ region of the cluster, showed discrete hybridization bands that slightly increased their intensity (around 2-fold) upon treatment with TPEN, whereas all4725 to all4721, in the 3′ half of the cluster, showed wide-range (from 0.5 to 7-kb upon overexposure) smeared hybridization signals with some prominent discrete bands that, when quantified as a whole, dramatically increased in intensity (almost 100-fold) a few hours after the addition of TPEN to the cultures (Fig. 2). This indicated cotranscription of genes all4725 to all4721 in long unstable polycistronic transcripts. To check this possibility, the existence of transcripts overlapping contiguous ORFs in the cluster was analyzed by reverse transcription-PCR (RT-PCR). As shown in Fig. 3A, transcripts overlapping contiguous ORFs existed from all4727 to all4721; however, no transcript overlapping asl4728 and all4727 could be observed. Therefore, all4729 and asl4728 are transcribed independently of the remainder of the cluster.
Fig 2.
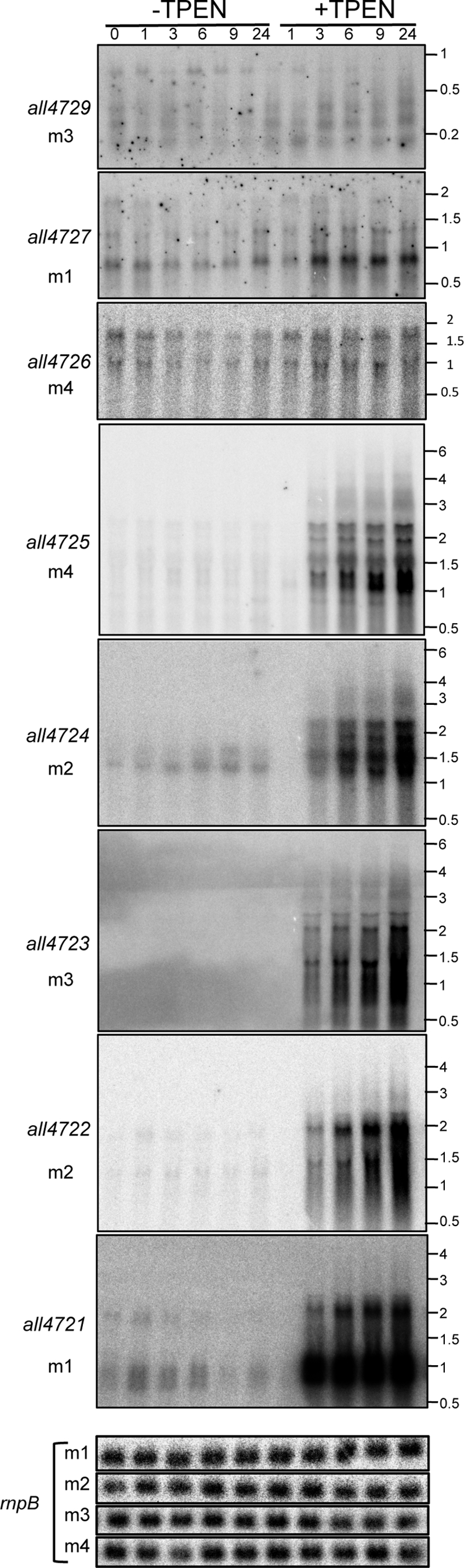
Transcriptional response of the genes in the all4729-all4721 cluster to the chelation of divalent metals. Anabaena cells growing in BG11 medium were split into two cultures; one of them was supplemented with TPEN at a final concentration of 20 μM, and both were further cultured for 24 h. Aliquots were extracted at the times indicated at the top of the panels; RNA was extracted and subjected to Northern hybridization with probes for the genes indicated at the left. m1, m2, m3, and m4 refer to the membranes used for the hybridization. Numbers on the right indicate the positions of RNA molecular weight markers. Hybridization of the four membranes (m1, m2, m3, and m4) with the rnpB probe, used as a control for RNA loading, is shown at the bottom.
Fig 3.
Determination of the limits of the operon by RT-PCR and promoter mapping. (A) Products of RT-PCRs. + and −, reactions in which retrotranscriptase was added or omitted, respectively. C, control PCRs using genomic DNA as the template. The letters A to L identify the fragments on the electrophoresis and correspond to the fragments in panel B. (B) Fragments that were amplified by RT-PCR are depicted with solid lines, and those that failed to be amplified are depicted with gray lines. The primers used for retrotranscription are indicated by arrowheads. Bent arrows indicate the position of transcription start points, mapped by 5′-RACE. (C) Promoter mapping by 5′-RACE. RNA samples from Anabaena cells cultured for 24 h in the presence or absence of TPEN were treated with TAP or left untreated and subjected to 5′-RACE as described in Materials and Methods. The reverse oligonucleotides used in the PCR step annealed with the 5′ region of the ORF indicated to the left of each panel. Numbers indicate the positions of DNA size markers. Arrowheads indicate the positions of major differential bands.
Promoter mapping.
Transcription start points in the cluster were mapped by the 5′-RACE procedure described by Bensing et al. (4), suitable for the distinction of 5′ ends with a triphosphate group, corresponding to true transcription initiation sites, from those with a monophosphate group resulting from the processing of a longer transcript. Transcription start points were mapped upstream of all4727, all4725, all4723, all4722, and all4721 at positions −68, −19, −20, −118, and −90 from their respective ATG codons (Fig. 3B and C). A single transcription start point upstream of all4727 was detected when primers annealing with either all4727 or all4726 were used, indicating that these two ORFs cotranscribe from a single promoter. It is worth noting that the 5′-RACE products obtained with primers for all4725, all4723, all4722, and all4721 correlated in intensity with the signals observed by Northern blotting for samples that were not treated or treated with TPEN, suggesting that they correspond to regulated promoters (Fig. 3C).
