Abstract
Terminal sialic acid in the lipopolysaccharides (LPSs) of mucosal pathogens is an important virulence factor. Here we report the characterization of a Helicobacter sialyltransferase involved in the biosynthesis of sialylated LPS in Helicobacter bizzozeronii, the only non-pylori gastric Helicobacter species isolated from humans thus far. Starting from the genome sequences of canine and human strains, we identified potential sialyltransferases downstream of three genes involved in the biosynthesis of N-acetylneuraminic acid. One of these candidates showed monofunctional α,2,3-sialyltransferase activity with a preference for N-acetyllactosamine as a substrate. The LPSs from different strains were shown by SDS-PAGE and high-performance anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD) to contain sialic acid after neuraminidase treatment. The expression of this sialyltransferase and sialyl-LPS appeared to be a phase-variable characteristic common to both human and canine H. bizzozeronii strains. The sialylation site of the LPSs of two H. bizzozeronii strains was determined to be NeuAc-Hex-HexNAc, suggesting terminal 3′-sialyl-LacNAc. Moreover, serological typing revealed the possible presence of sialyl-Lewis X in two additional strains, indicating that H. bizzozeronii could also mimic the surface glycans of mammalian cells. The expression of sialyl-glycans may influence the adaptation process of H. bizzozeronii during the host jump from dogs to humans.
INTRODUCTION
Structural similarity between microbial and mammalian glycans (molecular mimicry) is a well-established phenomenon in many human mucosal pathogens, including Campylobacter jejuni and Helicobacter pylori (24, 36). C. jejuni expresses different types of lipooligosaccharides (LOSs), which resemble the ganglioside structures present on mammalian cells (24), and H. pylori expresses fucosylated Lewis and related antigens in the O chain on its lipopolysaccharides (LPSs) (36). A key enzyme for the ganglioside mimicry of C. jejuni LOSs is the sialyltransferase (ST) that belongs to glycosyltransferase family 42 (GT-42), according to the sequence-based classification of glycosyltransferases available in the CAZy database (10).
It has been shown that H. pylori LPSs express sialyl-Lewis X antigen (36), but the presence of sialylated LPSs has rarely been reported for this species, and the ST involved has not been characterized. Moreover, although putative GT-42 STs have been described for animal gastric Helicobacter spp. (H. acinonychis, H. mustelae, and H. felis), there are no studies that have reported a glycan containing sialic acid in these species, nor have any studies investigated the activity and substrate specificity of these enzymes.
H. pylori is the primary cause of gastritis and peptic ulceration in humans and is a major risk factor for mucosa-associated lymphoid tissue (MALT) lymphoma and adenocarcinoma (6). However, in gastric biopsy specimens of a minority of patients with upper gastrointestinal symptoms (0.17 to 2.3%), long, tightly coiled, spiral bacteria, ascribed to non-pylori Helicobacter spp., have been observed (5). Among the five non-pylori gastric Helicobacter species reported to be involved in human infections (19), only Helicobacter bizzozeronii has been successfully cultivated from human gastric biopsy specimens (27, 30). Humans may acquire the infections as a consequence of direct contact with dogs or cats (35), which are normally colonized by H. bizzozeronii (21). However, although virtually all dogs carry this Helicobacter species, its pathogenic significance in these animals remains unknown (19).
The role of H. bizzozeronii in human disease is limited compared to that of H. pylori, but both species persistently colonize the same niche and may exploit similar mechanisms to interact with the host, evade the immune system, and induce gastritis (45). Moreover, in order to colonize the stomach of the human host and induce gastritis, H. bizzozeronii undergoes an adaptation process (45). This process may involve changes to external structures, including the LPSs.
A genome analysis of human-derived H. bizzozeronii CIII-1 revealed the presence of a glycosyltransferase which appears to encode an ST belonging to the GT-42 family (44). We further identified two additional GT-42 STs cotranscribed with three genes involved in the biosynthesis of N-acetylneuraminic acid in the shotgun genome sequence of another H. bizzozeronii strain (StorkisT). The aim of this study was to characterize these STs in terms of activity and substrate specificity and to identify the nature of sialyl-glycans expressed by human- and canine-derived H. bizzozeronii strains.
MATERIALS AND METHODS
Bacterial strains, cultivation, and DNA extraction.
The bacterial strains and plasmids used in this study are listed in Table 1. Helicobacter strains were grown on HP medium (LabM, Ltd., Lancashire, United Kingdom) containing 5% bovine blood and Skirrow selective supplement (Oxoid, Ltd., Cambridge, United Kingdom) at 37°C in a microaerobic atmosphere (Thermo Forma series II water-jacketed incubator; Thermo Fisher Scientific, Waltham, MA). For DNA, RNA, and LPS extraction, Helicobacter strains were cultivated in brain heart infusion (BHI) broth (BD, Becton Dickinson and Co., Franklin Lakes, NJ) containing 10% fetal bovine serum (Gibco, Invitrogen, Carlsbad, CA), Skirrow selective supplement (Oxoid), and Vitox supplement (Oxoid) at 37°C in a jar with a microaerobic atmosphere. C. jejuni strains were cultivated on Mueller-Hinton agar (BD) with 5% bovine blood at 37°C in an incubator with a microaerobic atmosphere. Escherichia coli strains were cultivated on Luria-Bertani (LB) medium or 2× yeast extract-tryptone (2×YT) medium (Bio 101, Carlsbad, CA) supplemented with 150 mg/liter of ampicillin or 20 mg/liter of chloramphenicol when needed. DNA extraction was performed as described previously (30).
Table 1.
Bacterial species and plasmids used in this study
| Bacterial species or plasmid | Strain (GT-42 locus EMBL accession no.), relevant characteristic(s), or genotype | Reference or source |
|---|---|---|
| Bacterial species | ||
| H. bizzozeronii | StorkisT (CCUG 35545T) (HE653986) | 28 |
| 14 (CCUG 35546) (HE653982) | 28 | |
| 12a (HE653981) | 28 | |
| Heydar (HE653984) | 28 | |
| Yrjälä (HE653987) | 28 | |
| Emo (HE653983) | 28 | |
| CIII-1 | 30 | |
| Rigshospitalet 53 (R53) (HE653985) | 27 | |
| H. pylori | 26695 | 50 |
| P466 | 36 | |
| C. jejuni | 81-176 | 9 |
| 11168 | 15 | |
| Escherichia coli | ||
| JM109 | endA1 glnV44 thi-1 relA1 gyrA96 recA1 mcrB+ Δ(lac-proAB) e14− [F′ traD36 proAB+lacIqlacZΔM15] hsdR17(rK− mK+) | Promega |
| AD202 | F−ompT::Km Δ(argF lac)U169 araD139 relA1 rpsL150 flbB5301 deoC1 tonA21 thi ptsF25 | 1 |
| DH5α | F− endA1 glnV44 thi-1 recA1 relA1 gyrA96 deoR nupG ϕ80dlacZΔM15 Δ(lacZYA-argF)U169 hsdR17(rK− mK+) λ− | 20 |
| Plasmids | ||
| pCWmalE-T | pCWori+malE | 8 |
| pHBS01 | pCWmalE-T MalE-HBS01 | This study |
| pHBS02 | pCWmalE-T MalE-HBS02 | This study |
Shotgun genome sequencing and genome analysis.
The draft genome of H. bizzozeronii strain StorkisT (CCUG 35545T; EMBL WGS project CAGP00000000) was sequenced by use of the 454-pyrosequencing technique (Roche Diagnostics GmbH, Mannheim, Germany) with approximately 20-fold coverage and assembled into 147 contigs greater than 500 bp. Contigs were automatically annotated by use of the RAST Server (4).
Phylogenetic analysis.
Phylogenetic analyses of the amino acid sequences of the GT-42 ST were conducted with MEGA5 (48), using the minimum evolution (ME) method. The amino acid sequences were aligned by the use of MAFFT (29). The evolutionary distances were computed by the Dayhoff matrix-based method, and the ME tree was searched by using the close-neighbor-interchange (CNI) algorithm (level 1). All positions with less than 90% site coverage were eliminated.
PCR.
Oligonucleotides used for the DNA amplification of the GT-42 locus from H. bizzozeronii strains are listed in Table 2. Phusion high-fidelity DNA polymerase (Finnzymes, Oy, Espoo, Finland) was used to perform standard PCR, followed by sequencing. If needed, the PCR products were inserted into the pGEM-T Easy plasmid vector (Promega Co., Madison, WI) and cloned into E. coli JM109 cells, and the derivative plasmids were sequenced by the use of M13 primers. Sequences were analyzed with MEGA5 (48).
Table 2.
