Abstract
Bacterial flagellar filaments grow at their distal ends, from flagellin that travels through a central channel ∼2 nm in diameter. The flagellin is extruded from the cytoplasm by a pump powered by a proton motive force (PMF). We measured filament growth in cells near the mid-exponential-phase with flagellin bearing a specific cysteine-for-serine substitution, allowing filaments to be labeled with sulfhydryl-specific fluorescent dyes. We labeled filaments first with a green maleimide dye and then, following an additional period of growth, with a red maleimide dye. The contour lengths of the green and red segments were measured. The average lengths of red segments (∼2.3 μm) were the same regardless of the lengths of the green segments from which they grew (ranging from less than 1 to more than 9 μm in length). Thus, flagellar filaments do not grow at a rate that decreases exponentially with length, as formerly supposed. If flagellar filaments were broken by viscous shear, the broken filaments continued to grow. Identical results were obtained whether flagellin was expressed from fliC on the chromosome under the control of its native promoter or on a plasmid under the control of the arabinose promoter.
INTRODUCTION
A bacterial flagellum has three parts: a basal body embedded in the cell wall (a rotary motor) and two external structures, a short proximal hook (a universal joint) and a long helical filament (a propeller, polymerized from the protein flagellin). A flagellum is assembled from the inside out, beginning with the MS ring in the cytoplasmic membrane and the C ring in the cytoplasm. A rod connects the MS ring to the hook, and hook-associated proteins connect the hook to the filament (12). The axial components are exported by a type III secretion system, with each new component added at the end of the growing structure (3, 13). This export is powered by a proton motive force (PMF) (14, 15). Two of the hook-associated proteins couple the hook and the filament, allowing the hook to flex and the filament to rotate rigidly (4), while a third protein caps the distal end of the filament, preventing loss of flagellin and orchestrating its polymerization (7, 20). The hook-associated proteins are exported continuously, appearing in the culture medium, even of wild-type strains, suggesting that caps lost from broken filaments can be replaced (6).
In seminal work summarized in 1974, Iino (8) used electron microscopy to determine the numbers and lengths of flagellar filaments on cells in a culture of Salmonella growing in the exponential phase. He concluded that the rate of elongation decreases exponentially with length. Support for this idea was obtained in vivo by dark-field microscopy by Aizawa and Kubori (1), but the data were limited, and only long filaments could be seen. Having shown that flagellin moves through the filament rather than via the external medium, Iino assumed that transport efficiency decreases with filament length, like the flow of liquid through a pipe driven by a constant pressure head. This idea has been reinforced by recent theoretical work (17).
Several years ago, we found that flagellar filaments of Gram-negative bacteria could be labeled with fluorescent dyes in a manner that did not interfere with their motility (18). The original work was done with amino-specific dyes (succinimidyl esters), which also labeled outer cell membranes. A milder and more specific procedure was developed by adding cysteine to flagellin and using sulfhydryl-specific (maleimide) dyes (19). These methods enabled us to visualize flagellar filaments all the way to the cell surface. By using dyes of different colors and measuring the lengths of segments of different colors on individual filaments, we now have monitored filament growth. We have found, to our surprise, that Escherichia coli filaments grow in a manner that is independent of their length, not at a rate that is an exponentially decreasing function of length, as commonly supposed (1, 8, 17). We also have confirmed that broken filaments continue to grow, a proposition that has remained in doubt (16).
MATERIALS AND METHODS
Strains, growth, and labeling.
