Abstract
Transcription-coupled repair (TCR) is a cellular process by which some forms of DNA damage are repaired more rapidly from transcribed strands of active genes than from nontranscribed strands or the overall genome. In humans, the TCR coupling factor, CSB, plays a critical role in restoring transcription following both UV-induced and oxidative DNA damage. It also contributes indirectly to the global repair of some forms of oxidative DNA damage. The Escherichia coli homolog, Mfd, is similarly required for TCR of UV-induced lesions. However, its contribution to the restoration of transcription and to global repair of oxidative damage has not been examined. Here, we report the first direct study of transcriptional recovery following UV-induced and oxidative DNA damage in E. coli. We observed that mutations in mfd or uvrA reduced the rate that transcription recovered following UV-induced damage. In contrast, no difference was detected in the rate of transcription recovery in mfd, uvrA, fpg, nth, or polB dinB umuDC mutants relative to wild-type cells following oxidative damage. mfd mutants were also fully resistant to hydrogen peroxide (H2O2) and removed oxidative lesions from the genome at rates comparable to wild-type cells. The results demonstrate that Mfd promotes the rapid recovery of gene expression following UV-induced damage in E. coli. In addition, these findings imply that Mfd may be functionally distinct from its human CSB homolog in that it does not detectably contribute to the recovery of gene expression or global repair following oxidative damage.
INTRODUCTION
Xeroderma pigmentosum (XP) and Cockayne syndrome (CS) are heritable diseases with distinct clinical outcomes stemming from defects in nucleotide excision repair. Both diseases are typically associated with sunlight sensitivity. Yet, XP patients are predisposed to skin cancers, whereas CS patients have severe neurological and developmental defects (reviewed in reference 35). Classical XP results from defects in any one of 7 genes, XPA to XPG, and is characterized by an inability to remove bulky lesions from the overall genome, sometimes referred to as a defect in global genome repair (GGR) (2, 12, 30, 31, 55, 61, 71). In contrast, mutations in CSA and CSB are associated with CS and are specifically defective in removing bulky adducts from the template strand of actively transcribed genes, a process termed transcription-coupled repair (TCR) (34, 60, 64, 70). TCR also requires the participation of 5 of the 7 XP genes, XPA, XPB, XPD, XPF, and XPG.
While CS is sometimes considered a deficiency in a subpathway of nucleotide excision repair, the clinical outcome for individuals with this disease is, in many ways, more severe than for those with XP, which affects both pathways of nucleotide excision repair. Based on this apparent contradiction, a number of researchers have speculated that CSA and CSB may have additional functions beyond TCR of bulky lesions. Some clues to these additional roles have come from the observation that CS-B cell lines are hypersensitive to oxidizing agents, such as ionizing radiation, and are deficient in the global repair of DNA lesions induced by oxidizing agents (21, 24, 47, 65, 66). In addition, microarray analyses suggest that the absence of CSB leads to an impaired transcriptional response following H2O2 treatment (32). These observations have led researchers to propose that an underlying cause of the developmental and neurological deficiencies in CS patients may result from an abnormal accumulation of oxidative lesions, which are normally repaired by the base excision repair pathway, rather than by nucleotide excision repair (reviewed in references 13 and 27).
A series of initial reports claiming to directly demonstrate that oxidative DNA lesions in human cells are subject to TCR in a CS-dependent manner have been retracted (16, 26, 37). However, indirect evidence for TCR of oxidative lesions in mammalian cells has come from studies demonstrating that in comparison to wild-type cells, CS cell lines exhibit an increased sensitivity to H2O2 and fail to restore gene expression on plasmids containing the oxidative lesions 8-oxoguanine and thymine glycol (57). In addition, a role for oxidative damage in the etiology of CS has been strengthened by the recent identification of a CS-A patient exhibiting UV hypersensitivity correlating with impaired TCR of UV lesions but normal sensitivity to oxidizing agents. In this individual, mutation of W361 in CSA manifests in the mild clinical outcome of UV-sensitive syndrome without any of the severe neurological defects associated with CS (46).
TCR of bulky lesions is highly conserved and well established in Escherichia coli (52). Similar to humans, TCR in E. coli is initiated by the arrest of RNA polymerase at DNA damage and depends upon the nucleotide excision repair pathway genes (44, 54). RNA polymerase that is blocked by DNA damage serves as a signal for the coupling factor Mfd to recruit the nucleotide excision repair proteins UvrABC and initiate TCR (54). Cells lacking mfd or any of the nucleotide excision repair genes are defective in transcription-coupled repair of UV-induced damage (43, 53). However, how the absence of TCR or Mfd affects the rate that transcription recovers after UV-induced damage in E. coli has not been directly examined.
Whereas UV-induced lesions are strong blocks to RNA polymerases, only a subset of the lesions induced by reactive oxygen species are known to arrest prokaryotic DNA and RNA polymerases (28, 29, 49, 62, 63). In addition, base excision repair (BER) rather than nucleotide excision repair (NER) is the predominant mechanism that removes oxidative base damage, raising the possibility that BER may be recruited to repair transcriptional blocks (51). Indirect evidence for TCR at oxidative damage in E. coli has been inferred from studies using nongrowing cells in which inactivation of Mfd, UvrA, or Fpg glycosylase increased transcriptional mutagenesis (mutations in transcripts) opposite 8-oxoguanine lesions (8, 11). Curiously, 8-oxoguanine does not arrest RNA polymerase in vitro (62). Taken together, these studies would suggest a cellular role for TCR at oxidative lesions in E. coli. However, whether Mfd contributes to transcriptional recovery or global repair after oxidative DNA damage, similar to its CSB homolog, has not been investigated.
In this study, we measured the rate that transcription recovered in TCR-proficient and -deficient E. coli following UV-induced DNA damage and H2O2-induced oxidative damage and characterized the rate that oxidative damage was repaired in mfd mutants relative to wild-type cells. We found that uvrA and mfd cells were deficient in the recovery of gene expression following UV irradiation, consistent with a role for TCR in rapid restoration of transcription following UV-induced DNA damage. In contrast, the absence of Mfd did not detectably affect the transcriptional recovery following H2O2-induced oxidative DNA damage or contribute to the rate that oxidative lesions were removed from the genome, suggesting that Mfd may be functionally distinct from its CSB homolog in these aspects.
MATERIALS AND METHODS
Bacterial strains.
The parent of all strains used in this study is SR108, a thyA36 deoC2 derivative of W3110 (44). Isogenic strains lacking uvrA (HL952), polB dinB umuDC (CL646), fpg (CL1778), and nth (CL1006) were constructed using standard P1 transduction methods and have been described previously (18, 51). SR108 recG (CL559) was constructed by P1 transduction of the recG6201::kan allele from TP539 (45) into SR108. SR108 mfd (HL939, D. J. Crowley and P. C. Hanawalt, unpublished strain) was constructed by P1 transduction of the mfd::kan allele from WU3610-45 (53) into SR108. SR108 mfd fpg (CL1955) was constructed by P1 transduction of the fpg::tet allele from CL1778 into HL939. A list of the strains constructed and used in this study is shown in Table 1.
Table 1.