Identification of regulatory factors.
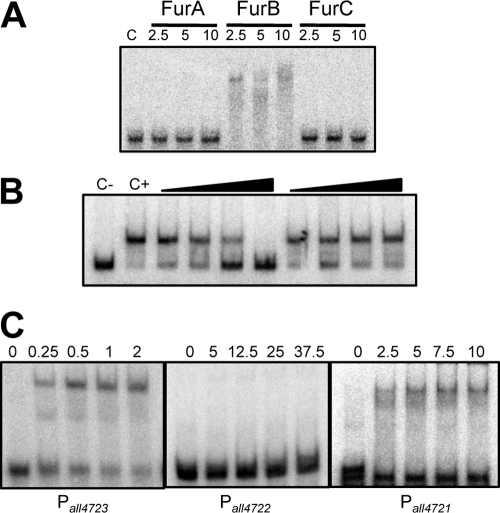
In the Anabaena genome, 3 ORFs (all1691, all2473, and alr0957, also known as furA, furB, and furC) encode proteins of the Fur family (27). To test the possible involvement of any of these three factors in the regulation of the operon, the binding of recombinant purified proteins to a DNA fragment from the promoter region of all4725 was assayed by electrophoresis mobility shift assays (EMSA) (Fig. 4). Only FurB was able to produce a retarded band (Fig. 4A). Binding of FurB to this DNA fragment was demonstrated to be specific in competition assays: an excess of the unlabeled all4725 promoter fragment but not an equivalent amount of an unrelated DNA fragment was able to abolish binding (Fig. 4B). Binding of FurB to other promoter regions in the all4725-all4721 operon was also tested. FurB was able to bind in EMSAs to the promoter regions of all4723 and all4721 but not to that of all4722 (Fig. 4C). To confirm regulation of the operon by FurB, an insertion-deletion mutant was generated by gene replacement. A segregated mutant (i.e., a strain carrying the mutated version in all chromosomes copies) was selected for further studies. In this mutant, expression of the genes in the operon was found to be high under noninducing conditions (e.g., in the absence of TPEN) (Fig. 5A), consistent with a repressor role for FurB in these genes. all4727 was not upregulated in the furB mutant, suggesting that it does not belong to the same transcriptional unit as all4725-all4721. Maximal noninhibitory concentrations of several divalent metals (Cu, Zn, Ni, and Co) for WT Anabaena were determined, and growth of the WT and the furB mutant were tested under these conditions (Fig. 5B). Growth of the furB mutant was severely affected by the presence of zinc but not other metals (Fig. 5B), indicating a major role for FurB in the regulation of zinc homeostasis in Anabaena. To corroborate the specificity of FurB for zinc, EMSA reaction mixtures containing FurB were treated with increasing concentrations of the TPEN chelator, and it was observed that concentrations above 500 μM mostly inhibited the binding of FurB to DNA (Fig. 5C, lane 3). DNA binding was restored when reaction mixtures were later supplemented with zinc (Fig. 5C). These results identified FurB as Zur, the zinc uptake regulator of Anabaena (referred to here as Zur).
Fig 4.
Binding of Fur proteins to the promoter regions of genes in the all4725-all4721 operon. (A) EMSA of FurA, FurB, and FurC with a DNA fragment from the promoter region of all4725. C, control assay in which no protein was added to the reaction mixture. The amounts (pmol) of dimer protein in 15-μl assay mixtures are indicated at the top. (B) Binding to 0.25 fmol of a labeled fragment of the promoter region of all4725 generated with primers all4725_2F and all4725_2R was assayed with no protein in the reaction mixture (C−) or with 0.1 pmol FurB, in the absence (C+) or presence (C+) of a 10-, 50-, 100-, or 200-fold excess of cold all4725 promoter fragment (left) or cold unrelated competitor DNA (right) generated by PCR with primers all4726_2F and all4726_1R. (C) EMSA of FurB with DNA fragments of the promoter regions of all4723, all4722, and all4721. The amount (pmol) of FurB dimer in each reaction is indicated on top of each lane.
Fig 5.
Phenotype of the zur mutant. (A) Northern hybridizations of RNA from cells of the WT or the zur mutant treated with TPEN for 24 h or left untreated. Probes used for hybridization are shown above the panels. Hybridizations of the membranes with the rnpB gene used as a RNA loading control are shown at the bottom of each panel. (B) Growth curves of the WT (squares) and the zur mutant (circles) in the presence of 25 μM ZnSO4, 25 μM CuSO4, 25 μM CoCl2, or 25 μM NiSO4. The data are representative of three independent repeats. (C) EMSA reaction mixtures (15 μl) containing 0.1 pmol of FurB dimer were incubated in the presence (+) or absence (−) of 500 μM TPEN for 1 h at 25°C and subsequently supplemented with increasing concentrations (100, 250, or 500 μM) of ZnSO4, incubated 30 min at 25°C, and resolved in a native acrylamide gel.
Characterization of the Zur-binding sequence.