E. coli strain AW405 (2) is wild type for chemotaxis and swims vigorously. Strain HCB1737 is isogenic with AW405, except for a single cysteine substitution in fliC (S219C). The wild-type fliC allele of strain AW405 was replaced by the mutant allele using the λ-Red system, described previously (22). This strain also swims vigorously. Strain HCB1668 is a Tn5 fliC null derivative of AW405 in which fliC(S353C) is expressed on the plasmid pBAD33 under the control of the arabinose promoter, maintained by addition to the growth medium of kanamycin (50 μg/ml) and chloramphenicol (34 μg/ml). This strain swims and also swarms (19) vigorously. Details of the serine-to-cysteine replacements are given under “Strain Construction” in the supplemental material. Strains HCB1737 and HCB1668 were cultured from single-colony isolates from frozen stocks (−75°C), placed in 10 ml LB broth (10 g Bacto tryptone, 5 g yeast extract, and 5 g NaCl per liter) in 125-ml Erlenmeyer flasks, and grown to saturation at 30°C with gyration at 150 rpm. Dilutions (1/100) of the LB cultures were made in 10 ml of T broth (10 g Bacto tryptone and 5 g NaCl per liter) and grown in 125-ml Erlenmeyer flasks at 30°C with aeration by gyration at 150 rpm for 4 h to a cell density of ∼4.1 × 108 cells/ml. For HCB1668, arabinose at a 0.5% final concentration was added to the T broth. Cultures were checked for motility using a phase-contrast microscope (Nikon Optiphot), and washed with 10-ml aliquots of motility buffer (MB) (0.01 M potassium phosphate [pH 7.0], 10−4 M EDTA, 0.067 M NaCl) containing 0.0001% Tween 20 (MBT) by 12 min of centrifugation at 1,400 × g followed by gentle resuspension, 4 times, the manipulations spanning a period of 68 min. Then the cells were labeled with Alexa Fluor 488 maleimide dye, as described in reference 19, for ∼90 min rather than 60 min with double the amount of dye, so that even very short filaments could easily be seen by fluorescence. Once labeled, 2 ml of MBT was added to the cells, and the suspension was divided into two 1-ml aliquots in separate centrifuge tubes. One aliquot was sheared by passing it 5 times in and out of a 3-ml syringe equipped with a 22-gauge needle. The extent of shearing was checked by microscopy. A small sample (diluted 10-fold) was imaged by fluorescence (loose filaments were seen stuck to the coverslips), by phase contrast (cell bodies appeared dark), and by combined illumination (filament stubs could be seen on cell bodies). After shearing, the two aliquots (unsheared and sheared) were treated in parallel. Cell aliquots were brought to 10-ml volumes with MBT, washed by centrifugation twice (as described above) and then centrifuged a final time, and resuspended in 5 ml of T broth, the manipulations spanning a period of 51 min. The entire 5 ml of T broth was used as an inoculant for continued culturing in 250-ml Erlenmeyer flasks containing 20 ml of T broth (with antibiotics and arabinose, when required). The culture flasks were stored overnight (∼14 h) at 7°C and then incubated at 30°C with aeration by gyration at 150 rpm for 3 h to a cell density of ∼3.1 × 108 cells/ml. After growth, 10 ml of each culture was washed into MBT by 12 min of centrifugation at 1,400 × g followed by gentle resuspension, 4 times (as described above), and then labeled with Alexa Fluor 546 maleimide dye, using the procedure for the 488-nm dye (as described above). Excess dye was removed by repeated centrifugation and resuspension in MBT (as described above). The final suspension of the pellets was in 2.5 ml of motility buffer (MB), without Tween 20.
Growth curves.
The growth and labeling procedure described above was repeated in precisely the same way with more cells and flasks, and 1.5-ml samples were withdrawn from the different flasks at different times for measurements of optical density at 610 nm (OD610) and construction of growth curves. The conversion from OD610 to cells/ml was based upon a calibration of the spectrometer (Perkin Elmer Lambda EZ142) by serial dilution and plating.
Microscopy.
Microscope slides and coverslips were used out of the box. Tunnel slides were constructed using Scotch brand double-sided sticky tape, by placing two layers of tape about 7 mm apart across the width of the microscope slide and placing a 22- by 22-mm no. 1 coverslip on top of the tape. Poly-l-lysine (0.01%) (Sigma P4707) was added to the tunnels, which were allowed to sit for ∼5 min in a humidity chamber and then rinsed with ∼0.5 ml MB. The cell suspensions, described in the previous section, were diluted with an equal volume of MB and then added to fill the tunnels. The tunnel slides were inverted and placed in the humidity chamber for ∼5 min and then rinsed with MB. The phase-contrast microscope was used to monitor the density of cells stuck to the coverslips, to determine if the final cell density was suitable for fluorescence imaging of the flagellar filaments. If too many cells were stuck, a new preparation was made using a more dilute cell suspension. Glutaraldehyde (0.25% in MB) was added to the tunnel slides for 2 to 3 min to stop flagellar rotation and to cross-link the filaments to the surface of the coverslip. Tunnel-slide preparations were rinsed with MB and then stored in the humidity chamber prior to microscopy. Fluorescence imaging was performed with a Nikon inverted microscope (Diaphot 200) equipped with a 100-W mercury arc, with a dual filter cube (fluorescein isothiocyanate [FITC] and Texas Red; Chroma Technologies no. 51006), a 100× oil immersion objective, a 2.5× relay lens, and a Nikon D-70 camera. Images were captured using Nikon Image Capture software running on a PC laptop. Exposure times were sample dependent and usually between 2 and 10 s.