E. coli K-12 strains used
| Strain | Relevant genotype | Reference or construction |
|---|---|---|
| SR108 | λ−thyA36deoC2IN(rrnD-rrnE)1 rph | 44 |
| TP539 | recG6201::kan | 45 |
| WU3610-45 | mfd-1::kan | 53 |
| Strains isogenic to SR108 | ||
| HL939 | mfd-1::kan | SR108 × P1 (WU3610-45), Crowley and Hanawalt, unpublished |
| HL952 | uvrA::Tn10 | 18 |
| CL559 | recG6201::kan | SR108 × P1 (TP539) |
| CL646 | polB::Ω Sm-Sp dinB::kan umuDC595::cat | 17 |
| CL1006 | nth::kan | 51 |
| CL1778 | fpg::tet | 51 |
| CL1955 | mfd-1::kan fpg::tet | HL939 × P1 (CL1778) |
RNA synthesis.
UV irradiation used a 15-watt germicidal lamp (254 nm) at an incident dose of 0.9 J/m2/s. For experiments using UV irradiation, overnight cultures were diluted 1:100 and grown at 37°C in Davis medium supplemented with 0.4% glucose, 0.2% Casamino Acids, and 10 μg/ml thymine (DGCthy medium) to an optical density at 600 nm (OD600) of precisely 0.3, at which point one-third of the cells were mock irradiated, while the remaining culture was divided equally and irradiated at an incident dose of 50 or 100 J/m2.
For experiments using H2O2 as a DNA-damaging agent, overnight cultures were diluted 1:100 and grown at 37°C in Luria Bertani medium supplemented with 10 μg/ml thymine (LBthy medium) to an OD600 of precisely 0.3, at which point half of the culture was mock treated, while the other half was exposed to 10 mM H2O2 (Fisher Scientific) for 5 min at 37°C. Following either mock or H2O2 treatment, catalase (Fisherbrand) was added directly to the culture to a final concentration of 200 μg/ml to remove excess H2O2 from the medium.
For both UV irradiation and H2O2 treatment, cultures were returned immediately to 37°C following exposure to allow recovery and continued growth. Duplicate 0.5-ml aliquots of culture were pulse-labeled with 0.2 μCi/ml [3H]uridine for 2 min at 37°C at the times indicated. Cells were then lysed, and the DNA precipitated in cold 5% trichloroacetic acid (TCA; Fisherbrand). The precipitate was collected on Millipore glass fiber filters and the amount of 3H on each filter was determined by scintillation counting.
Lesion frequency and repair rates.
For UV irradiation, overnight cultures were diluted 1:100 and grown in DGCthy medium to an OD600 of 0.3, at which point they were UV irradiated with 50 J/m2 and then returned to 37°C to allow recovery. For H2O2 challenge, overnight cultures were diluted 1:100 and grown in LBthy medium to an OD600 of 0.3 and treated with 10 mM H2O2 for 5 min at 37°C. The cells were then collected on Millipore 0.45-μm membranes, resuspended in fresh, prewarmed LBthy medium and returned to 37°C for the duration of the time course. At the times indicated, a 0.75-ml aliquot of culture was transferred to an equal volume of ice-cold NET (100 mM NaCl, 10 mM Tris [pH 8.0], 20 mM EDTA [pH 8.0]), centrifuged for 60 s, resuspended in 140-μl lysis buffer (1 mg/ml lysozyme, 0.5 mg/ml RNase A in 10 mM Tris, 1 mM EDTA [pH 8.0]), and incubated at 37°C for 30 min. Ten microliters of 10 mg/ml proteinase K and 10 μl of 20% Sarkosyl were then added to the samples, and incubation continued for a further 30 min. Samples were then extracted with four volumes of phenol-chloroform-isoamyl alcohol (25:24:1), followed by four volumes of chloroform-isoamyl alcohol (24:1) and then dialyzed against 200 ml of 1 mM Tris (pH 8.0), 2 mM EDTA (pH 8.0) for 45 min using 47-mm Millipore 0.025-μm-pore-size disks. For UV-irradiated samples, 15 μl of each DNA sample was then treated in reaction buffer (12.5 mM sodium phosphate [pH 6.8], 5 mM EDTA [pH 8.0], 50 mM NaCl, 0.5 mM dithiothreitol [DTT], 0.005% Triton X-100, 0.1 mg/ml bovine serum albumin [BSA]) supplemented with either no enzyme or 2 U T4 endonuclease V (T4 endo V; Trevigen) for 1 h at 37°C. For H2O2-exposed samples, 15 μl of each DNA sample was then treated in reaction buffer (30 mM EDTA [pH 8.0], 22.5 mM NaCl, 5 mM Tris [pH 8.0]) supplemented with either no enzyme or 0.53 μM Fpg glycosylase for 1 h at 37°C. Enzyme preparations were titrated using purified undamaged genomic DNA as a template. The highest enzyme concentration which did not exhibit nonspecific activity on the undamaged DNA was used. For the preparations in our lab, this corresponded to 0.53 μM Fpg glycosylase and 2 U T4 endo V.
Treated samples were then electrophoresed on a 0.5% alkaline agarose gel in 30 mM NaOH, 1 mM EDTA at 30 V for 16 h, stained and visualized with ethidium bromide. The intensity of each high-molecular-weight band was determined using ImageQuant software (GE Healthcare). The fraction of lesion-free DNA fragments was quantified as a ratio of high-molecular-weight DNA in the T4 endo V- or Fpg-treated band to the band with no enzyme treatment at each time point. To normalize for any nicks or AP sites present in the DNA before UV or H2O2 exposure, the ratio at each time point was divided by the ratio at the pretreatment time point, resulting in the following formula: (EnzT/NoEnzT)/R0. Where EnzT is the band intensity for T4 endo V- or Fpg-treated DNA at time, T. NoEnzT is the band intensity for DNA with no enzyme treatment at time, T. R0 is EnzT/NoEnzT at the time immediately preceding UV irradiation or the addition of H2O2.
UV survival assays.
Fresh overnight cultures were diluted 1:100 and grown at 37°C in DGCthy medium to an OD600 between 0.4 and 0.5 (approximately 6 × 108 cells/ml). Ten-μl aliquots of serial 10-fold dilutions were spotted in triplicate on LBthy plates and UV irradiated at the indicated doses. Viable colonies were counted following overnight incubation at 37°C.
H2O2 survival assays.
Fresh overnight cultures were diluted 1:100 and grown at 37°C in LBthy medium to an OD600 of 0.4 (approximately 6 × 108 cells/ml) and then treated with H2O2 at a final concentration of 10 mM. At the times indicated, 0.1-ml aliquots of each culture were removed and serially diluted in 10-fold increments. Triplicate 10-μl aliquots of each dilution were then spotted on LBthy plates. Viable colonies were counted following overnight incubation at 37°C.
RESULTS
Mfd contributes to the rapid recovery of gene expression following UV-induced DNA damage.