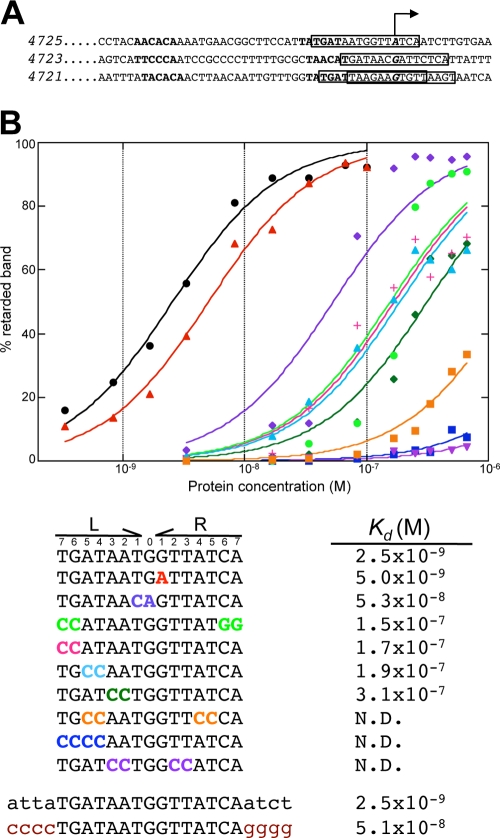
The promoter sequences of all4725, all4723, and all4721 were compared searching for the putative binding sequence for Zur. Sequences with a dyad symmetry axis with some similarity to 7-1-7 sequences reported as targets for proteins of the Fur family (33) were observed in the promoters of all4725 and all4723. In the promoter of all4721, two overlapping imperfect sequences were observed (Fig. 6A). The affinity constant (Kd) of Zur calculated by EMSAs was estimated to be 2.5 × 10−9 and 7 × 10−9 M for the target sequences at all4725 and all4723, respectively (the affinity for the sequences at the all4721 promoter was very low, and the Kd could not be calculated). The affinity of Zur for the sequence at the all4725 promoter was on the same order of magnitude as that of other Fur family factors for their consensus binding sequences (22, 33). To determine the relative contribution of the DNA nucleotides to the interaction with Zur, the effect of mutations in the sequence at the all4725 promoter on the affinity for Zur was analyzed. Except for the mutation at position 1R (Fig. 6B), which makes the sequence perfectly symmetric and does not significantly alter the Kd, any other of the introduced changes had a severe effect on the Kd, indicating that Zur binding to DNA is highly sequence specific. It is be noticed, however, that not all changes had a similar effect, with the mutation of nucleotides 2 and 3 and nucleotides 4 and 5 giving a greater effect than the mutation of nucleotides 6 and 7. Changes at symmetric positions had additive effects in the case of nucleotides 2-3 and 4-5 but not in the case of 6-7. Sequences flanking the symmetric 7-1-7 motif also contribute to Zur binding, since their mutation decreases the affinity for Zur (Fig. 6B).
Fig 6.
Mapping and characterization of Zur-binding sequences. (A) The sequences of the promoter regions of all4725, all4723, and all4721 are depicted. The bent arrow indicates the position of the transcription start point, bold indicates putative −35 and −10 sequences, and putative Zur-binding sequences are boxed. (B) The binding of FurB to different DNA sequences was assayed by EMSA. The plot shows the curves resulting from the adjustment of the percentage of retarded band to a simplified version of the Hill equation. The colors correspond to the sequences at the bottom. The top sequence is that of the all4725 promoter. Mutations introduced to generate the other DNA fragments are indicated by colors corresponding to the graph. The Kd of Zur for each fragment is indicated. The curve for the sequence at the very bottom is not shown.
Insight into the Zur regulon in Anabaena.
As a first approach to identify genes in the Zur regulon in Anabaena, the two sequences with lower Kds in our affinity analyses (Fig. 6B) were used as a seed to search for similar sequences in the Anabaena genome using the Genolist Web server at the Pasteur Institute (http://genodb.pasteur.fr/cgi-bin/WebObjects/GenoList.woa/) allowing a maximum of 2 mismatches. Since only sequences properly located in the promoter (i.e., those overlapping the RNA polymerase binding region) are expected to affect transcription, the search was restricted to sequences located between positions −400 and + 30 with respect to the translational start codon. Thirty-three putative Zur-binding sites were retrieved from this search. Downstream adjacent genes were tested for expression in the WT and the zur mutant by quantitative real-time PCR (Q-PCR) (all4723 and all4725 were retrieved and included in the analysis as controls). When one site was between two divergent ORFs, both were tested. The expression of genes not retrieved in our search but previously reported to be involved in zinc homeostasis in Anabaena, including aztA, aztR, znuABC, and bmtA, was also analyzed (8, 36). Of the 41 genes analyzed (see Table S1 in the supplemental material), 14 showed a higher level of expression in the zur mutant than in the WT, suggesting a direct regulation by Zur (Table 1; a 2-fold difference was established as the minimal threshold). No gene showed a higher level in the WT than in the mutant, and 27 had no change or showed a barely detectable expression level (see Table S1). Comparison using the WebLogo program of the putative Zur-binding sequences upstream of the regulated genes allowed the definition of a consensus binding sequence (TGATAATNATTATCA) for the Anabaena Zur protein (Table 1). Sequences flanking this 7-1-7 palindrome are not highly conserved, but there seems to be some preference for A or T.
Table 1.