Image analysis.
Red-green-blue (RGB)-colored images (Fig. 1A) were transferred to a Macintosh computer and analyzed using ImageJ software. Each image file was 3,872 by 2,592 pixels at 300 pixels/inch. The image magnification was 40 pixels/μm. For analysis, each RGB image was split via the standard ImageJ command “Split Channels” to recover the green images in black and white, with green appearing white and other colors black (Fig. 1B). Green filament lengths, indicating old growth, were identified on these black and white images. Red filament lengths, indicating new growth, were identified on the original RGB images (Fig. 1C). The ImageJ free-form line tool was used to trace filament contours. Contour lengths were smoothed using the ImageJ cubic spline fit function, drawn on the image, recorded as a length measurement on a spreadsheet, and then analyzed and graphed using KaleidaGraph software. Rough estimates of errors for measurements of contour lengths were ∼5% when filaments were short and ∼2.5% when filaments were long. If a green filament had no red filament section, then a zero red segment length was recorded. Every in-focus image was analyzed, and every green segment in each image was measured until a total of 1,250 filaments had been logged.
Fig 1.
Examples of flagellar filaments of strain HCB1737 imaged by fluorescence microscopy. Cells were labeled with Alexa Fluor 488 dye (green), cultured for an additional period of time, and then labeled with Alexa Fluor 546 dye (red). (A) Full-color image. (B) An image of the same scene in which the green pixels appear white and other colors appear black, a technique used to more clearly distinguish green from red. The green segments were traced and are shown in white over white. (C) An image of the same scene in which the red segments of green and red filaments were traced and are shown in white over red.
RESULTS
Filament growth by protocol A.
To learn how growth of flagellar filaments depends upon initial length, we used two derivatives of the wild-type strain AW405 with flagellin bearing a serine-to-cysteine substitution. Expression was either from the chromosome under the control of the native promoter or from a plasmid under the control of the arabinose promoter. The results for the two strains were essentially identical. The experimental protocol is shown in Fig. 2A. Cells were cultured to the late exponential phase, checked for maximal motility, and then labeled to saturation with a sulfhydryl-reactive green fluorescent dye. The flagellar filaments of these cells were all green, creating a population of filaments that could be traced through subsequent generations. A second growth period was initiated. After 3 h of growth, the cells were labeled a second time with a red fluorescent dye, so new growth was red. A total of 99.85% of the cells of strain HCB1737 showed new (red) flagellar growth, indicating that the green labeling procedure did not kill cells. Three types of colored filaments were seen by fluorescence (Fig. 1A), as indicated in the cartoon (Fig. 2): type a, filaments that were both green and red; type b, filaments that were entirely green; and type c, filaments that were entirely red. About 14% of filaments bearing green dye were entirely green, showing no evidence of growth during the second culture period, although red filaments grew elsewhere on the same cells. The other 86% of the filaments bearing green dye were green at their proximal ends and red at their distal ends, indicating new filament growth during the second culture period. Many filaments were entirely red, indicating that their growth was initiated after cells had been labeled green. These filaments were not considered in the analysis, because the time for their initiation could not be determined. However, lengths of red-only filaments were measured on a subset of 200 unsheared cells of strain HCB1737, yielding a mean of 2.52 μm with a standard error of the mean (SEM) of 0.09, values not significantly different from those from red growth on green filaments.
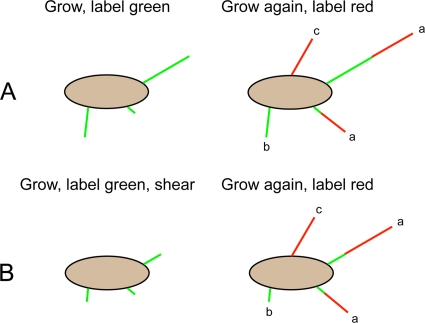
Fig 2.