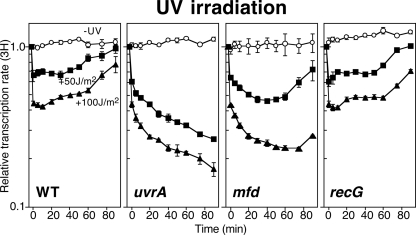
In mammalian cells, it has been demonstrated that a primary function of TCR is to allow cells to rapidly express genes needed to respond and recover following acute exposures to DNA damage (36, 41). The mammalian Mfd homolog, CSB, is required for the rapid recovery of transcription following UV-induced DNA damage (64). To determine whether TCR by Mfd similarly contributes to the rate that gene expression recovers after UV-induced DNA damage in E. coli, we monitored the rate of RNA synthesis after UV irradiation. To this end, duplicate aliquots of cultures were pulse-labeled with [3H]uridine for 2 min at various times following irradiation with 50 or 100 J/m2 of UV and the rate of transcription at each time (3H incorporation/2 min) was then determined. These irradiation conditions generate approximately one cyclobutane-pyrimidine dimer (CPD) per 4.8 kb and 2.4 kb of single-strand DNA, respectively, as measured by T4 endonuclease V (T4 endo V)-sensitive sites in the DNA (19) (see Fig. 3). To ensure that any observed differences in RNA synthesis were due to UV irradiation and not culture conditions, a mock-irradiated control was included for comparison in each experiment. Under these assay conditions, the rate of synthesis in the mock-irradiated cells increased approximately 2-fold, reflecting cell growth over the course of the experiment. In contrast, the rate of RNA synthesis in UV-irradiated wild-type cells initially decreased by approximately 35% and 55% following 50- and 100-J/m2 UV irradiation, respectively (Fig. 1). Following the initial inhibition, the recovery of transcription occurred in two distinct phases. An early phase was observed that involved a modest but reproducible increase in RNA synthesis approximately 5 min after the 50-J/m2 dose and 20 min after the 100-J/m2 dose. A late, more robust phase of the recovery occurred around 40 min after the 50-J/m2 dose and 70 min after the 100-J/m2 dose.
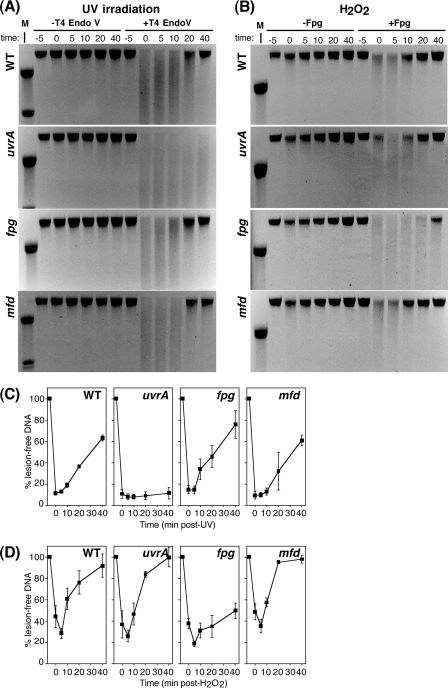
Fig 3.
Mfd does not affect the global rate of repair following either UV irradiation or H2O2 treatment. (A) wild-type (WT; first panel), uvrA (second panel), fpg (third panel), and mfd (last panel) cells were UV irradiated at 50 J/m2 and allowed to recover. At the indicated times, genomic DNA was purified and either treated with T4 endonuclease V (T4 endo V) or no T4 endo V for 1 h at 37°C and then analyzed on alkali-agarose gels. A representative gel is shown for each analyzed strain. (B) WT (first panel), uvrA (second panel), fpg (third panel), and mfd (last panel) cells were exposed to 10 mM H2O2 for 5 min and allowed to recover. At the indicated times, genomic DNA was purified and either treated with Fpg or no glycosylase for 1 h at 37°C and then analyzed on alkali-agarose gels. A representative gel is shown for each analyzed strain. (C) The percentage of lesion-free, high-molecular-weight DNA in T4 endo V-treated samples is plotted for each time point relative to mock-treated samples. (D) The percentage of lesion-free, high-molecular-weight DNA in Fpg-treated samples is plotted for each time point relative to mock-treated samples. Graphs represent an average of at least three independent experiments. Error bars represent one standard error of the mean. M, lambda HindIII molecular-weight marker, top two bands represent 23.1 and 9.4 kb, respectively.
Fig 1.
The transcription-coupling factor Mfd is required for rapid transcriptional recovery following UV irradiation. [3H]uridine was added to cultures for 2 min at the indicated times following either 50- or 100-J/m2 UV irradiation (filled symbols) or mock irradiation (open symbols) at time zero. The amount of RNA synthesis/2 min (3H) is plotted. Graphs represent an average of at least two independent experiments. Error bars represent one standard error of the mean.
In UV-irradiated uvrA cultures, which are defective in both TCR and GGR, the rate of transcription was initially inhibited to a similar extent as in wild-type cultures; however, no subsequent recovery of RNA synthesis was observed (Fig. 1). Instead, the rate of RNA synthesis continued to gradually decline for the remainder of the 90-min time course for both of the UV doses that were examined.
In UV-irradiated mfd cultures, which are specifically deficient in TCR but proficient in GGR, we observed a distinct absence of the early phase of the transcription recovery compared to wild-type cells (Fig. 1). After the initial inhibition of transcription, the rate of RNA synthesis continued to decrease gradually in mfd mutants, similar to what was observed in UV-irradiated uvrA mutants. The gradual decrease continued until 75 min and 90 min following 50- and 100-J/m2 treatments, respectively, at which time, the second phase of transcriptional recovery was observed.
As an additional control, we also examined the rate at which gene expression recovered in recG mutants. Similar to mfd, recG encodes a DNA helicase that shares a homologous ATP-dependent translocation domain and renders cells hypersensitive to UV-irradiation (39). However, unlike mfd, recG is not required for TCR (1, 4, 9, 23, 59). Following UV irradiation, we observed that recG cells recovered gene expression with wild-type kinetics and in a similar biphasic manner (Fig. 1). The normal transcriptional recovery observed in recG mutants despite the elevated levels of cell lethality argues that the defect in recovering transcription in mfd mutants is specific to the absence of TCR and not due to the elevated levels of lethality that occur in these UV-irradiated populations.
The time at which the early and late phases of transcriptional recovery is observed correlates with the approximate period in which repair by TCR and GGR occurs, respectively (20, 44). Consistent with this, mfd mutants are specifically deficient in the early but not the late phase of transcriptional recovery, whereas uvrA mutants are defective in both phases of the recovery. These results demonstrate that Mfd in E. coli contributes functionally to the rapid recovery of transcription following UV-induced damage, similar to its mammalian CSB homolog.
The absence of Mfd does not delay the recovery of transcription following H2O2-induced DNA damage.