Oligonucleotides used in this study
| Function and oligonucleotide | Sequence |
|---|---|
| Expression of recombinant proteins | |
| Hb-ST1 | |
| MJS14 | GGAGGAGAATTCCATATGAAACCCTTAATCATCGCCGGT |
| MJS15 | GGAGGAGTCGACTTATTACCCCCCAATATCAGGCGCATCTGG |
| Hb-ST2 | |
| MJS18 | GGAGGAGAATTCCATATGCCACTTAAGCCCCTCATCGTTGCCGGC |
| MJS19 | GAGGAGTCGACTTATTACCCTCCCGTAGCCCTATGGCGTTTAAAC |
| Amplification and sequencing of the GT-42 locus and RT-PCR | |
| NeuAE1 | AAATGGGTGTTAGGCGGTGGTTG |
| NeuAE2 | TAAGCTGGTGTTGGGCGTTAGGG |
| NeuAE3 | AAGTGGCTCTCAGGGCGTGAAAC |
| NeuAE4 | CACCATTAGCGGGCTTTTAGAAGC |
| NeuAE5 | CAGCGTGCATTTGGTTACAGAGC |
| NeuAE6 | ACCAAGTAGAAACCCTAACGCCCAACACC |
| NeuAE7 | ACATCCACCAAGTAGAAACCCTAACGC |
| NeuBE1 | GCCCGGAGCGATGTTAGCAATAG |
| NeuBE2 | GCCCTTAGTGGTGGCTGAAGTGG |
| NeuBE1rc | TTGCTATTGCTAACATCGCTCCG |
| NeuBE3 | AGACTTTGGGACGGGTGGCTAAGGTG |
| NeuCE1 | CGGCTACCCAAATTCAAGCAAGG |
| NeuCE2 | TTTGCCCCGTGCCAAAATGATAG |
| NeuCE3 | TTGTGTGTGGCATGCACCTTTTG |
| NeuCE4 | CACCACTTTCTTGCCCTGCAACG |
| NeuCE5 | TATGACGGATGCTCTCATCTAAAGTCC |
| NeuCE6 | GAGCTTAGCGGGACTTTAGATGAGAG |
| hbs1-a | TAGCCACCTAGTTGCGTGCGATG |
| hbs1-b | TCCAATATGGTCGTTTCTTTCTTGTTGG |
| hbs1-c | CTCTCTGCAATCTTACCCTTGAGCC |
| hbs1-c(rc) | GGCTCAAGGGTAAGATTGCAGAGAG |
| hbs1-d | AATCCGGTGGGAATAAACTATAATCGAGG |
| hbs1-e | TATAGCCTGCATGTCTAGCTCTTGG |
| hbs2-a | AGCGCATAAAGCTGGGCGTATTG |
| hbs2-b | AAGGCTGTAATCTAAATCTTTAATGCTCGG |
| hbs1-fw | CGCAAAGATGGCGGATCGCTTAAC |
| hbs1-rw | TTGTGTGCAGGTCTAAAAAGGGTTTTAGAGTGG |
| promSAB1F | TCTTTACTTGTCCATTGGTTTTGGT |
| promSAB1R | TCTAAACTCCCATTATGGTTGATCC |
| promSAB2F | GATCGAAAATAGAGGCGTTCTTGTA |
| promSAB2R | CTTGGCAATCTCTAAACTCCCATTA |
| RTHBS2rw1 | CCATGTTCAAACCTTTGTAAATGAGATTGTC |
| SSCP/heteroduplex analysis | |
| SSCPNeuCE3b | TTATGCGATCTTTGTGTGTGGCATGCACCTTTTGG |
| SSCPNeuCE5b | ATTTAGACACGCTGTGGCGGATACTCTCATCTAAAGTTC |
Single-strand conformation polymorphism (SSCP)/heteroduplex analysis.
PCR was performed by using Phusion high-fidelity DNA polymerase (Finnzymes) and primers SSCPneuCE3b and SSCPneuCE6b to amplify a ∼240 bp fragment containing the homopolymeric tract. The PCR products were mixed with the same volume of formamide loading buffer (80% formamide in 10 mM EDTA, 1 mg/ml of xylene, and 1 mg/ml of bromophenol blue), incubated at 90°C for 3 min, cooled on ice, loaded onto 10% Criterion Tris-boric acid-EDTA (TBE) gels (Bio-Rad Laboratories, Hercules, CA), and run at 150 V at 4°C for 4 h. The gels were then stained with ethidium bromide and imaged by use of an Alphaimager instrument (Alpha Innotech, San Leandro, CA).
RNA extraction and RT-PCR.
For RNA extraction, H. bizzozeronii strains were cultivated as described above for 24 h with continuous shaking (150 rpm). After incubation, the optical density at 600 nm (OD600) was measured, and the same amount of cells was treated with RNAprotect Bacteria reagent (Qiagen GmbH, Hamburg, Germany). RNA was extracted by use of an RNeasy minikit (Qiagen) and treated with Turbo DNase (Applied Biosystems/Ambion, Austin, TX). cDNAs were synthesized from 1 μg of total RNA with SuperScript III reverse transcriptase (Invitrogen), using random hexamers (Finnzymes). Reverse transcription (RT)-PCRs (40 cycles) were conducted with a total volume of 50 μl, using Phusion high-fidelity DNA polymerase (Finnzymes), 1 μl of cDNA and 1 μl of RNA without reverse transcriptase as a negative control, and 20 pmol primers specific for the GT-42 locus (Table 2).
Expression, purification, and characterization of recombinant Hb-ST1 and Hb-ST2.
Primer pairs (Table 2) were designed to amplify the complete sequences of the Hb-ST1 and Hb-ST2 genes using Phusion high-fidelity DNA polymerase (Finnzymes). Both genes were inserted into the NdeI and SalI restriction sites of pCW-malET (8). The resulting expression constructs were sequenced and confirmed to be Hb-ST1 and Hb-ST2, respectively. Both expression constructs were used to transform E. coli AD202 (1). The expression of recombinant proteins was induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 24 h at 25°C in 2×YT medium supplemented with 150 mg/liter of ampicillin and 0.2% of glucose. Larger protein preparations started with 3 g of frozen cell extract resuspended in 30 ml of cold 50 mM HEPES (pH 7.0) with 10% glycerol and 1× Protease Inhibitor solution (Sigma-Aldrich, Inc., St. Louis, MO); the suspensions were further disrupted by use of an Emulsiflex C5 instrument (Avestin, Inc., Ottawa, Ontario, Canada). After cell disruption, the debris was pelleted at 21,100 × g for 30 min at 4°C. The supernatant was then decanted, and the pellet was resuspended in 3 ml of cold 50 mM HEPES (pH 7.0) with 10% glycerol to assess how much enzyme was in the pellet fraction. A further centrifugation at 100,000 × g for 1 h at 4°C was performed to assess the solubility of the proteins. Recombinant proteins were purified by affinity chromatography on amylose as previously described (8). The eluted fractions were analyzed on a 10% acrylamide gel, and the pure fractions were pooled and tested for ST activity. The activity was examined by using a 0.5 mM solution of one of the following 6-(5-fluorescein-carboxamido)-hexanoic acid succimidyl ester (FCHASE)-labeled acceptors: FCHASE-GM3, FCHASE-lactose, FCHASE-LacNAc, FCHASE-lacto-N-biose, or FCHASE-T antigen. The reaction was performed with a final volume of 10 μl in the presence of 50 mM HEPES (pH 7.5), 10 mM MgCl2, 0.1 mg/ml acetylated bovine serum albumin (AcBSA), 1 mM CMP-N-acetyl-neuraminic acid (CMP-Neu5Ac), and 5 μl of extract diluted in 1 mg/ml AcBSA, and it was optimized according to pH, temperature, and the metal cofactor. The reaction mixtures were incubated at room temperature, reactions were stopped with and equal volume of stop mix (50% acetonitrile, 1% SDS, 10 mM EDTA), and the reaction mixtures were then diluted to 1 μM and analyzed by capillary electrophoresis as previously described (51). Kinetic analysis of the purified protein was performed by using various concentrations of FCHASE-LacNAc, from 0.0201 to 1.415 mM, in the presence of 2 mM CMP-Neu5Ac under the optimized reaction conditions. Once the Km for FCHASE-LacNAc was determined, we repeated the kinetic assay using concentrations of CMP-Neu5Ac from 0.01476 to 1.5236 mM with 1 mM FCHASE-LacNAc.
Dot blot hybridization with cholera toxin and anti-sialyl-Lewis X antibody.
For dot blot hybridization, H. bizzozeronii strains were cultivated in BHI broth as described above for 48 h. The bacterial cell mass was harvested in phosphate-buffered saline (PBS) (pH 7.2), and the optical density (OD600) was adjusted to 1. Bacterial cells were treated with proteinase K (0.4 mg/ml) for 1 h at 56°C and then boiled for 5 min. For neuraminidase treatment, cells were incubated with 6.7 U/ml neuraminidase of Clostridium perfringens (Sigma-Aldrich) (pH 6) for 20 h at 37°C. The bacterial suspension (2 μl) was spotted onto a nylon membrane; the membrane was then probed with 0.1% peroxidase-conjugated cholera toxin (CT) subunit B (po-CT; Sigma-Aldrich) or anti-sialyl-Lewis X antibodies (1:1,000) (purified mouse anti-human CD15s, catalog no. 551344; BD Pharmingen), as previously described (2, 25, 43), and developed with enhanced chemiluminescence (ECL) (18). E. coli DH5α was used as a negative control, while C. jejuni 11168 and H. pylori P466 were used as positive controls for po-CT and anti-sialyl-Lewis X antibodies, respectively.