Experimental protocol. Time flows from left to right. (Top row) Protocol A. Cells were grown to the late exponential phase, labeled to saturation with a green fluorescent dye, grown again to the late exponential phase, and labeled with a red fluorescent dye. Three kinds of filaments were observed: type a, filaments with green proximal ends and red distal ends (common); type b, filaments that were entirely green (rare); type c, filaments that were entirely red (common). Initiation of growth of filaments of the latter type occurred after cells had been labeled green. The filaments in the experiments were normal and helical; for simplicity, they are shown here as straight. (Bottom row) Protocol B. Protocol B is the same as protocol A, except an aliquot of cells that had been labeled green was subjected to viscous shear; two of the filaments shown were shortened, and one was not. The parent population for the cells that were sheared was the population studied in protocol A, so a record was made of cells both before and after shearing.
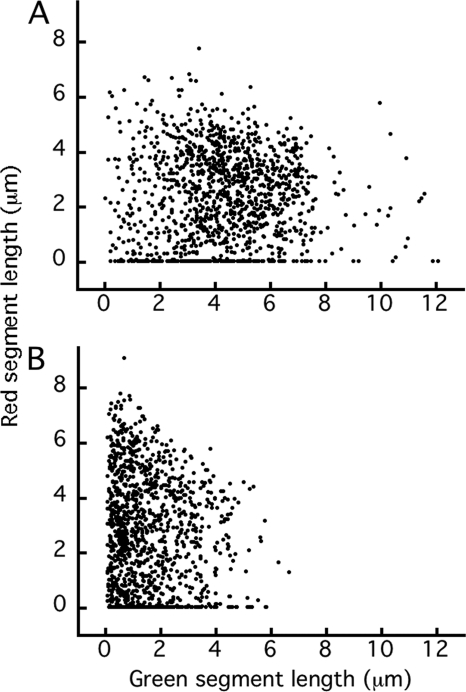
The contour lengths of 1,250 filaments were measured for each strain, with two measurements for each filament: the length of the green segment and the length of the corresponding red segment. The complete data set for strain HCB1737 is shown in Fig. 3A. This is a scatter plot in which the lengths of red segments are plotted versus the lengths of green segments from which they grew. The cloud of points is broad and roughly rectangular, centered around 4.5 μm in the green dimension and around 2.5 μm in the red dimension, indicating a varied range of growth for each filament length. The cloud does not skew downward to the right as expected were filament growth to decrease with length. There are several red points of value zero spread along the green axis (the 14% of green filaments that did not grow further).
Fig 3.
Plots of the lengths of red segments versus the lengths of the green segments from which they grew, for a single culture of strain HCB1737. (A) Filaments of unsheared cells. (B) Filaments of sheared cells. The corresponding plots for strain HCB1668 are shown in Fig. S1 in the supplemental material.
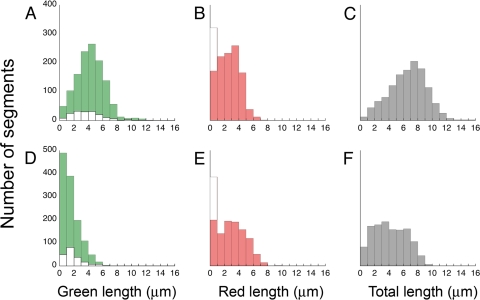
More could be learned about these data by examining distributions binned according to segment lengths, as shown in Fig. 4 (top row). The distribution of green segments (Fig. 4A) was roughly normal, with a mean of ∼4.3 μm, but included a few filaments as long as 13 μm. Following the second period of growth, the distribution of red segments (Fig. 4B) had a mean of ∼2.3 μm. The distribution of total lengths (Fig. 4C) was nearly normal, with a mean of 6.6 μm. These results are summarized in the first column of Table 1.
Fig 4.
Distributions of segment lengths of filaments on cells of strain HCB1737, binned at 1-μm intervals. Green segments are shown in green, red segments are shown in red, and total lengths (green and red) are shown in gray. The distribution of the subset of green segments that failed to produce red extensions is shown by the white bars in panels A and D, while the total numbers of such green segments, which equals the numbers of red segments of zero length, are shown by the white bars in panels B and E. The top row (A to C) represents cells that were not sheared, and the bottom row (D to F) represents cells that were sheared. The corresponding plots for strain HCB1668 are shown in Fig. S2 in the supplemental material.