Genes derepressed in the zur mutant
Expressed as the ratio of the expression level in the zur mutant to that in the WT in Q-PCR experiments. Data are means ± standard deviations from three independent experiments.
Putative Zur-binding sites on a gray background were not detected in our search at GenoList. A WebLogo sequence built with the sequences in the table is shown at the bottom.
The gene context of the Zur-regulated genes was analyzed, and 7 genes were found clustered with neighbor genes that could potentially be cotranscribed (Table 2). The expression of 15 additional genes clustered with Zur-regulated genes was analyzed by Q-PCR (see Table S1 in the supplemental material). Six were upregulated in the zur mutant, indicating that they likely form transcriptional units with Zur-regulated genes. Thus, in addition to the all4725-all4721 operon (named operon 1), five new Zur-regulated operons (named operons 2 to 6 [Table 2]) have been uncovered. In summary, our analyses have revealed 23 genes regulated by Zur, 17 organized in 6 operons and 6 in single transcriptional units.
Table 2.
Zur-regulated operonsa
Zur-regulated genes are depicted in arbitrary colors and indicated by a bracket. Other neighboring genes and genetic elements that encode putative metalloproteins or that may be functionally related to genes in the operon are in white. Other details are as in Table 1. An asterisk indicates that the induction of alr3243 could not be calculated since its expression was not detected in the WT.
DISCUSSION
Regulation of the all4725-all4721 operon.
Results obtained by RT-PCR (Fig. 3A) showed that transcripts encompassing two or three contiguous ORFS from all4727 to all4721 were detected; however, all4727 and all4726 seemed to be mostly transcribed independently, as deduced from their expression pattern, which is very different from downstream genes (Fig. 2). Indeed, all4727 does not seem to be under the control of zur (Fig. 5A), unlike genes in the all4725-all4721 operon. Four promoters were mapped in the operon, and three were shown to be controlled by zur. It is difficult to determine whether prominent bands hybridizing with probes of the operon in Fig. 2 were due to transcription initiation from internal promoters, to premature termination, or both. However, for some transcripts, the promoter from which they originate could have been ascertained with some precision. For instance, the 1-kb and 2-kb bands hybridizing with probes of all4721 or all4721 and all4722, respectively (Fig. 2), were still visible in a mutant with a polar insertion in all4723 (M. Napolitano, unpublished observations), indicating that the corresponding transcripts originate from promoters located downstream of the insertion. The 2-kb band hybridizes with all4722 (1,062 bp) and all4721 (651 bp) and probably corresponds to a dicistronic transcript originating at Pall4722, and the 1-kb band probably corresponds to a monocistronic transcript originating at Pall4721. The size of the 2-kb band hybridizing to all4723 is similar to the distance between the promoter of this gene and a transcriptional terminator located at the 5′ end of all4722, suggesting that it may correspond to a monocistronic transcript originating at Pall4723.
Although TPEN is often referred to as a zinc-specific chelator, it has been reported to bind other metals (6), and in E. coli it has been shown to alter the expression of genes responding not only to zinc but also to a variety of other divalent metals (56). In our experiments, TPEN altered the expression of the isiA and the petE genes of Anabaena that respond to iron and copper, respectively (see Fig. S2 in the supplemental material). Therefore, in Anabaena, TPEN seems to induce a general divalent metal deficiency, which is consistent with the observations in E. coli (56).
The Zur regulator and the Zur-binding sequence in Anabaena.
Zur is a major regulator of zinc homeostasis, which although well characterized in a variety of bacteria has received little attention in cyanobacteria. The high degree of conservation between different members of the Fur family and the presence of multiple homologues per genome hinders the determination of the metal specificity of these proteins from sequence data alone. Moreover, overall sequence similarity does not always correlate with metal specificity. In this study, all2473 (also named furB) has been identified as the zur gene of Anabaena sp. strain PCC 7120 based on experimental data. All2473/FurB was previously proposed to be a protein that binds unspecifically to DNA and protects it from oxidative damage (38). In contrast to this, results presented in this study show that All2473/FurB binds specifically, with high affinity (∼2.5 × 10−9 M) and in a zinc-dependent manner, to palindromic sequences in the promoter of regulated genes. Furthermore, a deletion mutant is shown to be impaired in the regulation of zinc homeostasis, and the genes under its regulation appear to be involved in the adaptation of Anabaena to zinc deficiency (see below). Altogether, these results strongly indicate that All2473/FurB is the Zur regulator that controls zinc homeostasis in Anabaena. If under particular circumstances in vivo this protein binds unspecifically to DNA, as proposed by López-Gomollón et al. (38), All4723/FurB/Zur would be a “moonlighting” protein with a dual role in the physiology of Anabaena.
Binding to 7-1-7 palindromic sequences has been previously reported for other Fur-family proteins from a variety of bacteria (2, 3, 33, 59). Nucleotides flanking the 7-1-7 sequences also contribute to the interaction with Anabaena Zur (Fig. 6B), and this is consistent with what has been reported for the Zur protein of Bacillus subtilis (22). Zur binding and dissociation from DNA are governed by the occupancy of a regulatory coordination site. Recent crystal structures have revealed three potential metal-coordinating sites in Zur from distinct bacteria (39, 55). The role of site 3 is controversial (41, 55), and it has been proposed that in vivo, zinc never binds to this site (41). Site 1 is a structural site, whereas site 2 is postulated to be the regulatory on-off switch (39, 41, 55). Consistently, the affinity of zinc for this site is in the predicted range of the buffered concentration of zinc in the cytoplasm (pico- to femtomolar) (41, 45). Importantly, regulatory site 2 is missing in the protein identified as the Anabaena Zur; only one (His78) of the four coordinating residues is conserved. In contrast, structural site 1 and site 3 are conserved (see Fig. S1 in the supplemental material), although amino acids at positions 75 and 96 of site 3 are permuted with respect to other Zur proteins (see Fig. S1). The absence of the regulatory site 2 in Anabaena Zur raises the question of which site functions as the on-off switch. Further work is required to elucidate the structural basis for the responsiveness of Anabaena Zur to zinc.