LPS extraction.
LPSs were extracted from biomass obtained after 48 h of incubation in broth as described above. Crude LPS was extracted from an equal amount of bacteria (as measured by the OD600) using the hot phenol-water method, and it was subsequently purified by enzymatic treatments (RNase A, DNase II, and proteinase K) as described previously (26). After enzymatic treatment, the LPS was precipitated at −20°C overnight in 10 volumes of pure ethanol in 0.03 M sodium acetate and then resuspended in water.
For structural analysis, the LPSs from H. bizzozeronii CIII-1 and StorkisT were extracted by the phenol-chloroform-light-petroleum method as previously described (14), with the modification of LPS precipitation by acetone-diethyl ether (6:1, vol/vol). The crude LPS was purified by ultracentrifugation (75,000 × g at 4°C for 16 h) (TL-100 ultracentrifuge; Beckmann Coulter), resulting in 15.6 mg and 6.1 mg of LPS, respectively. O-deacylated LPS (LPS-OH) was obtained as previously described (22). Briefly, LPS (1 mg) was stirred in anhydrous hydrazine (110 μl) at 40°C for 4 h. The reaction mixture was cooled, and cold acetone (1 ml) was added dropwise at 0°C. LPS-OH was allowed to precipitate at 0°C for 1 h before collection by centrifugation. The pellet was washed twice with cold acetone and then dissolved in water and lyophilized.
Neuraminidase treatment and SDS-PAGE profile.
The concentration of LPS purified with hot phenol-water was measured by using the purpald assay as described previously (42). Two micrograms of LPS from each sample was treated at pH 6 with 6.7 IU of neuraminidase from Clostridium perfringens (Sigma-Aldrich) overnight at 37°C and then boiled for 5 min. The patterns of LPS isolated from different strains before and after neuraminidase treatment were determined after fractionation in 15% Tris-HCl SDS-PAGE gels (Bio-Rad) and staining with silver (26).
Analytical methods.
LPS-OH was treated with aqueous 0.1 M HCl at 80°C for 1 h to release the Neu5Ac as previously described (7). The sample was evaporated, diluted in water, and subjected to high performance anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD). HPAEC-PAD was performed with a Dionex series 4500i chromatography system (Dionex, Sunnyvale, CA), using a CarboPac PA20 column (3 by 150 mm) (7). The samples (20 μl) were measured in duplicates and were eluted by using a linear gradient of 4 to 60% 0.5 M sodium acetate (NaOAc) in 0.1 M NaOH for 15 min at a flow rate of 0.5 ml/min. Peak areas were compared to a standard calibration curve (5 to 20 pg), using Neu5Ac as a standard (Sigma-Aldrich).
Capillary electrophoresis electrospray ionization mass spectrometry (CE-ESI-MS) was performed by using a Prince CE system (Prince Technologies, Netherlands) coupled to a 4000 QTRAP mass spectrometer (AB Sciex, Canada) with a MicroIon spray interface, as described previously (33).
RESULTS
Identification of sialyltransferases in the H. bizzozeronii genome belonging to the GT-42 family.
The H. bizzozeronii CIII-1 genome (EMBL accession no. FR871757) (44) revealed a cluster of three genes potentially involved in the biosynthesis of sialic acid (Neu5Ac) (HBZC1_02520 [neuB], HBZC1_02530/HBZC1_02540 [fragmented, neuC], and HBZC1_02550 [neuA]). The de novo synthesis of Neu5AC in prokaryotes requires two steps: NeuC produces N-acetyl-d-mannosamine from UDP N-acetylglucosamine, and NeuB condenses this N-acetylhexosamine with the three-carbon molecule pyruvate, forming the nine-carbon Neu5Ac. In addition, Neu5Ac is activated by the action of NeuA, resulting in CMPNeu5Ac, which is the required substrate for STs that complete the assembly of sialo-glycoconjugates (32). In the genome of H. bizzozeronii CIII-1, adjacent to the neuA gene is a glycosyltransferase (HBZC1_02560), which has been classified as a member of the glycosyltransferase family GT-42 (10). HBZC1_02560 showed 38.2% amino acid sequence identity to Campylobacter bifunctionalα,2,8/2,3-ST (UniProt identification no. Q9LAK3) and 32.5 to 80.1% identity to putative GT-42 STs found in other Helicobacter species.
A similar cluster was also found in the shotgun genome sequence of H. bizzozeronii strain StorkisT, isolated from a dog. However, in the canine strain, a tandem of GT-42 STs (denominated Hb-ST1 and Hb-ST2) was found downstream of the putative neuA gene. These two StorkisT GT-42 STs shared only 48.8% amino acid sequence identity and HBZC1_02560 showed 49.6% and 97.8% identities to StorkisT Hb-ST1 and Hb-ST2, respectively.
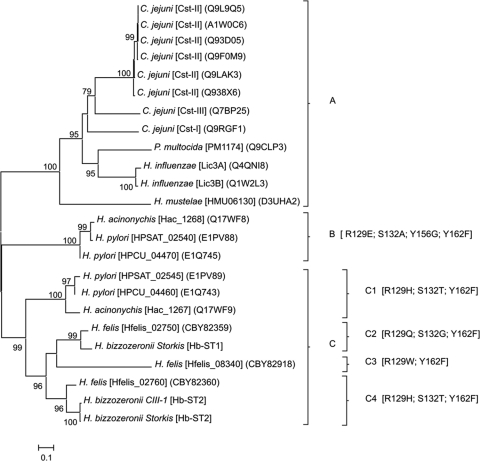
Phylogenetic analysis of GT-42 sialyltransferases and identification of critical amino acid substitutions in Hb-ST1 and Hb-ST2.
To investigate the evolution of Helicobacter STs and to predict functional activities of Hb-ST1 and Hb-ST2 (HBZC1_02560), a phylogenetic analysis of GT-42 STs was performed. The minimum evolution (ME) tree based on amino acid sequence alignments divided the GT-42 STs into three clades (Fig. 1). Clade A includes monofunctional α,2,3-STs and bifunctional α,2,3/α,2,8-STs from Campylobacter jejuni (CstI [11], CstII [12], and CstIII [16]) and Haemophilus influenzae (Lic3A [23] and Lic3B [13]) as well as the α,2,3-STs from Pasteurella multocida (PM1174 [49]) and Helicobacter mustelae (HMU06130 [41]). Clade B is comprised of putative STs identified in Helicobacter acinonychis (Hac_1268) and Helicobacter pylori (HPSAT_02540 and HPCU_04470), while the remaining Helicobacter STs, including those from H. bizzozeronii, form clade C.
Fig 1.
Unrooted tree based on complete amino acid sequences of sialyltransferases belonging to the family GT-42. The evolutionary history was inferred by use of the minimum evolution method, and the evolutionary distances were computed by use of the Dayhoff matrix-based method. The bar indicates the number of amino acid substitutions per position. Numbers at the nodes indicate support for the internal branches within the tree obtained by bootstrap analysis (≥70%; percentages of 500 bootstraps).
To detect amino acid positions potentially involved in the functional change, conserved sites of GT-42 STs were evaluated on the basis of structural data available for C. jejuni CstII (12) and CstI (11) and compared to the phylogenetic data. All the sequences belonging to clade A, both catalytic sites (Arg129 and His188 [CstII enumeration]), as well as the CMP-binding sites (Asn31, Ser132, Tyr156, and Tyr162) are conserved. In contrast, clades B and C are characterized by substitutions in one or more of the functional sites mentioned above. Moreover, these substitutions are able to further divide clade C into four branches (Fig. 1). The catalytic site His188 is conserved in all the sequences, but Arg129 is replaced with different amino acids (Glu, His, Gln, and Trp), which characterize each clade. The substitution Tyr162Phe is common in both clades B and C, while Tyr156 is conserved in clade C but is replaced with Gly in clade B. Although phylogenetically distant, subclades C1, which includes STs identified in H. acinonychis (Hac_1267) and H. pylori (HPSAT_02545 and HPCU_04460), and C4, comprising Hb-ST2 and an H. felis ST (Hfelis_02760), show the same substitutions: Arg129His and Ser132Thr. Finally, subclade C2, which includes Hb-ST1 and another H. felis ST (Hfelis_02750), has unique substitutions for Arg129 and Ser132 (Gln and Gly, respectively). Interestingly, in the hypervariable lid-like domain region described for CstII (between the two conserved sites Phe156 and His188), Helicobacter sequences belonging to both clades B and C showed similar deletions of 9 amino acids (data not shown).
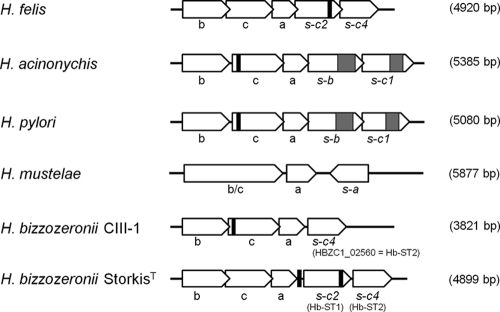
Conserved synteny of the GT-42 locus in different Helicobacter species and identification of hot spots of variation.