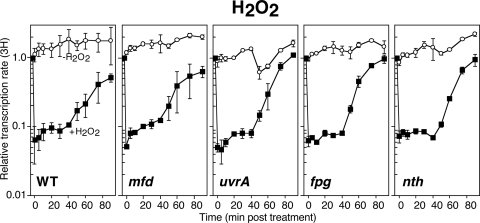
Mammalian cell lines lacking CSB fail to restore gene expression from plasmids containing 8-oxoguanine and thymine glycol lesions (33, 57). To determine whether Mfd detectably contributes to the recovery of gene expression following oxidative damage, we examined the rate of transcriptional recovery in cultures following treatment with H2O2 using the assay described above. To this end, cultures were treated with 10 mM H2O2 for 5 min, before catalase was added to remove excess H2O2 from the medium and allow recovery to occur. Then, duplicate aliquots of culture were pulse-labeled with [3H]uridine for 2 min at the indicated time points as before. To control for the effect of catalase and culture conditions on transcription, mock-treated cultures were included in each experiment.
By this assay, the rate of transcription increased by approximately 2-fold in mock-treated cultures of all strains over the time course of the experiment. In wild-type cultures treated with H2O2, we observed a pattern of transcriptional inhibition and a biphasic recovery that was similar to that seen in UV-irradiated cultures (Fig. 2). The rate of transcription initially decreased by ∼90% after H2O2 treatment. An initial, modest recovery in the rate of transcription occurred during the first 5 min, followed by a more rapid and robust recovery that began approximately 40 min after the H2O2 was removed.
Fig 2.
The absence of Mfd does not affect transcriptional recovery following H2O2-induced DNA damage. Cells were either exposed to 10 mM H2O2 for 5 min (filled symbols) or mock treated (open symbols) at time zero and then allowed to recover in the presence of 200 μg/ml catalase. At the indicated times, [3H]uridine was added to cultures for 2 min. The amount of RNA synthesis/2 min (3H) is plotted. Graphs represent an average of at least two independent experiments. Error bars represent one standard error of the mean.
When we examined H2O2-treated mfd cultures, we observed that the recovery of transcription was indistinguishable from that seen in wild-type cells (Fig. 2). Fpg and endonuclease III, encoded by fpg and nth, are two of the predominant glycosylases that remove oxidative base adducts in E. coli (7, 22, 51). Following H2O2 treatment of uvrA, fpg, or nth mutants, transcription also recovered at rates that were similar to wild-type cells. The results demonstrate that the absence of Mfd and TCR does not detectably impair the time or rate that transcription recovers following oxidative DNA damage in E. coli.
Mfd does not detectably contribute to the global repair of oxidative damage following H2O2 treatment.
Studies in mammalian cells have shown that the absence of CSB results in a deficiency in GGR of oxidatively induced lesions but not UV-induced lesions (21, 24, 47, 65–67). To determine whether Mfd affects the rate that oxidative lesions are repaired from the global genome, we monitored the removal of Fpg-sensitive sites from the DNA of H2O2-treated cultures over time. For comparison, the repair of UV-induced damage, as monitored by the removal of T4 endo V-sensitive sites, was also examined. To this end, cultures were either irradiated with 50 J/m2 of UV or treated with 10 mM H2O2 for 5 min before cells were filtered, resuspended in fresh, prewarmed medium, and allowed to recover at 37°C. High-molecular-weight genomic DNA was purified from both cultures at the indicated time points. The DNA from each time point was then incubated with T4 endo V or Fpg glycosylase and then electrophoresed in a denaturing alkali gel. The presence of T4 endo V- or Fpg-sensitive sites in the DNA is indicated by the loss of high-molecular-weight DNA.
In all UV-irradiated cultures examined in the absence of T4 endo V treatment, purified genomic DNA remained high molecular weight at all times throughout the recovery period (Fig. 3A). DNA fragments averaged greater than 40 kb, which was the approximate limit of resolution of our agarose gels. In wild-type cultures, incubation of the DNA from preirradiated cultures with T4 endo V resulted in little or no loss of high-molecular-weight DNA. At times immediately following UV irradiation in wild-type cells, a loss of high-molecular-weight DNA was observed in samples incubated with T4 endo V, indicating the presence of UV-induced lesions. Over time, the number of T4 endo V-recognized sites began to decrease, as lesion-free, high-molecular-weight DNA returned and by 40 min, more than 60% of the DNA had been restored (Fig. 3C). In contrast, in UV-irradiated uvrA cultures, which are defective in nucleotide excision repair, lesions persisted throughout the 40-min time course (Fig. 3A and C). As expected, the absence of Fpg, which is not involved in the repair of UV-induced damage, did not affect the rate that UV-induced lesions were removed from the genome. When we measured the rate of global repair in mfd mutants after UV irradiation, we observed a repair profile closely following the repair profile of wild-type cells (Fig. 3C).
To assess repair in wild-type, fpg, uvrA, and mfd cells following oxidative damage, total genomic DNA was prepared at the indicated time points after H2O2 exposure, and the number of Fpg-sensitive sites remaining in the DNA was determined. In samples not incubated with Fpg, genomic DNA remained high molecular weight at all time points examined, both before and after H2O2 treatment for all strains examined (Fig. 3B).
In wild-type cultures, DNA purified from cultures prior to H2O2 exposure remained high molecular weight following incubation with Fpg, demonstrating that the enzyme is specific for oxidative lesions. At times immediately following H2O2 treatment, there was a loss of high-molecular-weight DNA, indicating that lesions recognized by Fpg were present in the DNA (Fig. 3B). The number of Fpg-sensitive sites increased modestly within the first 5 min of the recovery period, which is likely to be due to reactive oxygen species that continue to be generated in the cell after the H2O2 has been removed (10, 40). Based on the average DNA fragment size in glycosylase-treated samples, the lesion frequency never exceeded more than one Fpg substrate per ∼40-kb strand of DNA at any given time. However, the continued induction of DNA lesions during the repair period, the high rate of glycosylase repair, and the observed 90% inhibition of transcription suggest that the total number of lesions generated by this treatment was significantly higher than that induced after UV treatment. Despite the higher number of lesions, repair occurred rapidly. The number of Fpg-sensitive sites in H2O2-treated wild-type cells began to decrease by 10 min, and by 40 min, greater than 90% of the fragments were resistant to cleavage by Fpg and migrated as a high-molecular-weight band (Fig. 3D).
In cells lacking UvrA, which is not involved in the repair of oxidative DNA damage (51), Fpg-sensitive sites were removed from the genome with kinetics that were similar to wild-type cultures (Fig. 3B and D). In contrast, in fpg cultures exposed to H2O2, the lesions were seen to persist, and at 40 min, less than 50% of the DNA was resistant to Fpg treatment, consistent with Fpg's role in GGR of oxidative lesions (51). To determine if Mfd plays a role in oxidative lesion removal from the chromosome, we next examined the repair rate of Fpg-sensitive sites in mfd cells. After treatment with H2O2, mfd strains removed oxidative damage at a rate that was not detectably different from either wild-type or uvrA cultures (Fig. 3B and D).
