Abstract
Synthesis of the compatible solute glycine betaine confers a considerable degree of osmotic stress tolerance to Bacillus subtilis. This osmoprotectant is produced through the uptake of the precursor choline via the osmotically inducible OpuB and OpuC ABC transporters and a subsequent two-step oxidation process by the GbsB and GbsA enzymes. We characterized a regulatory protein, GbsR, controlling the transcription of both the structural genes for the glycine betaine biosynthetic enzymes (gbsAB) and those for the choline-specific OpuB transporter (opuB) but not of that for the promiscuous OpuC transporter. GbsR acts genetically as a repressor and functions as an intracellular choline sensor. Spectroscopic analysis of the purified GbsR protein showed that it binds the inducer choline with an apparent KD (equilibrium dissociation constant) of approximately 165 μM. Based on the X-ray structure of a protein (Mj223) from Methanococcus jannaschii, a homology model for GbsR was derived. Inspection of this GbsR in silico model revealed a possible ligand-binding pocket for choline resembling those of known choline-binding sites present in solute receptors of microbial ABC transporters, e.g., that of the OpuBC ligand-binding protein of the OpuB ABC transporter. GbsR was not only needed to control gbsAB and opuB expression in response to choline availability but also required to genetically tune down glycine betaine production once cellular adjustment to high osmolarity has been achieved. The GbsR regulatory protein from B. subtilis thus records and integrates cellular and environmental signals for both the onset and the repression of the synthesis of the osmoprotectant glycine betaine.
INTRODUCTION
One of the challenges Bacillus subtilis faces in its soil habitat is frequent fluctuation in osmolarity (9). Desiccation of the soil creates a high-osmolarity surrounding that requires a highly integrated and timely cellular response to avoid cytoplasmic dehydration and a concomitant drop in turgor (62) to physiologically nonsustainable values (11, 63). The B. subtilis cell accomplishes this task by dynamically increasing the osmotic potential of its cytoplasm to promote water retention and reentry in tune with the prevailing osmolarity of the habitat. Initially, the cell takes up K+ ions as an emergency reaction (31, 62), after which it accumulates compatible solutes such as proline (13, 61) and glycine betaine (4, 5, 35–37) for its long-term physiological adjustment to sustained high-osmolarity growth conditions (9, 11, 38).
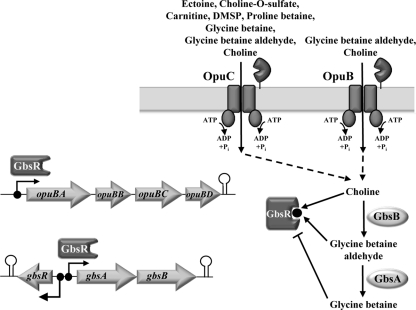
Glycine betaine, a trimethylated derivative of the amino acid glycine, is without doubt the most widely used compatible solute in nature (64). B. subtilis makes extensive use of glycine betaine to promote growth when it is challenged by sustained high-osmolarity conditions. It possesses three high-affinity and osmotically inducible uptake systems for this compound: the two ABC-type transporters OpuA and OpuC and the BCCT-type transporter OpuD (Fig. 1) (35–37). In addition to taking up extracellular glycine betaine, the B. subtilis cell can also synthesize it (4–6), an osmoadaptive process that is the focus of this study.
Fig 1.
Systems for the uptake of compatible solutes by B. subtilis and the osmoadaptive choline-to-glycine betaine biosynthetic pathway. The substrate profiles of the OpuB and OpuC ABC transporters shown were compiled from the literature (9, 30), except those for proline betaine and DMSP, whose import systems are assigned based on unpublished data from our laboratory (T. Hoffmann, A. Bashir, C. Chen, and E. Bremer, unpublished data). The dots and arrows shown within the context of the opuB and gbsAB operons and the gbsR gene represent the experimentally mapped promoters (5, 36). The lollipops indicate predicted factor-independent transcriptional terminators.