The genomic organizations of the GT-42 loci in different Helicobacter species were compared. With the exception of H. mustelae, all other species showed collinearity in the gene order: the neuBCA cluster is followed by one copy (in H. bizzozeronii CIII-1) or two copies (in all others) of STs in the same transcriptional direction (Fig. 2). In H. mustelae, the NeuB and NeuC domains are fused in a single gene (HMU06150), and the ST follows neuA in the opposite direction. The synteny and the overlap between the different genes observed suggest that the GT-42 cluster is potentially expressed as a single transcription unit (operon) in all the non-mustelae Helicobacter species.
Fig 2.
Schematic representation and comparison of the GT-42 loci of different Helicobacter species. Empty arrows represent the direction of transcription of the open reading frames within each locus. Black boxes correspond to the positions of simple sequence repeats (SSRs) of cytosine or guanine. Gray boxes indicate repeat regions of 8 to 15 units composed of six (KELERQ) or 10 (FKNIEEKLLE) amino acids. In parentheses are the dimensions of the GT-42 locus for each species. b, N-acetylneuraminic acid synthetase (neuB); c, UDP-N-acetylglucosamine-2-epimerase (neuC); a, CMP-sialic acid synthetase (neuA); s, sialyltransferase (with the corresponding phylogenetic group indicated).
In H. bizzozeronii CIII-1, neuC is fragmented due to the presence of a 5′ poly(C) tract involved in slipped-strand mispairing. A modification of the number of single nucleotides in the 5′ poly(G) tract in silico allowed the restoration of neuC in frame, indicating that in this strain, the epimerase may be affected by phase variation. The distribution and the location of single simple repeats (SSRs) in the GT-42 cluster differ across species and also between the two H. bizzozeronii strains. A 5′ poly(C) or poly(G) tract in neuC is present in both H. pylori and H. acinonychis, but it is missing in H. bizzozeronii strain StorkisT and in H. felis. On the contrary, H. bizzozeronii strain StorkisT showed a poly(C) tract in the 5′ and a poly(G) tract in the 3′ region of Hb-ST1. The 3′ poly(G) tract is also present in the Hb-ST1 orthologues of H. felis (Hfelis_02750), but it lacks the 5′ homopolymeric run. In silico, the 5′ poly(C) tract observed for the neuC genes of both H. pylori and H. acinonychis may be involved in switching the gene on and off. Nevertheless, the modification of the number of single nucleotides in the SSRs of Hb-ST1 did not affect the frame of the gene. Finally, the STs of both H. acinonychis and H. pylori are characterized in the C-terminal part by the presence of a repeat region of 8 to 15 units composed of six (KELERQ) or 10 (FKNIEEKLLE) amino acids. This type of region was not found in any other GT-42 STs, but repeats of this type are common in the fucosyltransferases of H. pylori (40).
Characterization of the GT-42 locus in H. bizzozeronii strains.
PCR was used to amplify the GT-42 locus from human H. bizzozeronii strain R53 as well as from five canine strains (Table 1). Partial sequences of the GT-42 locus obtained from each individual H. bizzozeronii strain revealed that the number of STs and the positions of SSRs differed across the strains (Fig. 3), indicating a different modulation in the expression of glycans containing sialic acid in H. bizzozeronii.
Fig 3.
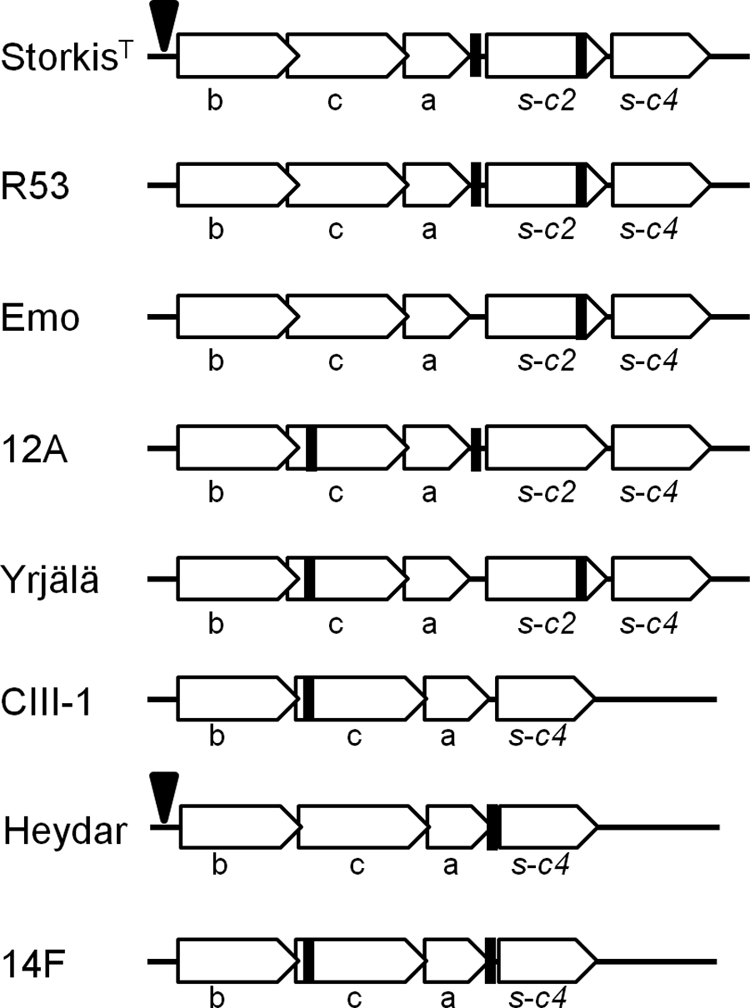
Schematic representation and comparison of the GT-42 loci of different Helicobacter bizzozeronii strains. Empty arrows represent the direction of transcription of the open reading frames within each locus. Black boxes correspond to the positions of simple sequence repeats (SSRs) of cytosine or guanine. Black arrows indicate the insertion in the promoter region of the mini-IS. b, N-acetylneuraminic acid synthetase (neuB); c, UDP-N-acetylglucosamine-2-epimerase (neuC); a, CMP-sialic acid synthetase (neuA); s, sialyltransferase (with the corresponding phylogenetic group indicated).
Single-strand conformation polymorphism (SSCP)/heteroduplex analysis was performed to verify the presence of heterogeneous homopolymeric tracts in the putative UDP-N-acetylglucosamine 2-epimerase (neuC) in different human H. bizzozeronii isolates from a patient (30) as well as in canine strains 12, Yrjälä, and 14F (data not shown). The SSCP analysis of the 240-bp PCR product containing the SSRs from all the samples showed multiple bands, indicating the presence of heterogeneous homopolymeric tracts. The presence of different numbers of cytosines in the DNA preparation of human strain CIII-1 was also confirmed by sequencing. The 240-bp PCR product containing the SSR was inserted into a pGEM vector and cloned into E. coli JM109. The sequences of plasmids obtained from three independent transformants showed different numbers of cytosines, confirming the SSCP results and supporting the contingency of the putative neuC gene (data not shown).
Finally, the putative promoter region of the GT-42 operon upstream of neuB was also amplified and sequenced from all the samples. In two H. bizzozeronii strains (StorkisT and Heydar), the promoter region is interrupted 50 bp upstream of the predicted start codon of neuB by an identical 233-bp insertion sequence (IS), which corresponds to a mini-IS previously described for the genome of H. bizzozeronii CIII-1 (45).
Transcription of the GT-42 locus of H. bizzozeronii in vitro.
The transcription of the GT-42 locus in different H. bizzozeronii strains was evaluated after 24 h of incubation. This corresponded to approximately the middle of the exponential phase. RT-PCR showed that all the genes belonging to the GT-42 locus were transcribed in all the strains as a single operon (data not shown).
Expression, purification, and biochemical characterization of recombinant Hb-ST1 and Hb-ST2.
The expression constructs containing the H. bizzozeronii STs were sequenced, and the expected sequences were obtained, except for an R310C substitution in Hb-ST1, which appears not to be in a conserved region of enzymes in CAZy GT-42. Hb-ST2 was well expressed as a fusion protein at the expected molecular mass of ∼80 kDa, whereas the ∼79-kDa band for Hb-ST1 was not as prominent. The enzyme assays showed that Hb-ST1 had no measurable enzyme activity on any acceptor we tested, but Hb-ST2 showed good activity on the synthetic acceptor FCHASE-aminophenyl-β-LacNAc. Differential centrifugation revealed that the Hb-ST2 fusion protein was mostly soluble, as only 30% of it was found in the pellet fraction after a spin at 100,000 × g. It was purified from the supernatant from the 27,000 × g spin by affinity chromatography on amylose. The Hb-ST2 enzyme was enriched 7-fold during purification. The protein was found to be stable for over 50 days at −70°C. We tested several acceptors with both 3- or 4-linked β-galactosidase (β-Gal) derivatives as well as a 3′-sialylated acceptor to determine α,2,8-ST activity. We observed no α2,8-ST activity, indicating that Hb-ST2 is a monofunctional ST with a strong preference for LacNAc over Lac and very low levels of activity on 3-linked Gal. The apparent Km [Km(app)] for FCHASE-aminophenyl-β-LacNAc was determined to be 108.5 μM, and the Km for CMP-Neu5Ac was 125.5 μM.