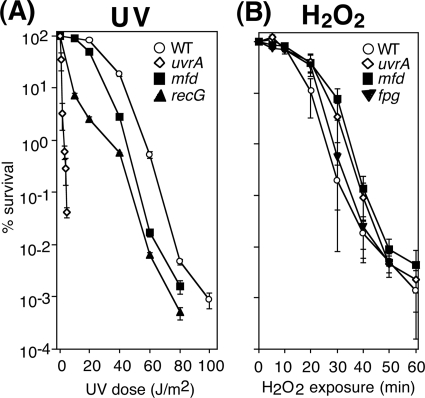
In mammals, the CSB defect in GGR of oxidative damage renders cells hypersensitive to H2O2 (24, 47, 65, 66). To determine the effect of the mfd mutation on H2O2 sensitivity, the relative survival of each strain was also examined (Fig. 4). Consistent with previous reports, the absence of mfd rendered cells modestly hypersensitive to UV-induced damage, although to a lesser extent than that of a recG mutant. In contrast, mfd mutants appeared as resistant as wild-type cultures when exposed to H2O2 (Fig. 4B). Similarly, neither fpg nor uvrA mutants were hypersensitive to H2O2, in agreement with previous reports (3, 50, 51). Taken together, the results indicate that the absence of mfd does not detectably impair the global repair of oxidative damage, in contrast to what is seen with CSB in mammalian cells.
Fig 4.
Mfd is hypersensitive to UV irradiation but not H2O2 treatment. (A) The survival rates of wild-type (○), mfd (■), recG (▲), and uvrA (♢) cultures are shown after UV irradiation at the indicated doses. (B) The survival rates of wild-type (○), mfd (■), fpg (▼), and uvrA (♢) cultures are shown after exposure to 10 mM H2O2 for the indicated amount of time. All graphs represent an average of at least three independent experiments. Error bars represent one standard error of the mean.
Base excision repair does not contribute to the restoration of transcription in mfd mutants nor is translesion DNA synthesis involved in transcriptional recovery following H2O2-induced DNA damage.
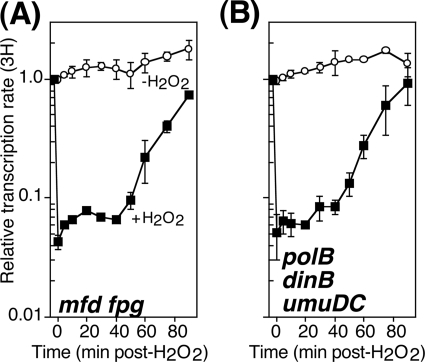
The lack of a phenotype for mfd mutants in the recovery of gene expression after oxidative challenge could reflect rapid turnover of oxidative lesions by base excision repair enzymes. In such a scenario, Fpg would efficiently and rapidly remove oxidative lesions before they arrested the transcription machinery, thus masking any effect of Mfd, and by extension TCR, on the recovery of RNA synthesis after oxidative DNA damage. Analogous to this, the rapid repair of 6-4 photoproducts by GGR has been shown to obscure TCR of this lesion in mammalian cells that express wild-type XPC (69). Also consistent with the idea that efficient GGR may be masking oxidative-TCR events is the observation that transcriptional mutagenesis at 8-oxoguanine lesions is significantly higher in fpg mutants (∼43-fold) than in mfd mutants (∼12-fold) and synergistically increases when both fpg and mfd are inactivated (8). To examine this possibility directly, we constructed an mfd fpg double mutant and determined the rate of transcriptional recovery in these cultures following treatment with H2O2 using the assay described above. We observed that the recovery of transcription in H2O2-treated mfd fpg cultures was indistinguishable from that seen in either single mutant or wild-type cells (Fig. 5A). This result indicates that the wild-type transcriptional recovery observed in mfd cells is not due to efficient removal of oxidative lesions by base excision repair enzymes.
Fig 5.
The lack of a requirement for mfd in restoring transcription after H2O2-induced DNA damage is not due to the high rate of base excision repair or translesion synthesis. Cells were either exposed to 10 mM H2O2 for 5 min (filled symbols) or mock treated (open symbols) at time zero and then allowed to recover in the presence of 200 μg/ml catalase. At the indicated times, [3H]uridine was added to cultures for 2 min. The amount of RNA synthesis/2 min (3H) is plotted for mfd fpg (A) and polB dinB umuDC (B) mutants. Graphs represent an average of at least two independent experiments. Error bars represent one standard error of the mean.
Our results suggested that the restoration of gene expression following oxidative challenge was not dependent on either mfd-mediated TCR or rapid fpg-mediated GGR. We therefore considered whether translesion DNA synthesis could be facilitating bypass of oxidative lesions on the transcribed strand. Transcription-coupled translesion synthesis has been suggested by physical and genetic interactions between the E. coli RNA polymerase modulator NusA and the translesion DNA polymerases Pol IV (dinB) and Pol V (umuDC) (14). This process has also been implicated in cell survival following nitrofurazone treatment in stationary cells (15). To test this possibility, we monitored the rate of RNA synthesis after H2O2 treatment in polB dinB umuDC cells, which lack all three translesion DNA polymerases. Similar to what we observed in wild-type cells, we found that gene expression was restored in a biphasic manner following oxidative challenge in polB dinB umuDC cells (Fig. 5B). This result indicates that translesion DNA polymerases are not contributing to the recovery of transcription following exposure to H2O2.
DISCUSSION
Mfd effect on UV-inactivated RNA synthesis.
It has been proposed that a primary function of transcription-coupled repair is to promote the rapid recovery of gene expression following DNA damage. In this sense, TCR is proposed to act as a triage mechanism by first restoring the integrity of genes needed for the cellular response to the stress. Several studies have shown that CSA and CSB function is required to rapidly restore transcription in eukaryotic cells (36, 41, 58, 64, 68). However, the effect that TCR has on the rate of transcriptional recovery had not been directly examined in E. coli. The results presented here demonstrate that TCR is essential for the early recovery of RNA synthesis following UV irradiation and supports the idea that Mfd functions similarly to its CSB homolog in this respect.
The level of transcriptional inhibition following UV irradiation in E. coli was dose dependent and consistent with the idea that transcription is arrested following a direct collision or encounter with the UV-induced photoproduct. Immediately following UV irradiation at 50 J/m2 (1 lesion/4.8 kb single-stranded DNA [ssDNA]) and 100 J/m2 (1 lesion/2.4 kb ssDNA), RNA synthesis was inhibited by 35% and 55%, respectively. Assuming an average transcript length of ∼1,200 bp in E. coli, this level is within a reasonable approximation of the 25% and 50% inhibition one would be expected to observe.
A previous study by Li and Bockrath (38) examined how the absence of Mfd affected the production of β-galactosidase, encoded by lacZ, and galactoside acetyl-transferase, encoded by lacA, after UV (38). The authors demonstrated that the absence of Mfd impaired the inducibility of these proteins after UV. At the level of protein expression, the authors found that both Mfd and nucleotide excision repair mutants were impaired to a similar extent, leading the authors to conclude that nucleotide excision repair appears to make no contribution beyond that associated with TCR in restoring gene expression. Our results complement and extend these observations in two aspects. First, using an assay to monitor the rate of transcription directly, we were able to observe distinct Mfd-dependent and -independent phases of recovery following UV-induced damage. The early phase of transcriptional recovery involved TCR and required both Mfd and nucleotide excision repair, whereas the late phase only depended on nucleotide excision repair. Second, the comparative levels of transcription inhibition and hypersensitivity between mfd, uvrA, and recG mutants establishes that the transcriptional defect in Mfd mutants is specific to its role in TCR and not due to an indirect product of the elevated levels of lethality that occur in these cells.