Table 1.
Lengths of colored segments in strains HCB1737 and HCB1668
| Color | Mean ± SE segment length (μm) in: |
|||
|---|---|---|---|---|
| HCB1737a |
HCB1668b |
|||
| Not sheared (n = 1,250) | Sheared (n = 1,250) | Not sheared (n = 1,250) | Sheared (n = 1,250) | |
| Green | 4.29 ± 0.05 | 1.60 ± 0.03 | 4.21 ± 0.06 | 1.85 ± 0.04 |
| Red | 2.33 ± 0.05 | 2.58 ± 0.06 | 2.09 ± 0.05 | 2.43 ± 0.05 |
| Green + red | 6.63 ± 0.07 | 4.18 ± 0.06 | 6.31 ± 0.08 | 4.27 ± 0.06 |
After the 2nd culture period, the not-sheared cell population had on average 4.76 ± 0.17 filaments/cell (n = 100) and the sheared cell population had on average 4.23 ± 0.18 filaments/cell (n = 100).
After the 2nd culture period, the not-sheared cell population had on average 4.87 ± 0.20 filaments/cell (n = 100) and the sheared cell population had on average 5.54 ± 0.24 filaments/cell (n = 100).
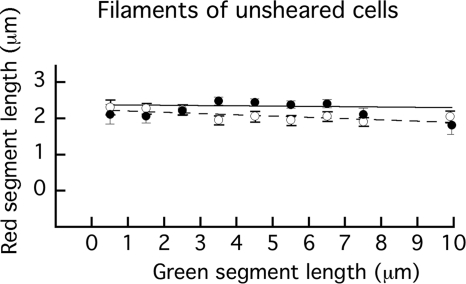
As expected from Fig. 3A, based on distributions of red segments binned in successive 1-μm intervals and sorted according to the lengths of the green segments from which they grew (data not shown), they were all fairly broad. Remarkably, they all had similar means, as plotted in Fig. 5. Means ranged between 1.8 μm and 2.5 μm, almost all within 3 standard errors of 2.16 μm, the mean of all points, and most within 1 standard error. This implies that new growth is much the same, regardless of the length of old growth, i.e., that the growth of flagellar filaments of E. coli does not depend upon filament length. This was true whether flagellin was expressed from the chromosome under the control of its native promoter (strain HCB1737) or from a plasmid under the control of the arabinose promoter (strain HCB1668).
Fig 5.
Plots showing the mean values and standard errors of the lengths of red segments as a function of the lengths of the green segments from which they grew (plotted at bin mid-values). Closed symbols (●) and the solid line represent filaments of unsheared cells of strain HCB1737. From left to right, n = 49, 103, 158, 240, 266, 207, 138, 56, and 33. The weighted line fit has a reduced χ2 value of 1.77. Open symbols (○) and the dashed line represent filaments of unsheared cells of strain HCB1668. From left to right, n = 72, 170, 178, 205, 155, 162, 142, 114, and 52. The weighted line fit has a reduced χ2 value of 0.63.
Growth curves.
As noted in Materials and Methods, the two culture periods were separated by washing and labeling steps that required cells to remain at room temperature in dilute phosphate buffers for about 4 h, followed by storage in growth medium at 7°C for about 14 h. The motility was checked along the way and found to be maximal, but it was important to know whether the two growth periods were equivalent. So we repeated the procedure with strain HCB1737 following all the same steps and measured the optical densities of the cultures during both growth periods. The results are shown in Fig. 6. During both periods, the cells grew exponentially, with a doubling time of ∼56 min up to the time point of 180 min. At later times, the rate of growth was smaller, as the cells entered late exponential phase. Although these results imply that labeling did not kill cells, we checked this directly by growing cells, dividing the culture into two parts, labeling one, and measuring growth curves for both without delay, using similar inocula. The two growth curves were identical (data not shown), indicating that labeling did not affect viability.
Fig 6.
Growth curves for the two culture periods. The logarithm of cell density (cells/ml) is plotted as a function of culture time (min). Open symbols (○) and the dashed line represent the first growth period; closed symbols (■) and the solid line represent the second growth period. Cells were harvested at log 8.6 for the first growth period (4.1 × 108 cells/ml) and at log 8.5 for the second growth period (3.1 × 108 cells/ml). During the exponential phase (straight-line fits), the doubling time was ∼56 min.