Microorganisms can synthesize glycine betaine by one of two different routes: (i) through a stepwise methylation of the amino acid glycine (39, 46, 59) or (ii) through the import and subsequent oxidation of choline using various combinations of enzymes (5, 15, 42, 49, 51). B. subtilis uses this latter route to produce glycine betaine and imports for this purpose the precursor choline into the cell via two osmotically inducible ABC-type transport systems: the aforementioned OpuC transporter and the OpuB system (Fig. 1). The two systems exhibit comparable kinetic parameters for choline uptake, with Km values in the low-μM range and similar Vmax values (36). However, OpuB is highly specific for the uptake of choline, whereas OpuC imports a broad range of osmoprotectants (Fig. 1) (9, 30). Once imported, choline is oxidized to glycine betaine in a two-step enzymatic reaction that involves the type III alcohol dehydrogenase GbsB and the glycine betaine aldehyde dehydrogenase GbsA, with glycine betaine aldehyde as the intermediate (Fig. 1) (5, 6).
It should be noted in this context that choline has no osmoprotective properties per se in B. subtilis and that neither choline nor glycine betaine can be used as a nutrient by this bacterium (4, 5). Hence, both the choline-dependent synthesis and the uptake of preformed glycine betaine serve exclusively for stress protection of the B. subtilis cell, either against high osmolarity (4, 5, 35–37) or against sustained high (52°C)- and low (13°C)-temperature challenges (12, 30, 31).
A previous study from our laboratory has shown that the transcription of the glycine betaine biosynthetic genes (gbsAB) from B. subtilis is upregulated in response to an exogenous supply of choline but is not stimulated per se in cells that are continuously cultivated at high osmolarity (5). However, sustained high osmolarity enhances gbsAB induction by choline (5), indicating that an increase in the intracellular choline pool generated via the osmotically inducible OpuB and OpuC systems (36) is beneficial for the induction of gbsAB expression. These findings suggest (i) that choline, or a metabolic derivative of it, serves as an inducer for gbsAB transcription and (ii) that a regulatory protein that monitors the intracellular pool of the inducer and then sets the level of gbsAB expression is likely to exist.
Here we report the identification of the long-suspected regulatory protein (5) for the osmoadaptive glycine betaine synthesis pathway of B. subtilis, the repressor protein GbsR. We demonstrate that GbsR functions as an intracellular choline sensor controlling the expression of both the structural genes for the substrate-specific choline import system OpuB and those encoding the glycine betaine biosynthetic enzymes GbsAB (Fig. 1). In addition, we found that the GbsR protein not only functions as a regulatory switch for the onset of glycine betaine biosynthesis when choline is present in the environment but also that GbsR is also required to downregulate glycine betaine production once the B. subtilis cell has achieved adjustment to the osmotic conditions prevailing in its ecosystem.
MATERIALS AND METHODS
Chemicals.
Choline, glycine betaine aldehyde, glycine betaine, carnitine, proline, and the chromogenic substrates for the TreA enzyme, para-nitrophenyl-α-d-glucopyranoside (α-PNPG), and the antibiotics chloramphenicol, kanamycin, erythromycin and spectinomycin were purchased from Sigma-Aldrich (Steinheim, Germany). Proline betaine (dimethyl-proline) was purchased from Atkins Chemicals (Chengdue, China); dimethylsulfoniopropionate (DMSP) and choline-O-sulfate were from laboratory stocks. Ectoine was a kind gift from G. Lentzen (Bitop AG, Witten, Germany) and [methyl-14C]choline (55 mCi mmol−1) was purchased from DuPont de Nemour GmbH (Neu-Isenburg, Germany). Avian myeloblastosis virus reverse transcriptase was obtained from Roche Diagnostics (Mannheim, Germany). The resin (Strep-Tactin Sepharose) for the affinity chromatography of the GbsR protein labeled with a Strep-Tag affinity peptide was purchased from IBA (Göttingen, Germany). The inducer for the tet promoter-mediated expression of the gbsR gene, anhydrotetracycline (AHT), was obtained from Acros Organics (Geel, Belgium).