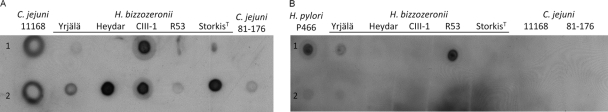
Typing of H. bizzozeronii strains with cholera toxin and anti-sialyl-Lewis X antibody.
To investigate the presence of surface glycan structures in H. bizzozeronii that may mimic host sialic acid-containing glycans, strains were typed by using peroxidase-conjugated cholera toxin subunit B (po-CT), which reacts to several ganglioside-like structures (2), and anti-sialyl-Lewis X antibodies. po-CT was able to bind to Yrjälä and CIII-1 (Fig. 4A). No binding to StorkisT, Heydar, and R53 was observed. After neuraminidase treatment, po-CT reacted with all the strains tested. Different results were observed when the strains were serotyped with anti-sialyl-Lewis X antibodies, which reacted strongly to R53 and weakly to Yrjälä, and no binding to other strains was observed (Fig. 4B). After neuraminidase treatment, anti-sialyl-Lewis X antibodies were not able to react subsequently to R53 and Yrjälä.
Fig 4.
Dot blot hybridization of a proteinase K extract from Helicobacter bizzozeronii strains untreated (line 1) or treated with neuraminidase from Clostridium perfringens (line 2). Dot blotting was done by using peroxidase-conjugated cholera toxin subunit B (A) and anti-sialyl-Lewis X antibodies (B).
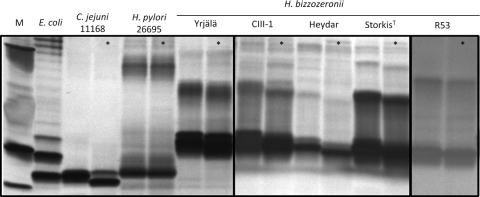
Detection of sialic acid in the LPS of H. bizzozeronii.
To determine whether sialic acid was present in the LPS of H. bizzozeronii, hot phenol-water extracts from different strains were subjected to neuraminidase treatment followed by fractionation by SDS-PAGE. All the strains tested showed low-molecular-weight LPS, and for some strains, a clear shift of the band appeared after neuraminidase treatment (Fig. 5), indicating that H. bizzozeronii could expresses sialylated LPS. The presence of Neu5Ac in the hot phenol-water extracts of four different strains was further analyzed by high-performance anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD). We determined Neu5Ac to be present in all the LPS preparations; there were 4.6, 3.7, 11.8, and 6.0 pmol Neu5Ac/μg of sample from H. bizzozeronii strain Heydar, R53, StorkisT, and CIII-1, respectively (data not shown).
Fig 5.
SDS-PAGE analysis of LPSs from different Helicobacter bizzozeronii strains treated (indicated with asterisks) or not treated with neuraminidase from Clostridium perfringens. M, marker.
Structural characterization of LPS by CE-ESI-MS.
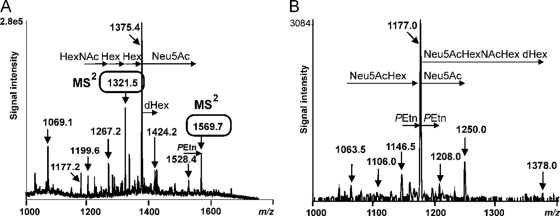
LPS-OHs from H. bizzozeronii strains CIII-1 and StorkisT were analyzed by CE-ESI-MS and capillary electrophoresis electrospray ionization tandem mass spectrometry (CE-ESI-MS/MS). The results are summarized in Tables 3 and 4. H. bizzozeroni contains a core oligosaccharide linked via one 3-deoxy-d-manno-oct-2-ulosonic acid (Kdo) molecule to a lipid A molecule, as observed for H. pylori LPS (38). Mass spectrometry showed that the core oligosaccharide of H. bizzozeronii is composed of heptoses (Hep), hexoses (Hex), hexosamines (HexNAc), and deoxysugars (dHex). In more detail, the CE-ESI-MS spectrum of LPS-OH from H. bizzozeronii strain CIII-1 (Fig. 6A) revealed triply deprotonated ions at m/z 1,199.6, 1,267.2, 1,321.5, 1,375.4, and 1,424.2, corresponding to compositions of dHex3·HexNAc2·Hex5·Hep3·PEtn·Kdo·lipid A-OH, dHex3·HexNAc3·Hex5-7·Hep3·PEtn·Kdo·lipid A-OH, and dHex4·HexNAc3·Hex7·Hep3·PEtn·Kdo·lipid A-OH. Ions at m/z 1,226.6, 1,280.3, 1,334.4, and 1,382.7 indicated similar glycoform compositions but lacked the PEtn residue. Sialylated glycoforms were observed at m/z 1,528.4 and 1,569.7 ([M − 4H]4−), corresponding to the respective composition of Neu5Ac2·dHex3·HexNAc3·Hex7·Hep3·PEtn0-1·Kdo·lipid A-OH. The presence of sialylated glycoforms in LPS-OH from H. bizzozeronii strain CIII-1 was confirmed by precursor ion monitoring tandem mass spectrometry experiments scanning for a loss of m/z 290 (Neu5Ac, negative-ion mode) (Fig. 6B). As this is a very sensitive MS/MS technique, ions resembling minor-abundance sialylated glycoforms, not identified in the full ESI-MS spectra, were observed. Quadruply charged ions at m/z 1,063.5, 1,146.5, 1,177.0, 1,250.0, and 1,378.0 corresponded to the glycoforms Neu5Ac·dHex3·HexNAc3·Hex6·Hep3·PEtn·Kdo·lipid A-OH, Neu5Ac2·dHex3·HexNAc3·Hex7·Hep3·PEtn0-2·Kdo·lipid A-OH, and Neu5Ac3·dHex4·HexNAc4·Hex8·Hep3·PEtn·Kdo·lipid A-OH, respectively, and indicated that LPS-OH from H. bizzozeronii strain CIII-1 contains up to three Neu5Ac residues.
Table 3.
CE-ESI-MS in negative-ion mode of LPS-OH from H. bizzozeronii CIII-1 and StorkisTa
| Proposed composition | Observed ion m/z |
Molecular mass (Da) |
Relative abundance (%) |
|||||
|---|---|---|---|---|---|---|---|---|
| [M − 6H]6− | [M − 5H]5− | [M − 4H]4− | [M − 3H]3− | Observed | Calculated | CIII-1 | StorkisT | |
| dHex3·HexNAc2·Hex5·Hep3·PEtn·Kdo·lipid A-OH | 1,199.6 | 3,601.8 | 3,602.47 | 4 | ||||
| dHex3·HexNAc3·Hex5·Hep3·Kdo·lipid A-OH | 1,226.6 | 3,682.8 | 3,683.61 | 2 | ||||
| dHex3·HexNAc3·Hex5·Hep3·PEtn·Kdo·lipid A-OH | 1,267.2 | 3,804.6 | 3,806.66 | 7 | ||||
| dHex3·HexNAc3·Hex6·Hep3·Kdo·lipid A-OH | 1,280.3 | 3,843.9 | 3,845.75 | 4 | ||||
| dHex3·HexNAc3·Hex6·Hep3·PEtn·Kdo·lipid A-OH | 1,321.5 | 3,967.5 | 3,968.80 | 18 | ||||
| dHex3·HexNAc3·Hex7·Hep3·Kdo·lipid A-OH | 1,334.4 | 4,006.2 | 4,007.89 | 9 | ||||
| dHex3·HexNAc3·Hex7·Hep3·PEtn·Kdo·lipid A-OH | 1,375.4 | 4,129.2 | 4,130.94 | 35 | ||||
| Neu5Ac2·HexNAc4 Hex3·dHex·Hep3·Kdo·PEtn2·P·lipid A-OH | 695.6 | 834.7 | 1,043.6 | 1,391.9 | 4,178.8 | 4,178.84 | 93 | |
| dHex4·HexNAc3·Hex7·Hep3·PEtn·Kdo·lipid A-OH | 1,069.1 | 1,424.2 | 4,275.6 | 4,277.08 | 5 | |||
| Neu5Ac2·HexNAc4·Hex3·dHex2·Hep3·Kdo·PEtn2·P·lipid A-OH | 1,080.3 | 4,325.2 | 4,324.98 | 7 | ||||
| Neu5Ac2·dHex3·HexNAc3·Hex7·Hep3·Kdo·lipid A-OH | 1,528.4 | 4,588.2 | 4,590.41 | 3 | ||||
| Neu5Ac2·dHex3·HexNAc3·Hex7·Hep3·PEtn·Kdo·lipid A-OH | 1,177.2 | 1,569.7 | 4,712.5 | 4,713.46 | 13 | |||
For the calculation of molecular masses, the following values (in daltons) were used: 1,028.31 for lipid A-OH {based on a β-(1→6)-linked d-glucosaminodisaccharide backbone acylated by (R)-3-hydroxyoctadecanoic acid [18:0(3-OH)] at positions 2 and 2′, carrying a phosphoethanolamine group at position 1), 220.10 for Kdo, 192.17 for Hep, 162.14 for Hex, 146.14 for dHex (deoxyhexose), 203.19 for HexNAc, 291.26 for Neu5Ac, 123.05 for PEtn, and 79.98 for P.