TCR effect on H2O2-inactivated RNA synthesis.
In contrast to UV damage, the absence of mfd did not have a detectable effect on either the transcriptional recovery, the removal of lesions, or cell survival following oxidative DNA damage and differs from the phenotypes observed for CSB in mammalian cells. H2O2 is known to induce several different classes of oxidative lesions on DNA, including 8-oxoguanine, thymine glycol, and 4,6-diamino-5-formamidopyrimidine (6). Of these, a subset such as thymine glycol has been shown to arrest prokaryotic DNA and RNA polymerases in vitro, while others such as 8-oxoguanine do not or only partially arrest the RNA polymerase (28, 29, 49, 56, 62, 63). Given the generally accepted model that an RNA polymerase stalling event is required for the activation of the TCR pathway (42), one possible reason we were unable to detect any effect of the mfd mutation following H2O2 treatment is that nonblocking oxidative lesions predominate. This type of model would predict that Mfd may still be required to repair the lesions in transcribed genes but would not prohibit transcription from occurring in its absence.
Another possibility is that these observed differences represent a true functional divergence between Mfd and CSB. Consistent with this, a number of studies have suggested CSB has secondary functions in chromatin remodeling and basal transcription that are independent from its role in processing arrested transcription complexes (5, 25, 48). How these potential functions relate to the repair of oxidative damage or whether Mfd has analogous functions in bacteria has not yet been examined. It is also worth noting that the processing of oxidative lesions is separable from TCR in eukaryotic cells as indicated in CSA W361C mutants, which have normal resistance to oxidizing agents but are defective in TCR and hypersensitive to UV lesions (46). These phenotypes closely mimic those reported here for the mfd mutant in E. coli.
A third possibility, which is not mutually exclusive, is that the absence of an effect by Mfd could reflect a growth phase-dependent activity for this protein following oxidative challenge. Consistent with this hypothesis is the indirect evidence that TCR does occur at 8-oxoguanine lesions in nongrowing E. coli (8). Using a novel assay to monitor mutations arising in transcripts of nonreplicating cells, Brégeon et al. found that the rate of transcriptional mutagenesis on DNA templates bearing 8-oxoguanine was elevated ∼12-fold in mfd mutants compared to wild-type cells. A more recent study by this group and using the same mutagenesis assay with nongrowing mfd uvrA and fpg uvrA found slightly elevated transcriptional mutagenesis rates at 8-oxoguanine lesions in these double mutants compared to their respective single mutant parents, suggesting that Mfd-mediated TCR might proceed through either base excision repair or nucleotide excision repair at 8-oxoguanine (11). The finding that nucleotide excision repair enzymes can function at 8-oxoguanine lesions in nongrowing cultures (11) contrasts with what we have found in our present and previous studies in actively growing E. coli (51), where we observe that UvrA mutants are able to remove oxidative lesions with wild-type kinetics, and further suggests an effect of growth phase on the repair of oxidative DNA damage. In a separate study, modulation of transcription by NusA was shown to alter cell viability in the presence of nitrofurazone-induced DNA damage under nongrowth conditions (15), an observation that would also be consistent with the presence of a DNA damage repair or tolerance mechanism at the level of transcription in stationary-phase E. coli cells. Finally, given the predominant neurological symptoms associated with CS patients (13, 27, 57), it seems possible that the effects of TCR at oxidative lesions may also be of consequence in nondividing human cells.
A somewhat unexpected observation in our previous study is that survival and recovery of replication and transcription are only minimally affected by the absence of Fpg, even though oxidative lesions persist in the genomes of these mutants (51). Equally surprising are the results presented here that show the absence of Mfd does not affect the survival or recovery of transcription in growing cells, even when global genomic repair of the prominent oxidative lesions is disrupted. The mechanism that allows replication and transcription to recover and continue in cells containing oxidative lesions remains an interesting question for further study.
ACKNOWLEDGMENTS
We thank David Crowley and Tony Poteete for providing strains.
This work was funded by National Institutes of Health/National Institute of Environmental Health research grants (R21ES018940) and (R15ES021594) and an American Heart Association predoctoral fellowship (11PRE6890009) to B.J.S.
Footnotes
Published ahead of print 16 March 2012
B.J.S. and C.T.C. contributed equally to this article.
REFERENCES
- 1. Al-Deib AA, Mahdi AA, Lloyd RG. 1996. Modulation of recombination and DNA repair by the RecG and PriA helicases of Escherichia coli K-12. J. Bacteriol. 178:6782–6789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arase S, Kozuka T, Tanaka K, Ikenaga M, Takebe H. 1979. A sixth complementation group in xeroderma pigmentosum. Mutat. Res. 59:143–146 [DOI] [PubMed] [Google Scholar]
- 3. Asad NR, de Almeida CE, Asad LM, Felzenszwalb I, Leitao AC. 1995. Fpg and UvrA proteins participate in the repair of DNA lesions induced by hydrogen peroxide in low iron level in Escherichia coli. Biochimie 77:262–264 [DOI] [PubMed] [Google Scholar]
- 4. Asai T, Kogoma T. 1994. Roles of ruvA, ruvC and recG gene functions in normal and DNA damage-inducible replication of the Escherichia coli chromosome. Genetics 137:895–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Balajee AS, May A, Dianov GL, Friedberg EC, Bohr VA. 1997. Reduced RNA polymerase II transcription in intact and permeabilized Cockayne syndrome group B cells. Proc. Natl. Acad. Sci. U. S. A. 94:4306–4311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Blakely WF, Fuciarelli AF, Wegher BJ, Dizdaroglu M. 1990. Hydrogen peroxide-induced base damage in deoxyribonucleic acid. Radiat. Res. 121:338–343 [PubMed] [Google Scholar]
- 7. Boiteux S, Gajewski E, Laval J, Dizdaroglu M. 1992. Substrate specificity of the Escherichia coli Fpg protein (formamidopyrimidine-DNA glycosylase): excision of purine lesions in DNA produced by ionizing radiation or photosensitization. Biochemistry 31:106–110 [DOI] [PubMed] [Google Scholar]
- 8. Brégeon D, Doddridge ZA, You HJ, Weiss B, Doetsch PW. 2003. Transcriptional mutagenesis induced by uracil and 8-oxoguanine in Escherichia coli. Mol. Cell 12:959–970 [DOI] [PubMed] [Google Scholar]