The Zur regulon.
These data have revealed 23 genes regulated by Zur, organized in 6 operons and single transcriptional units. The presence of internal promoters in operon 1 indicates that distinct genes may require some degree of independent regulation. Distinct Zur-regulated promoters have also been observed in the yciABC operon of Bacillus subtilis, correlating with different expression levels of the genes (20). Most proteins upregulated in the zur mutant of Anabaena can be classified into 4 categories, described below, that outline the strategy of this cyanobacterium for adaptation to zinc limitation.
Category 1: paralogues of zinc metalloproteins.
It has been shown that three genes in operon 1 encode proteins with essential housekeeping functions (All4725/HemE, All4723/ThrS and All4721/FolE). Their paralogues Alr4380/HemE, Alr0335/ThrS, and Alr5287/FolE, which are highly expressed under standard growth conditions, are all putative zinc metalloproteins based on the conservation of essential liganding residues. Very importantly, All4725/HemE is a porphobilinogen synthase with an aspartate-rich active site (DvALDpFtthGHDG) that fits the consensus sequence DxALDx(Y/F)xxxG(H/Q)DG for Mg- or K-dependent enzymes. In contrast, the active site of the paralogous Alr4380/HemE protein conforms to the consensus sequence of zinc-dependent enzymes [DxCxCx(Y/F)x3G(H/Q)CG] (31). Thus, the derepression of all4725 under zinc limitation would probably lead to the replacement of a zinc-dependent porphobilinogen synthase (All4380) by a zinc-independent one (All4725), under conditions in which the former is most probably inactive. Replacement of zinc-dependent proteins by zinc-independent isoforms seems to be a common strategy in a variety of prokaryotes (5, 16, 21, 31, 50). Less intuitive is the model for All4723/ThrS and All4721/FolE, both of which do conserve the deduced coordination residues for zinc. Adaptation to zinc limitation via the replacement of a zinc-dependent enzyme by another which is also zinc dependent does not appear to be an effective strategy, unless the alternative enzyme is somehow more efficient at the recruitment of zinc cations. Another possibility is that All4723/ThrS and All4721/FolE could be functional with a metal cofactor other than zinc. In any case, All4723/ThrS (and probably All4721/FolE) must somehow better fulfill the housekeeping function under zinc-limiting conditions and is essential for survival under these conditions (Napolitano et al., unpublished).
Category 2: genes encoding putative metallochaperones.
all4722 and all1751 encode G3E P-loop GTPases of the COG0523 family. Like other members of the family, they show a GTPase motif in the N terminus, a conserved putative metal-binding CXXC motif, and a C-terminal domain with a His-rich stretch. However, All4722 contains 19 His residues, whereas All1751 contains only 2. all1197 is also in this category although its product has a deletion in the N-terminal domain and a single histidine in the C-terminal domain. A recent exhaustive bioinformatic analysis classified members of the COG0523 family into 15 subfamilies (24). That study proposed that several of these subfamilies have a role in survival under conditions of deficient zinc nutrition and are frequently associated with Zur-binding sites in diverse bacteria (24). Consistent with this, Bacillus subtilis mutants defective in a protein of the COG0523 family exhibit a growth defect in low-zinc medium (19). Although the exact role of All4722 is still to be determined, it seems important for the survival of Anabaena under metal limitation (Napolitano et al., unpublished). Thus, our results further support a role for proteins of the COG0523 family in the adaptation to zinc limitation.
Category 3: genes encoding components of plasma membrane ABC transport systems.
Several genes regulated by Zur encode subunits of ABC transport systems of the plasma membrane. In general, these transporters are constituted by a substrate-binding periplasmic protein, an intrinsic membrane component, and an ATPase subunit in the cytoplasmic site (28). all0833, all0832, and all0830 in operon 6 were previously identified as the genes encoding the ZnuABC zinc uptake system of Anabaena based on sequence similarity (36). Two other genes, alr3243 in operon 4 and alr4031 in operon 5, encode the periplasmic protein of ABC transporters. In both cases, neighboring genes not regulated by Zur (alr3240 and alr3241 in operon 4 and alr4032 and alr4033 in operon 5) encode the membrane-intrinsic and the ATPase subunits, suggesting that each cluster may encode an individual ABC transport system (Table 2). The substrate specificity of these transport systems will need to be determined by specific approaches.
Category 4: genes encoding proteins of the outer membrane.