Bacterial strains.
The Escherichia coli K-12 strain DH5α (Invitrogen, Carlsbad, CA) was used for routine cloning purposes and maintenance of cloning vectors and recombinant plasmids. The E. coli B strain BL21 (Stratagene, Heidelberg, Germany) was used for the overproduction of recombinant proteins. These E. coli strains were grown and maintained on Luria-Bertani (LB) agar plates, and solid and liquid media contained, when necessary, antibiotics to select for the presence of plasmids. The B. subtilis strain JH642 (trpC2 pheA1) and its mutant derivatives were used throughout this study (Table 1). JH642 is a member of the domesticated 168 lineage of B. subtilis laboratory strains (54) and was a kind gift of J. Hoch (Scripps Research Institute, CA).
Table 1.
B. subtilis strains used in this study
| Strain | Relevant genotypea | Reference/source |
|---|---|---|
| JH642 | trpC2pheA1 | J. Hoch; BGSC 1A96b |
| JBB8 | (gbsR::neo)1 | 5 |
| RMKB25 | opuC-20::Tn10(spc) | 36 |
| RMKB26 | opuB-20::Tn10(spc) | 36 |
| BKB30 | (yvbF::tet)1 | This study |
| BKB31 | (yvaV::spc)1 | This study |
| GNB25 | (gbsR::neo)1opuC-20::Tn10(spc) | This study |
| GNB26 | (gbsR::neo)1opuB-20::Tn10(spc) | This study |
| GNB37 | Δ(treA::erm)2 | This study |
| GNB40 | Δ(treA::erm)2 (gbsR::neo)1 | This study |
| GNB45 | Δ(treA::erm)2 [amyE::(ΔgbsR)2-Φ(gbsA′-treA)1] | This study |
| GNB46 | Δ(treA::erm)2Δ(gbsAB::neo)2 [amyE::(ΔgbsR)2- Φ(gbsA′-treA)1] | This study |
| GNB48 | Δ(treA::erm)2 (gbsR::neo)1 [amyE::(ΔgbsR)2-Φ(gbsA′-treA)1] | This study |
| GNB55 | Δ(treA::erm)2 (yvbF::tet)1 [amyE::(gbsA′-treAΔ(gbsR)2)1] | This study |
| GNB56 | Δ(treA::erm)2 (yvaV::spc)1 [amyE::(gbsA′-treAΔ(gbsR)2)1] | This study |
| GNB67 | Δ(treA::erm)2 [amyE::yvbF-Φ(opuCA′-treA)2] | This study |
| GNB69 | Δ(treA::erm)2 (gbsR::neo)1 [amyE::yvbF-Φ(opuCA′-treA)2] | This study |
| GNB74 | Δ(treA::erm)2 [amyE::yvaV-Φ(opuBA′-treAyvaV+)2] | This study |
| GNB76 | Δ(treA::erm)2 (gbsR::neo)1 [amyE::yvaV-Φ(opuBA′-treA)2] | This study |
The designation amyE::Φ(gbsA′-treA) indicates that the gbsA-treA operon fusion is stably integrated via a double-recombination event into the chromosomal amyE gene of B. subtilis as a single copy, thereby rendering the fusion strains defective in the extracellular AmyE α-amylase. The Φ(gbsA′-treA)1 reporter fusion is linked to a chloramphenicol resistance gene (cat), thereby conferring resistance to the antibiotic chloramphenicol to all strains carrying the amyE::Φ(gbsA′-treA) construct. This is true also for Φ(opuCA′-treA) and Φ(opuBA′-treA) reporter fusion strains. The designation (ΔgbsR)2-Φ(gbsA′-treA)1 indicates that the DNA fragment used for the construction of the Φ(gbsA′-treA) reporter fusion (Fig. 5) does not carry an intact gbsR gene.