- 9. Chambers AL, Smith AJ, Savery NJ. 2003. A DNA translocation motif in the bacterial transcription—repair coupling factor, Mfd. Nucleic Acids Res. 31:6409–6418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chung FL, Chen HJ, Nath RG. 1996. Lipid peroxidation as a potential endogenous source for the formation of exocyclic DNA adducts. Carcinogenesis 17:2105–2111 [DOI] [PubMed] [Google Scholar]
- 11. Clauson CL, Saxowsky TT, Doetsch PW. 2010. Dynamic flexibility of DNA repair pathways in growth arrested Escherichia coli. DNA Repair (Amst) 9:842–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cleaver JE. 1968. Defective repair replication of DNA in xeroderma pigmentosum. Nature 218:652–656 [DOI] [PubMed] [Google Scholar]
- 13. Cleaver JE, Lam ET, Revet I. 2009. Disorders of nucleotide excision repair: the genetic and molecular basis of heterogeneity. Nat. Rev. Genet. 10:756–768 [DOI] [PubMed] [Google Scholar]
- 14. Cohen SE, Godoy VG, Walker GC. 2009. Transcriptional modulator NusA interacts with translesion DNA polymerases in Escherichia coli. J. Bacteriol. 191:665–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cohen SE, et al. 2010. Roles for the transcription elongation factor NusA in both DNA repair and damage tolerance pathways in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 107:15517–15522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cooper PK, Nouspikel T, Clarkson SG, Leadon SA. 1997. Defective transcription-coupled repair of oxidative base damage in Cockayne syndrome patients from XP group G. Science 275:990–993 [DOI] [PubMed] [Google Scholar]
- 17. Courcelle CT, Belle JJ, Courcelle J. 2005. Nucleotide excision repair or polymerase V-mediated lesion bypass can act to restore UV-arrested replication forks in Escherichia coli. J. Bacteriol. 187:6953–6961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Courcelle J, Crowley DJ, Hanawalt PC. 1999. Recovery of DNA replication in UV-irradiated Escherichia coli requires both excision repair and recF protein function. J. Bacteriol. 181:916–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Courcelle J, Donaldson JR, Chow KH, Courcelle CT. 2003. DNA damage-induced replication fork regression and processing in Escherichia coli. Science 299:1064–1067 [DOI] [PubMed] [Google Scholar]
- 20. Crowley DJ, Hanawalt PC. 1998. Induction of the SOS response increases the efficiency of global nucleotide excision repair of cyclobutane pyrimidine dimers, but not 6-4 photoproducts, in UV-irradiated Escherichia coli. J. Bacteriol. 180:3345–3352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dianov G, Bischoff C, Sunesen M, Bohr VA. 1999. Repair of 8-oxoguanine in DNA is deficient in Cockayne syndrome group B cells. Nucleic Acids Res. 27:1365–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dizdaroglu M, Laval J, Boiteux S. 1993. Substrate specificity of the Escherichia coli endonuclease III: excision of thymine- and cytosine-derived lesions in DNA produced by radiation-generated free radicals. Biochemistry 32:12105–12111 [DOI] [PubMed] [Google Scholar]
- 23. Donaldson JR, Courcelle CT, Courcelle J. 2004. RuvAB and RecG are not essential for the recovery of DNA synthesis following UV-induced DNA damage in Escherichia coli. Genetics 166:1631–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Foresta M, et al. 2010. Defective repair of 5-hydroxy-2′-deoxycytidine in Cockayne syndrome cells and its complementation by Escherichia coli formamidopyrimidine DNA glycosylase and endonuclease III. Free Radic. Biol. Med. 48:681–690 [DOI] [PubMed] [Google Scholar]
- 25. Fousteri M, Vermeulen W, van Zeeland AA, Mullenders LH. 2006. Cockayne syndrome A and B proteins differentially regulate recruitment of chromatin remodeling and repair factors to stalled RNA polymerase II in vivo. Mol. Cell 23:471–482 [DOI] [PubMed] [Google Scholar]
- 26. Gowen LC, Avrutskaya AV, Latour AM, Koller BH, Leadon SA. 1998. BRCA1 required for transcription-coupled repair of oxidative DNA damage. Science 281:1009–1012 [DOI] [PubMed] [Google Scholar]
- 27. Hanawalt PC, Spivak G. 2008. Transcription-coupled DNA repair: two decades of progress and surprises. Nat. Rev. Mol. Cell Biol. 9:958–970 [DOI] [PubMed] [Google Scholar]
- 28. Hatahet Z, Purmal AA, Wallace SS. 1994. Oxidative DNA lesions as blocks to in vitro transcription by phage T7 RNA polymerase. Ann. N. Y. Acad. Sci. 726:346–348 [DOI] [PubMed] [Google Scholar]
- 29. Ide H, Kow YW, Wallace SS. 1985. Thymine glycols and urea residues in M13 DNA constitute replicative blocks in vitro. Nucleic Acids Res. 13:8035–8052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Keijzer W, et al. 1979. A seventh complementation group in excision-deficient xeroderma pigmentosum. Mutat. Res. 62:183–190 [DOI] [PubMed] [Google Scholar]
- 31. Kraemer KH, et al. 1975. Five complementation groups in xeroderma pigmentosum. Mutat. Res. 33:327–340 [DOI] [PubMed] [Google Scholar]
- 32. Kyng KJ, et al. 2003. The transcriptional response after oxidative stress is defective in Cockayne syndrome group B cells. Oncogene 22:1135–1149 [DOI] [PubMed] [Google Scholar]
- 33. Leach DM, Rainbow AJ. 2011. Early host cell reactivation of an oxidatively damaged adenovirus-encoded reporter gene requires the Cockayne syndrome proteins CSA and CSB. Mutagenesis 26:315–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lehmann AR. 1982. Three complementation groups in Cockayne syndrome. Mutat. Res. 106:347–356 [DOI] [PubMed] [Google Scholar]
- 35. Lehmann AR. 2003. DNA repair-deficient diseases, xeroderma pigmentosum, Cockayne syndrome and trichothiodystrophy. Biochimie 85:1101–1111 [DOI] [PubMed] [Google Scholar]
- 36. Lehmann AR, Kirk-Bell S, Mayne L. 1979. Abnormal kinetics of DNA synthesis in ultraviolet light-irradiated cells from patients with Cockayne's syndrome. Cancer Res. 39:4237–4241 [PubMed] [Google Scholar]
- 37. Le Page F, et al. 2000. Transcription-coupled repair of 8-oxoguanine: requirement for XPG, TFIIH, and CSB and implications for Cockayne syndrome. Cell 101:159–171 [DOI] [PubMed] [Google Scholar]
- 38. Li BH, Bockrath R. 1995. Benefit of transcription-coupled nucleotide excision repair for gene expression in u. v.-damaged Escherichia coli. Mol. Microbiol. 18:615–622 [DOI] [PubMed] [Google Scholar]
- 39. Mahdi AA, Briggs GS, Sharples GJ, Wen Q, Lloyd RG. 2003. A model for dsDNA translocation revealed by a structural motif common to RecG and Mfd proteins. EMBO J. 22:724–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Marnett LJ. 1999. Lipid peroxidation-DNA damage by malondialdehyde. Mutat. Res. 424:83–95 [DOI] [PubMed] [Google Scholar]
- 41. Mayne LV, Lehmann AR. 1982. Failure of RNA synthesis to recover after UV irradiation: an early defect in cells from individuals with Cockayne's syndrome and xeroderma pigmentosum. Cancer Res. 42:1473–1478 [PubMed] [Google Scholar]