Table 4.
CE-ESI-MS/MS in negative-ion mode using precursor ion scanning on m/z 290, i.e., Neu5Ac, of LPS-OH from H. bizzozeronii CIII-1 and StorkisTa
| Proposed composition | Observed ion m/z |
Molecular mass (Da) |
Relative abundance (%) |
||||
|---|---|---|---|---|---|---|---|
| (M − 6H)6− | (M − 5H)5− | (M − 4H)4− | Observed | Calculated | CIII-1 | StorkisT | |
| Neu5Ac2·dHex·Hex3·HexNAc4·Hep3·Kdo·PEtn·P·lipid A-OH | 1,013.0 | 4,056.0 | 4,055.79 | 6 | |||
| Neu5Ac2·dHex·Hex3·dHexNAc4·Hep3·Kdo·PEtn2·P·lipid A-OH | 695.5 | 834.5 | 1,043.5 | 4,178.2 | 4,178.84 | 69 | |
| Neu5Ac2·dHex2·Hex3·dHexNAc4·Hep3·Kdo·PEtn2·P·lipid A-OH | 864.0 | 1,080.0 | 4,324.0 | 4,324.98 | 25 | ||
| Neu5Ac·dHex3·HexNAc3·Hex6·Hep3·PEtn·Kdo·lipid A-OH | 1,063.5 | 4,258.0 | 4,260.06 | 5 | |||
| Neu5Ac2·dHex3·HexNAc3·Hex6·Hep3·Kdo·lipid A-OH | 1,106.0 | 4,428.0 | 4,428.27 | 2 | |||
| Neu5Ac2·dHex3·HexNAc3·Hex7·Hep3·Kdo·lipid A-OH | 1,146.5 | 4,590.0 | 4,590.41 | 9 | |||
| Neu5Ac2·dHex3·HexNAc3·Hex7·Hep3·PEtn·Kdo·lipid A-OH | 1,177.0 | 4,712.0 | 4,713.46 | 64 | |||
| Neu5Ac2·dHex3·HexNAc3·Hex7·Hep3·PEtn2·Kdo·lipid A-OH | 1,208.0 | 4,836.0 | 4,836.51 | 5 | |||
| Neu5Ac3·dHex3·HexNAc3·Hex7·Hep3·PEtn·Kdo·lipid A-OH | 1,250.0 | 5,004.0 | 5,004.72 | 13 | |||
| Neu5Ac3·dHex4·HexNAc4·Hex8·Hep3·PEtn·Kdo·lipid A-OH | 1,378.0 | 5,516.0 | 5,516.19 | 2 | |||
For the calculation of molecular masses, the following values (in daltons) were used: 1,028.31 for lipid A-OH, 220.10 for Kdo, 192.17 for Hep, 162.14 for Hex, 146.14 for dHex, 203.19 for HexNAc, 291.26 for Neu5Ac, 123.05 for PEtn, and 79.98 for P.
Fig 6.
The O-deacylated LPS of H. bizzozeronii strain CIII-1 analyzed by CE-ESI-MS in negative-ion mode (A) and CE-ESI-MS/MS with precursor ion scanning (B) on the ion at m/z 290, i.e., Neu5Ac.
The CE-ESI-MS spectrum of LPS-OH from H. bizzozeronii strain StorkisT revealed abundant molecular peaks at m/z 1,043.6 and 1,080.3, corresponding to multiply deprotonated ions ([M − 5H]5− and [M − 4H]4−) and to the glycoforms Neu5Ac2·HexNAc4·Hex3·dHex·Hep3·PEtn2·P·Kdo·lipid A-OH and Neu5Ac2·HexNAc4·Hex3·dHex2·Hep3·PEtn2·P·Kdo·lipid A-OH, respectively. Precursor ion monitoring tandem mass spectrometry, scanning for the loss of m/z 290, showed quadruply charged ions at m/z 1,013.0, 1,043.5, and 1,080.0, corresponding to the glycoforms Neu5Ac2·HexNAc4·Hex3·dHex·Hep3·PEtn1,2·P·Kdo·lipid A-OH and Neu5Ac2·HexNAc4·Hex3·dHex2·Hep3·PEtn2·P·Kdo·lipid A-OH, respectively.
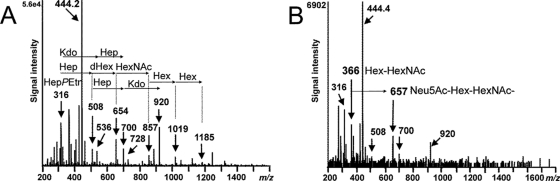
Information on the nature of the sialylated oligosaccharide extensions in the LPS-OHs of both H. bizzozeronii strains was obtained by MS/MS experiments on protonated ions in the positive mode. When the ion at m/z 1,571.5 (Neu5Ac2·dHex3·HexNAc3·Hex7·Hep3·PEtn·Kdo·lipid A-OH [strain CIII-1, positive mode]) (Fig. 7B) was fragmented, ions at m/z 366 (Hex·HexNAc) and m/z 657 were observed. The diagnostic fragment ion at m/z 657 corresponds to Neu5Ac-Hex-HexNAc, which indicates that sialyl-lacto-N-neotetraose is present (34). Additional structural information on the LPS-OH from H. bizzozeronii strain CIII-1 was obtained by MS/MS on m/z 1,323.2 (dHex3·HexNAc3·Hex6·Hep3·PEtn·Kdo·lipid A-OH [positive mode]). The resulting spectrum revealed fragment ions at m/z 316.0, 508.0, 700.2, and 920.2, corresponding to PEtn·Hep, PEtn·Hep2, PEtn·Hep3, and PEtn·Hep3·Kdo, respectively (Fig. 7A). Further fragment ions were observed at m/z 654.2, 857.2, 1,019.3, and 1,185.1, corresponding to PEtn·Hep2·Kdo, PEtn·Hep2·Kdo·HexNAc, PEtn·Hep2·Kdo·HexNAc·Hex, and PEtn·Hep2·Kdo·HexNAc·Hex2, respectively. Thus, the core part of the LPS of H. bizzozeronii is ostensibly composed of a triheptosyl moiety linked to Kdo. One heptose is replace by PEtn.
Fig 7.
The O-deacylated LPS of H. bizzozeronii strain CIII-1 analyzed in positive-ion mode. Shown are data for analysis by CE-ESI-MS/MS of the ion at m/z 1,323.2, corresponding to a composition of dHex3·HexNAc3·Hex6·Hep3·PEtn·Kdo·lipid A-OH, in the negative-ion mode (A) and CE-ESI-MS/MS on the ion at m/z 1,571.5, corresponding to a composition of Neu5Ac2·dHex3·HexNAc3·Hex7·Hep3·PEtn·Kdo·lipid A-OH (B).
DISCUSSION
In this study, we report the functional expression and characterization of a novel ST, belonging to glycosyltransferase family GT-42, identified in the zoonotic pathogen H. bizzozeronii; this is the first functionally characterized ST of the Helicobacter genus. Originally, the GT-42 family included monofunctional α,2,3-STs and bifunctional α,2,3/α,2,8-STs from C. jejuni (CstI, CstII, and CstIII) and H. influenzae (Lic3A and Lic3B) (3), but recently, sequenced genomes have revealed that many gastric Helicobacter species, including H. pylori, have multiple genes potentially coding for STs belonging to this family. Nevertheless, no members of GT-42 identified in Helicobacter spp. have been linked to specific sialylated structures.
In this study, we describe two novel STs from H. bizzozeronii (Hb-ST1 and Hb-ST2). Each of the Hb-STs showed a peculiar phylogenetic signal, and each is characterized by different substitutions in both functional and CMP-binding sites, suggesting that the two STs possibly acquired different substrate specificities. The duplication of STs appears to be a common event in gastric Helicobacter species except for H. mustelae, and the phylogenetic tree topology suggests that the different Helicobacter STs may have evolved by gene duplication from a common ancestral sequence, likely before the speciation event. The conserved synteny observed for different Helicobacter species and the transcription data for H. bizzozeronii indicate that the STs are cotranscribed with upstream genes (neuBCA) involved in the synthesis of sialic acid. We sequenced the ST operon of different H. bizzozeronii strains, showing that it is variable both in gene content and in the presence of hypervariable simple sequence repeats (SSRs). In particular, in three out of eight strains, only a single copy of ST is present, and in four strains, the UDP-N-acetylglucosamine-2-epimerase gene (neuC) is likely subjected to phase variation by slipped-strand mispairing associated with SSR. The hypervariable nature of SSR in the coding regions of the neuC genes of different H. bizzozeronii strains was demonstrated by SSCP/heteroduplex and sequence analyses. Interestingly, in several strains, the SSRs were also located in the intergenic region between neuA and the first ST and in the 3′-terminal part of the coding region of the first ST. Similar locations of the SSRs were also found for the ST operon of H. felis. How the instability of these regions could affect the transcription of the operon or the functions of the proteins is unknown, and further studies are needed. However, it has been shown that the physiological role of SSRs may not be limited to phase variation but can include a reorganization of the chromosome, an influence on protein structure or function, and possible antisense regulation (39, 46, 47). If this is the case, then the presence of multiple hypervariable SSRs in different locations of the ST operon may play roles in the modulation of expression of sialylated glycans by H. bizzozeronii, and this phenomenon could have important consequences for host adaptation.