Filament growth by protocol B.
To learn whether broken filaments continue to grow, half of the population of cells that had been labeled green were subjected to viscous shear and then were treated in the same way as the cells that were not sheared, as indicated in Fig. 2B.
A scatter plot of the lengths of red segments versus the lengths of the green segments from which they grew is shown in Fig. 3B. The cloud of points is no longer rectangular. It has shifted to the left: there are many more points near the left boundary, and the right boundary no longer extends beyond 7 μm. So it is evident that filaments were broken by shear. The upper edge of the cloud now slants downward to the right.
Again, more could be learned about such data by examining distributions binned according to segment lengths, as shown in Fig. 4 (bottom row). The distribution of green segments (Fig. 4D) was exponential, with the shortest lengths the most probable. The mean for this distribution is 1.6 μm, and the distribution only extends out to ∼7 μm. So, as asserted above, long filaments were broken. Following the second period of growth, the distribution of red segments (Fig. 4E) had a mean of 2.6 μm, including segments as long as 8 μm. So the new growth was roughly the same whether the filaments had been sheared or not. The distribution of total lengths (Fig. 4F) was more nearly normal, with a mean of 4.2 μm. These results are summarized in the second column of Table 1.
Evidence that filaments broken by shear continue to grow is contained in the length distributions of Fig. 4A, D, and F. The aliquot of cells that were sheared, with the green-segment length distribution shown in panel D, was taken from the suspension of cells that were not sheared, with the green segment length distribution shown in panel A. A comparison of these two distributions enables us to distinguish most of the filaments that were broken by shear from those that were too short to be broken by shear. There are only 46 filaments with a length of <1 μm in the initial distribution (Fig. 4A), but there were 483 in the final distribution (Fig. 4D). Therefore, approximately 437 of these filaments must originally have been longer than 1 μm (i.e., must have been broken). Following the second period of growth, only 82 filaments showed a total length of <1 μm (Fig. 4F). Hence, the majority of the 437 broken filaments must have grown. A formal extension of this analysis that includes data for filaments of length >1 μm is given in the supplemental material.
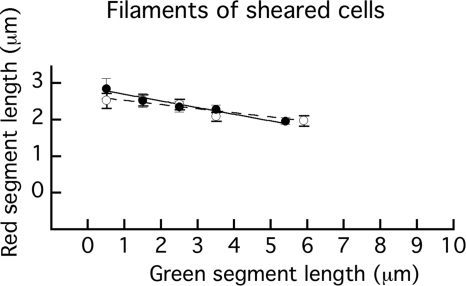
As expected from Fig. 3B, distributions of red segments binned in successive 1-μm intervals and sorted according to the lengths of the green segments from which they grew (data not shown) were all fairly broad, but now their means decreased somewhat with green segment length, as shown in Fig. 7. The average growth decreased approximately linearly from slightly less than 3 μm to slightly less than 2 μm over the entire range of initial lengths of less than 1 μm to 7 μm. This was true for both strains HCB1737 and HCB1668.
Fig 7.
Plots showing the mean values and standard errors of the lengths of red segments as a function of the lengths of the green segments from which they grew (plotted at bin midvalues). Closed symbols (●) and the solid line represent filaments of sheared cells of strain HCB1737. From left to right, n = 488, 389, 198, 105, and 70. The weighted line fit has a reduced χ2 value of 0.52. Open symbols (○) and the dashed line represent filaments of sheared cells of strain HCB1668. From left to right, n = 407, 370, 243, 132, and 98. The weighted line fit has a reduced χ2 value of 0.75.
DISCUSSION
We studied growth of flagellar filaments by a method that allowed us to distinguish new growth (segments labeled red) from old growth (segments labeled green) in individual filaments. We concluded that filament growth for samples that had not been sheared did not depend upon filament length (Fig. 5). For samples that had been sheared, shorter filaments grew somewhat longer extensions (Fig. 7). A comparison of length distributions between a sample of a cell culture that had not been sheared (Fig. 4A) with one that had been sheared (Fig. 4D) and then grown further (Fig. 4F) showed that broken filaments continue to grow.