- 42. Mellon I, Bohr VA, Smith CA, Hanawalt PC. 1986. Preferential DNA repair of an active gene in human cells. Proc. Natl. Acad. Sci. U. S. A. 83:8878–8882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mellon I, Champe GN. 1996. Products of DNA mismatch repair genes mutS and mutL are required for transcription-coupled nucleotide-excision repair of the lactose operon in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 93:1292–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mellon I, Hanawalt PC. 1989. Induction of the Escherichia coli lactose operon selectively increases repair of its transcribed DNA strand. Nature 342:95–98 [DOI] [PubMed] [Google Scholar]
- 45. Murphy KC, Campellone KG, Poteete AR. 2000. PCR-mediated gene replacement in Escherichia coli. Gene 246:321–330 [DOI] [PubMed] [Google Scholar]
- 46. Nardo T, et al. 2009. A UV-sensitive syndrome patient with a specific CSA mutation reveals separable roles for CSA in response to UV and oxidative DNA damage. Proc. Natl. Acad. Sci. U. S. A. 106:6209–6214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Osterod M, et al. 2002. A global DNA repair mechanism involving the Cockayne syndrome B (CSB) gene product can prevent the in vivo accumulation of endogenous oxidative DNA base damage. Oncogene 21:8232–8239 [DOI] [PubMed] [Google Scholar]
- 48. Proietti-De-Santis L, Drane P, Egly JM. 2006. Cockayne syndrome B protein regulates the transcriptional program after UV irradiation. EMBO J. 25:1915–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rouet P, Essigmann JM. 1985. Possible role for thymine glycol in the selective inhibition of DNA synthesis on oxidized DNA templates. Cancer Res. 45:6113–6118 [PubMed] [Google Scholar]
- 50. Saito Y, et al. 1997. Characterization of endonuclease III (nth) and endonuclease VIII (nei) mutants of Escherichia coli K-12. J. Bacteriol. 179:3783–3785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schalow BJ, Courcelle CT, Courcelle J. 2011. Escherichia coli Fpg glycosylase is nonrendundant and required for the rapid global repair of oxidized purine and pyrimidine damage in vivo. J. Mol. Biol. 410:183–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Selby CP, Sancar A. 1994. Mechanisms of transcription-repair coupling and mutation frequency decline. Microbiol. Rev. 58:317–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Selby CP, Witkin EM, Sancar A. 1991. Escherichia coli mfd mutant deficient in “mutation frequency decline” lacks strand-specific repair: in vitro complementation with purified coupling factor. Proc. Natl. Acad. Sci. U. S. A. 88:11574–11578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Selby CP, Sancar A. 1993. Molecular mechanism of transcription-repair coupling. Science 260:53–58 [DOI] [PubMed] [Google Scholar]
- 55. Setlow RB, Regan JD, German J, Carrier WL. 1969. Evidence that xeroderma pigmentosum cells do not perform the first step in the repair of ultraviolet damage to their DNA. Proc. Natl. Acad. Sci. U. S. A. 64:1035–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Smith AJ, Savery NJ. 2008. Effects of the bacterial transcription-repair coupling factor during transcription of DNA containing non-bulky lesions. DNA Repair (Amst) 7:1670–1679 [DOI] [PubMed] [Google Scholar]
- 57. Spivak G, Hanawalt PC. 2006. Host cell reactivation of plasmids containing oxidative DNA lesions is defective in Cockayne syndrome but normal in UV-sensitive syndrome fibroblasts. DNA Repair (Amst) 5:13–22 [DOI] [PubMed] [Google Scholar]
- 58. Spivak G, et al. 2002. Ultraviolet-sensitive syndrome cells are defective in transcription-coupled repair of cyclobutane pyrimidine dimers. DNA Repair (Amst) 1:629–643 [DOI] [PubMed] [Google Scholar]
- 59. Storm PK, Hoekstra WP, de Haan PG, Verhoef C. 1971. Genetic recombination in Escherichia coli. IV. Isolation and characterization of recombination-deficiency mutants of Escherichia coli K-12. Mutat. Res. 13:9–17 [DOI] [PubMed] [Google Scholar]
- 60. Tanaka K, Kawai K, Kumahara Y, Ikenaga M, Okada Y. 1981. Genetic complementation groups in Cockayne syndrome. Somatic Cell Genet. 7:445–455 [DOI] [PubMed] [Google Scholar]
- 61. Tang JY, Hwang BJ, Ford JM, Hanawalt PC, Chu G. 2000. Xeroderma pigmentosum p48 gene enhances global genomic repair and suppresses UV-induced mutagenesis. Mol. Cell 5:737–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tornaletti S, Maeda LS, Kolodner RD, Hanawalt PC. 2004. Effect of 8-oxoguanine on transcription elongation by T7 RNA polymerase and mammalian RNA polymerase II. DNA Repair (Amst) 3:483–494 [DOI] [PubMed] [Google Scholar]
- 63. Tornaletti S, Maeda LS, Lloyd DR, Reines D, Hanawalt PC. 2001. Effect of thymine glycol on transcription elongation by T7 RNA polymerase and mammalian RNA polymerase II. J. Biol. Chem. 276:45367–45371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Troelstra C, et al. 1992. ERCC6, a member of a subfamily of putative helicases, is involved in Cockayne's syndrome and preferential repair of active genes. Cell 71:939–953 [DOI] [PubMed] [Google Scholar]
- 65. Tuo J, Jaruga P, Rodriguez H, Bohr VA, Dizdaroglu M. 2003. Primary fibroblasts of Cockayne syndrome patients are defective in cellular repair of 8-hydroxyguanine and 8-hydroxyadenine resulting from oxidative stress. FASEB J. 17:668–674 [DOI] [PubMed] [Google Scholar]
- 66. Tuo J, et al. 2001. The Cockayne syndrome group B gene product is involved in general genome base excision repair of 8-hydroxyguanine in DNA. J. Biol. Chem. 276:45772–45779 [DOI] [PubMed] [Google Scholar]
- 67. van der Horst GT, et al. 1997. Defective transcription-coupled repair in Cockayne syndrome B mice is associated with skin cancer predisposition. Cell 89:425–435 [DOI] [PubMed] [Google Scholar]
- 68. van Hoffen A, Balajee AS, van Zeeland AA, Mullenders LH. 2003. Nucleotide excision repair and its interplay with transcription. Toxicology 193:79–90 [DOI] [PubMed] [Google Scholar]
- 69. van Hoffen A, Venema J, Meschini R, van Zeeland AA, Mullenders LH. 1995. Transcription-coupled repair removes both cyclobutane pyrimidine dimers and 6-4 photoproducts with equal efficiency and in a sequential way from transcribed DNA in xeroderma pigmentosum group C fibroblasts. EMBO J. 14:360–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Venema J, Mullenders LH, Natarajan AT, van Zeeland AA, Mayne LV. 1990. The genetic defect in Cockayne syndrome is associated with a defect in repair of UV-induced DNA damage in transcriptionally active DNA. Proc. Natl. Acad. Sci. U. S. A. 87:4707–4711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Venema J, et al. 1991. Xeroderma pigmentosum complementation group C cells remove pyrimidine dimers selectively from the transcribed strand of active genes. Mol. Cell. Biol. 11:4128–4134 [DOI] [PMC free article] [PubMed] [Google Scholar]