To further characterize the STs associated with H. bizzozeronii, both STs of strain StorkisT (Hb-ST1 and Hb-ST2) were expressed in E. coli, and their activities on several substrates were investigated. Hb-ST2 showed monofunctional α,2,3-ST activity with a strong preference for LacNAc over Lac and very low levels of activity on 3-linked Gal, which was not observed for the Campylobacter GT-42 enzymes CstI and CstII (11, 12). Another striking difference from C. jejuni GT-42 enzymes is the Km(app) of Hb-ST2 for FCHASE-LacNAc (108.5 μM), which is considerably lower than the 35 mM reported for CstII but similar to 500 μM reported for CstI (12). In contrast, recombinant Hb-ST1 showed no measurable enzyme activity on any acceptor tested, confirming the strong difference between the two H. bizzozeronii STs. Hb-ST2 is the only ST present in all the strains, suggesting the presence of a common sialyl structure in all the strains.
In C. jejuni, the cstI, cstII, and cstIII genes belong to the lipooligosaccharide (LOS) biosynthesis locus, and the presence of a functional ST is associated with the expression of sialylated LOS structures (17). In H. bizzozeronii and in the other Helicobacter species, the genes involved in the biosynthesis of LPS are scattered throughout the whole genome (45), hindering the prediction of the role of the ST operon. In addition, molecular tools for the genetic modification of this species are currently unavailable, which limits the functional analysis of these STs. However, our data strongly suggested that in H. bizzozeronii, Hb-ST2 acts as an LPS-ST. In fact, the LPSs of all the strains tested were sensitive to neuraminidase treatment, and the presence of Neu5Ac in the LPSs of four strains was confirmed by HPAEC-PAD. Structural data for the LPS-OHs of both strains StorkisT and CIII-1 indicate the presence of sialyl-N-acetyllactosamine, which is consistent with the activity of recombinant Hb-ST2 in vitro.
In C. jejuni, the sialyl-LOS mimics the ganglioside structures present on mammalian cells (24), and peroxidase-conjugated cholera toxin subunit B (po-CT) has been used in several studies for the serotyping of C. jejuni strains due to its affinity for ganglioside-like structures (2). In this study, po-CT reacted with the surface glycans of some H. bizzozeronii strains, especially CIII-1. However, structural data for CIII-1 LPS-OH did not suggest the presence of any ganglioside-like structures. After neuraminidase treatment of the bacterial extracts, po-CT reacted with all the strains, indicating that the binding did not require NeuAc. In fact, it has been shown that CT also has an affinity for asialo-GM1, and the Gal-GalNAc epitope is the critical part of the binding (31). Consequently, it is interesting that 84% of the observed ion m/z in the CE-ESI-MS spectrum (in negative-ion mode) of LPS-OH from CIII-1 corresponds to asialo-LPS, and compositional sugar analysis revealed the presence of galactose and glucosamine (data not shown). Thus, our data suggest that the binding observed between po-CT and surface glycans of some H. bizzozeronii strains was likely due to the presence of the asialo-Gal-GlcNAc epitope on their LPSs and not to the molecular mimicry of a ganglioside-like structure.
It was shown previously that H. pylori LPS expresses sialyl-Lewis X antigen (36), and we investigated whether H. bizzozeronii surface glycans also react with anti-sialyl-Lewis X antibodies. Two strains were positive for sialyl-Lewis X, although information on the structure of the LPSs of these strains is lacking and thus is needed to confirm the dot blot results. In contrast to what we observed, Hynes and colleagues previously reported the absence of molecular mimicry of Lewis and other blood groups in H. bizzozeronii R53 LPS by Western blotting (26). However, in that study, the LPSs of H. bizzozeronii were extracted from bacteria grown under different cultivation conditions, which could affect the expression of phase-variable structures (37). In H. bizzozeronii, the expression of sialyl-Lewis X is probably subject to phase variation, as described previously for H. pylori (37), due to the contingency nature of the putative fucosyltransferase and β-1,4-galactosyltransferase (45). For H. pylori, the phase-variable expression of the Lewis antigen was implicated in the evasion of the immune response upon initial infection, in influencing bacterial colonization and adhesion, and in autoimmunity that leads to gastric atrophy (37). A similar scenario could also be hypothesized for H. bizzozeronii, although further studies are necessary to confirm this assumption.
Our results show that H. bizzozeronii expresses a monofunctional LPS α,2,3-ST, which has a strong preference for LacNAc. The expression of this α,2,3-ST and sialyl-LPS appeared to be a phase-variable characteristic common to both human and canine H. bizzozeronii strains. The sialylation site of the LPSs of two H. bizzozeronii strains was determined to be NeuAc-Hex-HexNAc, suggesting terminal 3′-sialyl-LacNac. Moreover, serological typing revealed the possible presence of sialyl-Lewis X in two additional strains, indicating that H. bizzozeronii can also mimic the surface glycans of mammalian cells. The expression of sialyl-glycans may influence the adaptation process of H. bizzozeronii during the host jump from dogs to humans.
ACKNOWLEDGMENTS
This study was funded by the Academy of Finland (FCoE MiFoSa, grant no. 118602 and 141140). M.R. was supported by an Academy of Finland postdoctoral fellowship (grant no. 132940). P.K.K. was supported by a 2010 grant from the Research Foundation of the University of Helsinki and by a CIMO India fellowship (25.11.2009/FM-09-6586). In addition, this work was partially supported by a grant to W.W. from the Canadian Institutes of Health Research (grant no. MOP84272).
We acknowledge Thomas Boren (University of Umeå) for providing us with the H. pylori P466 strain.
Footnotes
Published ahead of print 9 March 2012
REFERENCES
- 1. Akiyama Y, Ito K. 1990. SecY protein, a membrane-embedded secretion factor of E. coli, is cleaved by the ompT protease in vitro. Biochem. Biophys. Res. Commun. 167:711–715 [DOI] [PubMed] [Google Scholar]
- 2. Ang CW, et al. 2002. Structure of Campylobacter jejuni lipopolysaccharides determines antiganglioside specificity and clinical features of Guillain-Barre and Miller Fisher patients. Infect. Immun. 70:1202–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Audry M, et al. 2011. Current trends in the structure-activity relationships of sialyltransferases. Glycobiology 21:716–726 [DOI] [PubMed] [Google Scholar]
- 4. Aziz RK, et al. 2008. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baele M, et al. 2009. Non-Helicobacter pylori helicobacters detected in the stomach of humans comprise several naturally occurring Helicobacter species in animals. FEMS Immunol. Med. Microbiol. 55:306–313 [DOI] [PubMed] [Google Scholar]
- 6. Basso D, Plebani M, Kusters JG. 2010. Pathogenesis of Helicobacter pylori infection. Helicobacter 15 (Suppl. 1):14–20 [DOI] [PubMed] [Google Scholar]
- 7. Bauer SH, et al. 2001. A rapid and sensitive procedure for determination of 5-N-acetyl neuraminic acid in lipopolysaccharides of Haemophilus influenzae: a survey of 24 non-typeable H. influenzae strains. Carbohydr. Res. 335:251–260 [DOI] [PubMed] [Google Scholar]
- 8. Bernatchez S, et al. 2007. Variants of the beta 1,3-galactosyltransferase CgtB from the bacterium Campylobacter jejuni have distinct acceptor specificities. Glycobiology 17:1333–1343 [DOI] [PubMed] [Google Scholar]
- 9. Black RE, Levine MM, Clements ML, Hughes TP, Blaser MJ. 1988. Experimental Campylobacter jejuni infection in humans. J. Infect. Dis. 157:472–479 [DOI] [PubMed] [Google Scholar]
- 10. Cantarel BL, et al. 2009. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37:D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chiu CP, et al. 2007. Structural analysis of the alpha-2,3-sialyltransferase Cst-I from Campylobacter jejuni in apo and substrate-analogue bound forms. Biochemistry 46:7196–7204 [DOI] [PubMed] [Google Scholar]
- 12. Chiu CP, et al. 2004. Structural analysis of the sialyltransferase CstII from Campylobacter jejuni in complex with a substrate analog. Nat. Struct. Mol. Biol. 11:163–170 [DOI] [PubMed] [Google Scholar]
- 13. Fox KL, et al. 2006. Identification of a bifunctional lipopolysaccharide sialyltransferase in Haemophilus influenzae: incorporation of disialic acid. J. Biol. Chem. 281:40024–40032 [DOI] [PubMed] [Google Scholar]