An earlier study of this kind was made by Iino (9), who grew cells at 37°C on minimal medium with glucose or on minimal medium with glucose followed by saline and then on minimal medium with glucose plus a complete mixture of l-amino acids, in which phenylalanine was replaced by p-fluorophenylalanine (FPA). This analogue had been shown by Kerridge (11) to promote growth of curly filaments. The number of flagella per cell decreased from ∼4.4 to ∼2.5, but newly grown curly waves were shorter in filaments with more normal waves and reached zero at about 5 normal waves (∼12 μm). However, it was not possible to distinguish normal waves newly grown on FPA from those present before treatment. In this work, Iino also concluded that broken filaments continue to grow. In a later study conducted without FPA, Iino (8) concluded that flagella grew at a rate that decreased exponentially with length. His analysis was based upon the assumption that the order of relative lengths of filaments remained fixed at successive time points during exponential growth; for example, that the longest filaments at one time point were derived from the longest filaments at an earlier time point. This is not true for our data. Pick a green length from the abscissa of Fig. 3 and note the wide distribution of red lengths denoting subsequent growth. These distributions look much the same regardless of green length. New growth is not correlated with old growth. We would expect results obtained with Salmonella to be similar to those obtained with E. coli, but our results are different.
If Iino's theory (8) is used to compute the times required for filaments to grow to the lengths shown on the abscissa of Fig. 5 (using the data Iino gives for a generation time of 56 min) and then the filaments are allowed to grow for an additional 3 h, we find the additional lengths to be much shorter at the 7.5-μm point than at the 0.5-μm point—only 39% as long. The plot for this (steep) decline is nearly linear (slightly concave upwards), as in Fig. 8 in Iino's study. Our data for unsheared filaments (Fig. 5) do not show such decline.
There is a decline, albeit slight, for sheared filaments (Fig. 7). It is linear but not steep, and appears with both strains. We do not understand the reason for this decline. Presumably, it has to do with replacement of the cap protein, FliD. However, if FliD moves slowly through sheared filaments, why does it not also move slowly through unsheared ones? Might the mode of operation of the pump change when the filament lacks a cap?
In vitro, filaments grow at a constant rate, with pauses thought to occur when defective flagellin monomers are transiently incorporated (10). In between pauses, the average rate of extension is proportional to the concentration of free flagellin, with a distribution of rates from filament to filament that is fairly broad. In our experiments, filaments continue to grow in length and number, suggesting that the supply of flagellin is adequate and sustained, but the distributions of rates from filament to filament also are broad. The results are the same whether the flagellin is expressed from the chromosome under the control of its native promoter (strain HCB1737) or from a plasmid under the control of the arabinose promoter (strain HCB1668). Note that we did not measure rates, per se, as one might do with a time-lapse movie of growing filaments, but rather the extension produced over a specified time span. Thus, we do not know how variable rates of growth might have been or whether there were pauses.
In vivo, flagellin monomers move along the axis of a flagellar filament through a pore only 2 nm in diameter (21), crystallizing out under the FliD cap at the distal end. To pass through this pore, the flagellin must be unfolded. The N- and C-terminal ends of the molecule are α-helical, so if we assume that the molecule goes through the pore as an extended α-helical chain, then it is ∼74 nm long! This is 14 times longer than the distance between subunits comprising flagellar protofilaments (5). Since there are 11 protofilaments, it takes 156 subunits (i.e., 11 × 14) to lengthen the filament by enough for the pore to contain one more flagellin α-helix. So if one pumps in 157 subunits, 156 will crystallize and 1 will remain in the pore. Because growth does not vary with length, on average, our results imply that this PMF-driven export pump (14, 15) is a constant-rate device, not one that runs at constant pressure. Under the conditions of our experiment, it extrudes one flagellin molecule every 2 s. Variations from flagellum to flagellum could arise from variations in PMF, in pump structure, or in rates at which flagellin monomers are delivered to the pump.
If pumps keep pumping, what limits the lengths of flagellar filaments? Long filaments probably break.
Supplementary Material
ACKNOWLEDGMENTS
We thank Yilin Wu and Gabriel Basarab Hosu for helpful discussions and Vedhavalli Nathan for expert assistance with strain construction.
This work was supported by the Rowland Institute at Harvard and grants AI016478 and AI066540 from the National Institutes of Health.