TonB-dependent receptors (TBDRs) are integral proteins of the outer membrane of Gram-negative bacteria which function as energy-dependent importers of extracellular molecules (44). Transport by TBDRs of iron complexes (siderophores, heme, and ferritin) and vitamin B12 (cobalamin) is well characterized (44). However, the involvement of the TBDR in the import of divalent metals other than iron was not reported until very recently (29, 51, 58). Twenty-two genes in the Anabaena genome encode putative TBDRs (43), suggesting that some of them may be involved in the transport of molecules other than iron complexes or vitamin B12. Genes encoding TBDRs have not been found in previous analyses of Zur regulons in Gram-negative bacteria (34, 46). Two genes, alr3242 and alr4028-4029 (the latter is a single ORF mistakenly annotated as two independent genes [43]), encoding putative TBDRs regulated by Zur in Anabaena are described here. alr3242 is highly induced (∼100-fold) and reaches a high level of expression in the zur mutant, suggesting that Alr3242 is probably a TBDR specific for the import of zinc complexes from the outer medium, which hitherto has been described only for human pathogens (58). The alr4028-4029 gene is also induced in the zur mutant but to a lower extent (∼10-fold) (Table 2). Interestingly, the two genes encoding TBDRs cotranscribe with genes for periplasmic proteins of ABC transporters (Table 2), suggesting the necessity of a coordinated regulation of transporters of the outer membrane and the plasma membrane.
The protein encoded by the all3515 gene is predicted by the PSORT.3b program (63) to be located in the outer membrane. This gene shows the highest induction level of the Zur regulon (more than 250 times) and reaches a very high level of expression in the zur mutant. All3515 contains a putative signal peptide in the N terminus, a PEP-CTERM domain in the C terminus, which is proposed to mediate secretion through the plasma membrane (25), and two conserved His-rich regions in the N-terminal part that may be involved in metal coordination. Although the exact role of the All3515 protein is unknown, its regulation by Zur, its peripheral localization in the cell, and its expected abundance suggest a role in scavenging zinc or zinc complexes under limiting conditions.
Other global analyses of the Zur regulon performed in Gram-positive and Gram-negative bacteria have unveiled a variety of genes directly or indirectly regulated by Zur (18, 20, 34, 42, 46, 52). Although our analysis of the Zur regulon of Anabaena would need to be completed with further studies, genes uncovered in our work outline how Anabaena rearranges its physiology to adapt to zinc limitation: transporters of the outer membrane and plasma membrane are induced for scavenging extracellular zinc or zinc complexes; putative metallochaperones, which may mediate the delivery of zinc or alternative metals to target proteins, are expressed in the cytoplasm; and some zinc-specific metalloproteins are replaced by paralogues able to fulfill essential functions during zinc limitation.
Supplementary Material
ACKNOWLEDGMENTS
We are indebted to Antonia Herrero for critical reading of the manuscript.
This work was supported by grants BFU2007-66589/BMC and BFU2010-19544 (Ministerio de Ciencia e Innovación & Fondo Social Europeo) and P07-CVI-03167 (Proyectos de Excelencia, Junta de Andalucía y FEDER).
Footnotes
Published ahead of print 2 March 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Ausubel FM, et al. 2010. Current protocols in molecular biology. Greene Publishing & Wiley-Interscience, New York, NY [Google Scholar]
- 2. Baichoo N, Helmann JD. 2002. Recognition of DNA by Fur: a reinterpretation of the Fur box consensus sequence. J. Bacteriol. 184:5826–5832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baichoo N, Wang T, Ye R, Helmann JD. 2002. Global analysis of the Bacillus subtilis Fur regulon and the iron starvation stimulon. Mol. Microbiol. 45:1613–1629 [DOI] [PubMed] [Google Scholar]
- 4. Bensing BA, Meyer BJ, Dunny GM. 1996. Sensitive detection of bacterial transcription initiation sites and differentiation from RNA processing sites in the pheromone-induced plasmid transfer system of Enterococcus faecalis. Proc. Natl. Acad. Sci. U. S. A. 93:7794–7799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Blaby-Haas CE, Furman R, Rodionov DA, Artsimovitch I, de Crecy-Lagard V. 2011. Role of a Zn-independent DksA in Zn homeostasis and stringent response. Mol. Microbiol. 79:700–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Blindauer CA, Razi MT, Parsons S, Sadler PJ. 2006. Metal complexes of N,N,N′,N′-tetrakis(2-pyridylmethyl)ethylenediamine (TPEN): variable coordination numbers and geometries. Polyhedron 25:513–520 [Google Scholar]
- 7. Blindauer CA. 2008. Zinc-handling in cyanobacteria: an update. Chem. Biodivers. 5:1990–2013 [DOI] [PubMed] [Google Scholar]
- 8. Blindauer CA, et al. 2002. Multiple bacteria encode metallothioneins and SmtA-like zinc fingers. Mol. Microbiol. 45:1421–1432 [DOI] [PubMed] [Google Scholar]
- 9. Borrelly GP, Rondet SA, Tottey S, Robinson NJ. 2004. Chimeras of P-type ATPases and their transcriptional regulators: contributions of a cytosolic amino-terminal domain to metal specificity. Mol. Microbiol. 53:217–227 [DOI] [PubMed] [Google Scholar]