- 14. Galanos C, Luderitz O, Westphal O. 1969. A new method for the extraction of R lipopolysaccharides. Eur. J. Biochem. 9:245–249 [DOI] [PubMed] [Google Scholar]
- 15. Gaynor EC, et al. 2004. The genome-sequenced variant of Campylobacter jejuni NCTC 11168 and the original clonal clinical isolate differ markedly in colonization, gene expression, and virulence-associated phenotypes. J. Bacteriol. 186:503–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gilbert M, et al. 2002. The genetic bases for the variation in the lipo-oligosaccharide of the mucosal pathogen, Campylobacter jejuni. Biosynthesis of sialylated ganglioside mimics in the core oligosaccharide. J. Biol. Chem. 277:327–337 [DOI] [PubMed] [Google Scholar]
- 17. Gilbert M, et al. 2000. Biosynthesis of ganglioside mimics in Campylobacter jejuni OH4384. Identification of the glycosyltransferase genes, enzymatic synthesis of model compounds, and characterization of nanomole amounts by 600-mhz (1)h and (13)c NMR analysis. J. Biol. Chem. 275:3896–3906 [DOI] [PubMed] [Google Scholar]
- 18. Haan C, Behrmann I. 2007. A cost effective non-commercial ECL-solution for Western blot detections yielding strong signals and low background. J. Immunol. Methods 318:11–19 [DOI] [PubMed] [Google Scholar]
- 19. Haesebrouck F, et al. 2009. Gastric helicobacters in domestic animals and nonhuman primates and their significance for human health. Clin. Microbiol. Rev. 22:202–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557–580 [DOI] [PubMed] [Google Scholar]
- 21. Hanninen ML, Hirvi U. 1999. Genetic diversity of canine gastric helicobacters, Helicobacter bizzozeronii and H. salomonis studied by pulsed-field gel electrophoresis. J. Med. Microbiol. 48:341–347 [DOI] [PubMed] [Google Scholar]
- 22. Holst O, Brade L, Kosma P, Brade H. 1991. Structure, serological specificity, and synthesis of artificial glycoconjugates representing the genus-specific lipopolysaccharide epitope of Chlamydia spp. J. Bacteriol. 173:1862–1866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hood DW, et al. 2001. Identification of a lipopolysaccharide alpha-2,3-sialyltransferase from Haemophilus influenzae. Mol. Microbiol. 39:341–350 [DOI] [PubMed] [Google Scholar]
- 24. Houliston RS, et al. 2011. Lipooligosaccharide of Campylobacter jejuni: similarity with multiple types of mammalian glycans beyond gangliosides. J. Biol. Chem. 286:12361–12370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hynes SO, Moran AP. 2000. Comparison of three serological methods for detection of Lewis antigens on the surface of Helicobacter pylori. FEMS Microbiol. Lett. 190:67–72 [DOI] [PubMed] [Google Scholar]
- 26. Hynes SO, et al. 2004. Comparative chemical and biological characterization of the lipopolysaccharides of gastric and enterohepatic helicobacters. Helicobacter 9:313–323 [DOI] [PubMed] [Google Scholar]
- 27. Jalava K, et al. 2001. A cultured strain of “Helicobacter heilmannii,” a human gastric pathogen, identified as H. bizzozeronii: evidence for zoonotic potential of Helicobacter. Emerg. Infect. Dis. 7:1036–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jalava K, et al. 1998. Isolation and identification of Helicobacter spp. from canine and feline gastric mucosa. Appl. Environ. Microbiol. 64:3998–4006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Katoh K, Asimenos G, Toh H. 2009. Multiple alignment of DNA sequences with MAFFT. Methods Mol. Biol. 537:39–64 [DOI] [PubMed] [Google Scholar]
- 30. Kivisto R, Linros J, Rossi M, Rautelin H, Hanninen ML. 2010. Characterization of multiple Helicobacter bizzozeronii isolates from a Finnish patient with severe dyspeptic symptoms and chronic active gastritis. Helicobacter 15:58–66 [DOI] [PubMed] [Google Scholar]
- 31. Kuziemko GM, Stroh M, Stevens RC. 1996. Cholera toxin binding affinity and specificity for gangliosides determined by surface plasmon resonance. Biochemistry 35:6375–6384 [DOI] [PubMed] [Google Scholar]
- 32. Lewis AL, et al. 2009. Innovations in host and microbial sialic acid biosynthesis revealed by phylogenomic prediction of nonulosonic acid structure. Proc. Natl. Acad. Sci. U. S. A. 106:13552–13557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li J, Cox AD, Hood D, Moxon ER, Richards JC. 2004. Application of capillary electrophoresis-electrospray-mass spectrometry to the separation and characterization of isomeric lipopolysaccharides of Neisseria meningitidis. Electrophoresis 25:2017–2025 [DOI] [PubMed] [Google Scholar]
- 34. Li J, et al. 2007. Structural characterization of sialylated glycoforms of H. influenzae by electrospray mass spectrometry: fragmentation of protonated and sodiated O-deacylated lipopolysaccharides. Rapid Commun. Mass Spectrom. 21:952–960 [DOI] [PubMed] [Google Scholar]
- 35. Meining A, Kroher G, Stolte M. 1998. Animal reservoirs in the transmission of Helicobacter heilmannii. Results of a questionnaire-based study. Scand. J. Gastroenterol. 33:795–798 [DOI] [PubMed] [Google Scholar]
- 36. Monteiro MA, et al. 2000. Lipopolysaccharide structures of Helicobacter pylori genomic strains 26695 and J99, mouse model H. pylori Sydney strain, H. pylori P466 carrying sialyl Lewis X, and H. pylori UA915 expressing Lewis B classification of H. pylori lipopolysaccharides into glycotype families. Eur. J. Biochem. 267:305–320 [DOI] [PubMed] [Google Scholar]
- 37. Moran AP. 2009. Molecular mimicry of host glycosylated structures by bacteria, p 847–870 In Moran AP, Holst O, von Itzstein M. (ed), Microbial glycobiology. Elsevier Academic Press, London, United Kingdom [Google Scholar]
- 38. Moran AP. 2007. Lipopolysaccharide in bacterial chronic infection: insights from Helicobacter pylori lipopolysaccharide and lipid A. Int. J. Med. Microbiol. 297:307–319 [DOI] [PubMed] [Google Scholar]
- 39. Mrazek J. 2006. Analysis of distribution indicates diverse functions of simple sequence repeats in Mycoplasma genomes. Mol. Biol. Evol. 23:1370–1385 [DOI] [PubMed] [Google Scholar]
- 40. Nilsson C, et al. 2006. An enzymatic ruler modulates Lewis antigen glycosylation of Helicobacter pylori LPS during persistent infection. Proc. Natl. Acad. Sci. U. S. A. 103:2863–2868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. O'Toole PW, et al. 2010. Comparative genomics and proteomics of Helicobacter mustelae, an ulcerogenic and carcinogenic gastric pathogen. BMC Genomics 11:164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Reeves EP, et al. 2008. Helicobacter pylori lipopolysaccharide interacts with TFF1 in a pH-dependent manner. Gastroenterology 135:2043–2054.e2 [DOI] [PubMed] [Google Scholar]
- 43. Rice BE, Lamichhane C, Joseph SW, Rollins DM. 1996. Development of a rapid and specific colony-lift immunoassay for detection and enumeration of Campylobacter jejuni, C. coli, and C. lari. Clin. Diagn. Lab. Immunol. 3:669–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schott T, Rossi M, Hanninen ML. 2011. Genome sequence of Helicobacter bizzozeronii strain CIII-1, an isolate from human gastric mucosa. J. Bacteriol. 193:4565–4566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schott T, Kondadi PK, Hanninen ML, Rossi M. 2011. Comparative genomics of Helicobacter pylori and the human-derived Helicobacter bizzozeronii CIII-1 strain reveal the molecular basis of the zoonotic nature of non-pylori gastric Helicobacter infections in humans. BMC Genomics 12:534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sharma CM, et al. 2010. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature 464:250–255 [DOI] [PubMed] [Google Scholar]
- 47. Snyder LA, et al. 2010. Simple sequence repeats in Helicobacter canadensis and their role in phase variable expression and C-terminal sequence switching. BMC Genomics 11:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tamura K, et al. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Thon V, Li Y, Yu H, Lau K, Chen X. 11 November 2011, posting date PmST3 from Pasteurella multocida encoded by Pm1174 gene is a monofunctional alpha2-3-sialyltransferase. Appl. Microbiol. Biotechnol. [Epub ahead of print.] doi:10.1007/s00253-011-3676-6 [DOI] [PubMed] [Google Scholar]
- 50. Tomb JF, et al. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539–547 [DOI] [PubMed] [Google Scholar]
- 51. Wakarchuk WW, Cunningham AM. 2003. Capillary electrophoresis as an assay method for monitoring glycosyltransferase activity. Methods Mol. Biol. 213:263–274 [DOI] [PubMed] [Google Scholar]