Footnotes
Published ahead of print 23 March 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Aizawa S-I, Kubori T. 1998. Bacterial flagellation and cell division. Genes Cells 3:625–634 [DOI] [PubMed] [Google Scholar]
- 2. Armstrong JB, Adler J, Dahl MM. 1967. Nonchemotactic mutants of Escherichia coli. J. Bacteriol. 93:390–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chevance FFV, Hughes KT. 2008. Coordinating assembly of a bacterial macromolecular machine. Nat. Rev. Microbiol. 6:455–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fahrner KA, Block SM, Krishnaswamy S, Parkinson JS, Berg HC. 1994. A mutant hook-associated protein (HAP3) facilitates torsionally-induced transformations of the flagellar filament of Escherichia coli. J. Mol. Biol. 238:173–186 [DOI] [PubMed] [Google Scholar]
- 5. Hasegawa K, Yamashita I, Namba K. 1998. Quasi- and nonequivalence in the structure of bacterial flagellar filament. Biophys. J. 74:569–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Homma M, Iino T. 1985. Excretion of unassembled hook-associated proteins by Salmonella typhimurium. J. Bacteriol. 164:1370–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Homma M, Iino T, Kutsukake K, Yamaguchi S. 1986. In vitro reconstitution of flaglellar filaments onto hooks of filamentless mutants of Salmonella typhimurium by addition of hook-associated proteins. Proc. Natl. Acad. Sci. U. S. A. 83:6169–6173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Iino T. 1974. Assembly of Salmonella flagellin in vitro and in vivo. J. Supramol. Struct. 2:372–384 [DOI] [PubMed] [Google Scholar]
- 9. Iino T. 1969. Polarity of flagellar growth in Salmonella. J. Gen. Microbiol. 56:227–239 [DOI] [PubMed] [Google Scholar]
- 10. Ishihara A, Hotani H. 1980. Micro-video study of discontinuous growth of bacterial flagellar filaments in vitro. J. Mol. Biol. 139:265–276 [DOI] [PubMed] [Google Scholar]
- 11. Kerridge D. 1959. The effect of amino acid analogues on the synthesis of bacterial flagella. Biochim. Biophys. Acta 31:579–581 [DOI] [PubMed] [Google Scholar]
- 12. Macnab RM. 2003. How bacteria assemble flagella. Annu. Rev. Microbiol. 57:77–100 [DOI] [PubMed] [Google Scholar]
- 13. Minamino T, Imada K, Namba K. 2008. Mechanisms of type III protein export for bacterial flagellar assembly. Mol. Biosyst. 4:1105–1115 [DOI] [PubMed] [Google Scholar]
- 14. Minamino T, Namba K. 2008. Distinct roles of the FliI ATPase and proton motive force in bacterial flagellar protein export. Nature 451:485–488 [DOI] [PubMed] [Google Scholar]
- 15. Paul K, Erhardt M, Hirano T, Blair DF, Hughes KT. 2008. Energy source of flagella type III secretion. Nature 451:489–492 [DOI] [PubMed] [Google Scholar]
- 16. Rosu V, Hughes KT. 2006. σ28-dependent transcription in Salmonella enterica is independent of flagellar shearing. J. Bacteriol. 188:5196–5203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tanner DE, Ma W, Chen Z, Schulten K. 2011. Theoretical and computational investigation of flagellin translocation and bacterial flagellum growth. Biophys. J. 100:2548–2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Turner L, Ryu WS, Berg HC. 2000. Real-time imaging of fluorescent flagellar filaments. J. Bacteriol. 182:2793–2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Turner L, Zhang R, Darnton NC, Berg HC. 2010. Visualization of flagella during bacterial swarming. J. Bacteriol. 192:3259–3267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yonekura K, et al. 2000. The bacterial flagellar cap as the rotary promoter of flagellin self-assembly. Science 290:2148–2152 [DOI] [PubMed] [Google Scholar]
- 21. Yonekura K, Maki-Yonekura S, Namba K. 2003. Complete atomic model of the bacterial flagellar filament by electron cryomicroscopy. Nature 424:643–650 [DOI] [PubMed] [Google Scholar]
- 22. Yuan J, Berg HC. 2008. Resurrection of the flagellar rotary motor near zero load. Proc. Natl. Acad. Sci. U. S. A. 105:1182–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.