- 10. Brocklehurst KR, et al. 1999. ZntR is a Zn(II)-responsive MerR-like transcriptional regulator of zntA in Escherichia coli. Mol. Microbiol. 31:893–902 [DOI] [PubMed] [Google Scholar]
- 11. Capone DG, Zehr J, Paerl HW, Bergman B, Carpenter EJ. 1997. Trichodesmium, a globally significant marine cyanobacterium. Science 276:1221–1229 [Google Scholar]
- 12. Cavet JS, Borrelly GP, Robinson NJ. 2003. Zn, Cu and Co in cyanobacteria: selective control of metal availability. FEMS Microbiol. Rev. 27:165–181 [DOI] [PubMed] [Google Scholar]
- 13. Coleman JE. 1992. Zinc proteins: enzymes, storage proteins, transcription factors, and replication proteins. Annu. Rev. Biochem. 61:897–946 [DOI] [PubMed] [Google Scholar]
- 14. Elhai J, Wolk CP. 1988. Conjugal transfer of DNA to cyanobacteria. Methods Enzymol. 167:747–754 [DOI] [PubMed] [Google Scholar]
- 15. Elhai J, Wolk CP. 1988. A versatile class of positive-selection vectors based on the nonviability of palindrome-containing plasmids that allows cloning into long polylinkers. Gene 68:119–138 [DOI] [PubMed] [Google Scholar]
- 16. El Yacoubi B, et al. 2006. Discovery of a new prokaryotic type I GTP cyclohydrolase family. J. Biol. Chem. 281:37586–37593 [DOI] [PubMed] [Google Scholar]
- 17. Escolar L, Perez-Martin J, de Lorenzo V. 1999. Opening the iron box: transcriptional metalloregulation by the Fur protein. J. Bacteriol. 181:6223–6229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Feng Y, et al. 2008. Functional definition and global regulation of Zur, a zinc uptake regulator in a Streptococcus suis serotype 2 strain causing streptococcal toxic shock syndrome. J. Bacteriol. 190:7567–7578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gaballa A, Helmann JD. 1998. Identification of a zinc-specific metalloregulatory protein, Zur, controlling zinc transport operons in Bacillus subtilis. J. Bacteriol. 180:5815–5821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gaballa A, Wang T, Ye RW, Helmann JD. 2002. Functional analysis of the Bacillus subtilis Zur regulon. J. Bacteriol. 184:6508–6514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gabriel SE, Helmann JD. 2009. Contributions of Zur-controlled ribosomal proteins to growth under zinc starvation conditions. J. Bacteriol. 191:6116–6122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gabriel SE, Miyagi F, Gaballa A, Helmann JD. 2008. Regulation of the Bacillus subtilis yciC gene and insights into the DNA-binding specificity of the zinc-sensing metalloregulator Zur. J. Bacteriol. 190:3482–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Goericke R, Welshmeyer NA. 1993. The marine prochlorophyte Prochlorococcus contributes significantly to phytoplankton biomass and primary production in the Sargasso Sea. Deep Sea Res. 40:2283–2294 [Google Scholar]
- 24. Haas CE, et al. 2009. A subset of the diverse COG0523 family of putative metal chaperones is linked to zinc homeostasis in all kingdoms of life. BMC Genomics 10:470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Haft DH, Paulsen IT, Ward N, Selengut JD. 2006. Exopolysaccharide-associated protein sorting in environmental organisms: the PEP-CTERM/EpsH system. Application of a novel phylogenetic profiling heuristic. BMC Biol. 4:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hantke K. 2005. Bacterial zinc uptake and regulators. Curr. Opin. Microbiol. 8:196–202 [DOI] [PubMed] [Google Scholar]
- 27. Hernández JA, López-Gomollón S, Bes MT, Fillat MF, Peleato ML. 2004. Three fur homologues from Anabaena sp. PCC7120: exploring reciprocal protein-promoter recognition. FEMS Microbiol. Lett. 236:275–282 [DOI] [PubMed] [Google Scholar]
- 28. Higgins CF. 2001. ABC transporters: physiology, structure and mechanism–an overview. Res. Microbiol. 152:205–210 [DOI] [PubMed] [Google Scholar]
- 29. Hohle TH, Franck WL, Stacey G, O'Brian MR. 2011. Bacterial outer membrane channel for divalent metal ion acquisition. Proc. Natl. Acad. Sci. U. S. A. 108:15390–15395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hudek L, et al. 2009. Bioinformatic and expression analyses of genes mediating zinc homeostasis in Nostoc punctiforme. Appl. Environ. Microbiol. 75:784–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jaffe EK. 2003. An unusual phylogenetic variation in the metal ion binding sites of porphobilinogen synthase. Chem. Biol. 10:25–34 [DOI] [PubMed] [Google Scholar]
- 32. Kloosterman TG, van der Kooi-Pol MM, Bijlsma JJ, Kuipers OP. 2007. The novel transcriptional regulator SczA mediates protection against Zn2+ stress by activation of the Zn2+-resistance gene czcD in Streptococcus pneumoniae. Mol. Microbiol. 65:1049–1063 [DOI] [PubMed] [Google Scholar]
- 33. Lee JW, Helmann JD. 2007. Functional specialization within the Fur family of metalloregulators. Biometals 20:485–499 [DOI] [PubMed] [Google Scholar]
- 34. Li Y, et al. 2009. Characterization of Zur-dependent genes and direct Zur targets in Yersinia pestis. BMC Microbiol. 9:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu HB, Nolla HA, Campbell L. 1997. Prochlorococcus growth rate and contribution to primary production in the equatorial and subtropical North Pacific Ocean. Aquatic Microb. Ecol. 12:39–47 [Google Scholar]
- 36. Liu T, Golden JW, Giedroc DP. 2005. A zinc(II)/lead(II)/cadmium(II)-inducible operon from the cyanobacterium Anabaena is regulated by AztR, an alpha3N ArsR/SmtB metalloregulator. Biochemistry 44:8673–8683 [DOI] [PubMed] [Google Scholar]
- 37. Llull D, et al. 2011. Lactococcus lactis ZitR is a zinc-responsive repressor active in the presence of low, nontoxic zinc concentrations in vivo. J. Bacteriol. 193:1919–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. López-Gomollón S, Sevilla E, Bes MT, Peleato ML, Fillat MF. 2009. New insights into the role of Fur proteins: FurB (All2473) from Anabaena protects DNA and increases cell survival under oxidative stress. Biochem. J. 418:201–207 [DOI] [PubMed] [Google Scholar]
- 39. Lucarelli D, et al. 2007. Crystal structure and function of the zinc uptake regulator FurB from Mycobacterium tuberculosis. J. Biol. Chem. 282:9914–9922 [DOI] [PubMed] [Google Scholar]
- 40. Luque I, Contreras A, Zabulon G, Herrero A, Houmard J. 2002. Expression of the glutamyl-tRNA synthetase gene from the cyanobacterium Synechococcus sp PCC 7942 depends on nitrogen availability and the global regulator NtcA. Mol. Microbiol. 46:1157–1167 [DOI] [PubMed] [Google Scholar]
- 41. Ma Z, Gabriel SE, Helmann JD. 2011. Sequential binding and sensing of Zn(II) by Bacillus subtilis Zur. Nucleic Acids Res. 39:9130–9138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Maciag A, et al. 2007. Global analysis of the Mycobacterium tuberculosis Zur (FurB) regulon. J. Bacteriol. 189:730–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mirus O, Strauss S, Nicolaisen K, von Haeseler A, Schleiff E. 2009. TonB-dependent transporters and their occurrence in cyanobacteria. BMC Biol. 7:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Noinaj N, Guillier M, Barnard TJ, Buchanan SK. 2010. TonB-dependent transporters: regulation, structure, and function. Annu. Rev. Microbiol. 64:43–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Outten CE, O'Halloran TV. 2001. Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science 292:2488–2492 [DOI] [PubMed] [Google Scholar]
- 46. Panina EM, Mironov AA, Gelfand MS. 2003. Comparative genomics of bacterial zinc regulons: enhanced ion transport, pathogenesis, and rearrangement of ribosomal proteins. Proc. Natl. Acad. Sci. U. S. A. 100:9912–9917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Partensky F, Hess WR, Vaulot D. 1999. Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol. Mol. Biol. Rev. 63:106–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rippka R. 1988. Isolation and purification of cyanobacteria. Methods Enzymol. 167:3–27 [DOI] [PubMed] [Google Scholar]
- 49. Robinson NJ, et al. 1990. Prokaryotic metallothionein gene characterization and expression: chromosome crawling by ligation-mediated PCR. Proc. Biol. Sci. 242:241–247 [DOI] [PubMed] [Google Scholar]
- 50. Sankaran B, et al. 2009. Zinc-independent folate biosynthesis: genetic, biochemical, and structural investigations reveal new metal dependence for GTP cyclohydrolase IB. J. Bacteriol. 191:6936–6949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schauer K, Gouget B, Carriere M, Labigne A, de Reuse H. 2007. Novel nickel transport mechanism across the bacterial outer membrane energized by the TonB/ExbB/ExbD machinery. Mol. Microbiol. 63:1054–1068 [DOI] [PubMed] [Google Scholar]
- 52. Schroder J, Jochmann N, Rodionov DA, Tauch A. 2010. The Zur regulon of Corynebacterium glutamicum ATCC 13032. BMC Genomics 11:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Segel IH. 1976. Biochemical calculations, 2nd ed John Wiley and Sons, Inc, New York, NY [Google Scholar]
- 54. Shafeeq S, Kloosterman TG, Kuipers OP. 2011. Transcriptional response of Streptococcus pneumoniae to Zn2+ limitation and the repressor/activator function of AdcR. Metallomics 3:609–618 [DOI] [PubMed] [Google Scholar]
- 55. Shin JH, et al. 2011. Graded expression of zinc-responsive genes through two regulatory zinc-binding sites in Zur. Proc. Natl. Acad. Sci. U. S. A. 108:5045–5050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sigdel TK, Easton JA, Crowder MW. 2006. Transcriptional response of Escherichia coli to TPEN. J. Bacteriol. 188:6709–6713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Smith KS, Ferry JG. 2000. Prokaryotic carbonic anhydrases. FEMS Microbiol. Rev. 24:335–366 [DOI] [PubMed] [Google Scholar]
- 58. Stork M, et al. 2010. An outer membrane receptor of Neisseria meningitidis involved in zinc acquisition with vaccine potential. PLoS Pathog. 6:e1000969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tottey S, et al. 2012. Cyanobacterial metallochaperone inhibits deleterious side reactions of copper. Proc. Natl. Acad. Sci. U. S. A. 109:95–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Vioque A. 1992. Analysis of the gene encoding the RNA subunit of ribonuclease P from cyanobacteria. Nucleic Acids Res. 20:6331–6337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Waldron KJ, Robinson NJ. 2009. How do bacterial cells ensure that metalloproteins get the correct metal? Nat. Rev. Microbiol. 7:25–35 [DOI] [PubMed] [Google Scholar]
- 62. Whitton BA, Potts M. 2002. Introduction to the cyanobacteria, p 1–10 In Whitton BA, Potts M. (ed), The ecology of cyanobacteria: their diversity in time and space. Kluwer Academic Publisher, New York, NY [Google Scholar]
- 63. Yu NY, et al. 2010. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26:1608–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.