Bacillus Genetic Stock Center, Columbus, Ohio.
Media and growth conditions.
B. subtilis strains were maintained on LB agar plates; liquid cultures were grown at 37°C in Spizizen's minimal medium (SMM) (25) supplemented with 0.5% (wt/vol) glucose as the carbon source, a solution of trace elements (26), and the amino acids l-tryptophan (20 mg liter−1) and l-phenylalanine (20 mg liter−1) to meet the auxotrophic needs of strain JH642 (trpC2 pheA1) and its derivatives (Table 1). Cultures of the E. coli strain BL21 used for the production of recombinant proteins were grown in minimal medium A (MMA) with glucose (0.5%) as the carbon source and supplemented with 0.5% Casamino Acids and thiamine (1 μg ml−1). The antibiotics chloramphenicol, kanamycin, erythromycin, and spectinomycin were used with B. subtilis strains at final concentrations of 5 μg ml−1, 10 μg ml−1, 1 μg ml−1, and 100 μg ml−1, respectively. Ampicillin and chloramphenicol were used for E. coli cultures at final concentrations of 100 μg ml−1 and 35 μg ml−1, respectively. Operon fusions of a promoterless treA reporter gene (22) to the regulatory regions of the gbsAB, opuB, and opuC gene clusters were constructed with the aid of plasmid pJMB1 (M. Jebbar and E. Bremer, unpublished data) (see the supplemental material). In this treA reporter fusion plasmid, a gene conferring resistance to chloramphenicol follows the operon fusion construct; the entire genetic construct is flanked by the 5′ and 3′ segments of the B. subtilis amyE gene. This allows the stable single-copy integration of the treA reporter gene fusion and of the cat gene via a double homologous recombination event into the chromosomal copy of the B. subtilis amyE locus by selecting for chloramphenicol-resistant colonies. This renders the resulting treA reporter fusion reporter strain defective in the production of the extracellular AmyE amylase.
Recombinant DNA techniques and construction of plasmids and B. subtilis strains.
The routine manipulations of plasmid DNA, the construction of recombinant DNA plasmids, the isolation of chromosomal DNA from B. subtilis, and transformation with plasmid or chromosomal DNA were carried out using standard procedures (26). The construction of recombinant DNA plasmids and their use for the generation of B. subtilis mutants carrying chromosomal gene disruptions or treA reporter fusions integrated at the chromosomal amyE locus are detailed in the supplemental material. The genotypes of the B. subtilis strains used in this study are described in Table 1 (also see Table 4 and Fig. 8).
Table 4.
Role of the GbsR-related YvaV and YvbF B. subtilis proteins on gbsA promoter activitya
| Strain | gbsRb | yvaV | yvbF | TreA activity (U/mg protein) |
|---|---|---|---|---|
| GNB45 | + | + | + | 10 ± 3 |
| GNB48 | − | + | + | 568 ± 13 |
| GNB55 | + | − | + | 16 ± 2 |
| GNB56 | + | + | − | 14 ± 2 |
| GNB57 | − | − | + | 574 ± 24 |
| GNB58 | − | + | − | 564 ± 33 |
| GNB59 | − | − | − | 565 ± 5 |
| GNB60 | + | − | − | 15 ± 4 |
Cells of the various gbsA-treA reporter fusion strains were cultivated in SMM until the cultures had reached on OD578 of 1.5; samples were then assayed for TreA reporter enzyme activity. The strains carry the same gbsA-treA reporter gene fusion and are present in a B. subtilis JH642 (trp phe) strain background.
The strains carrying mutations in the gbsR gene (gbsR::neo), the yvaV gene (yvaV::spc) or the yvbF gene (yvbF::tet) are indicated by −; the presence of wild-type alleles is indicated by +.
Fig 8.
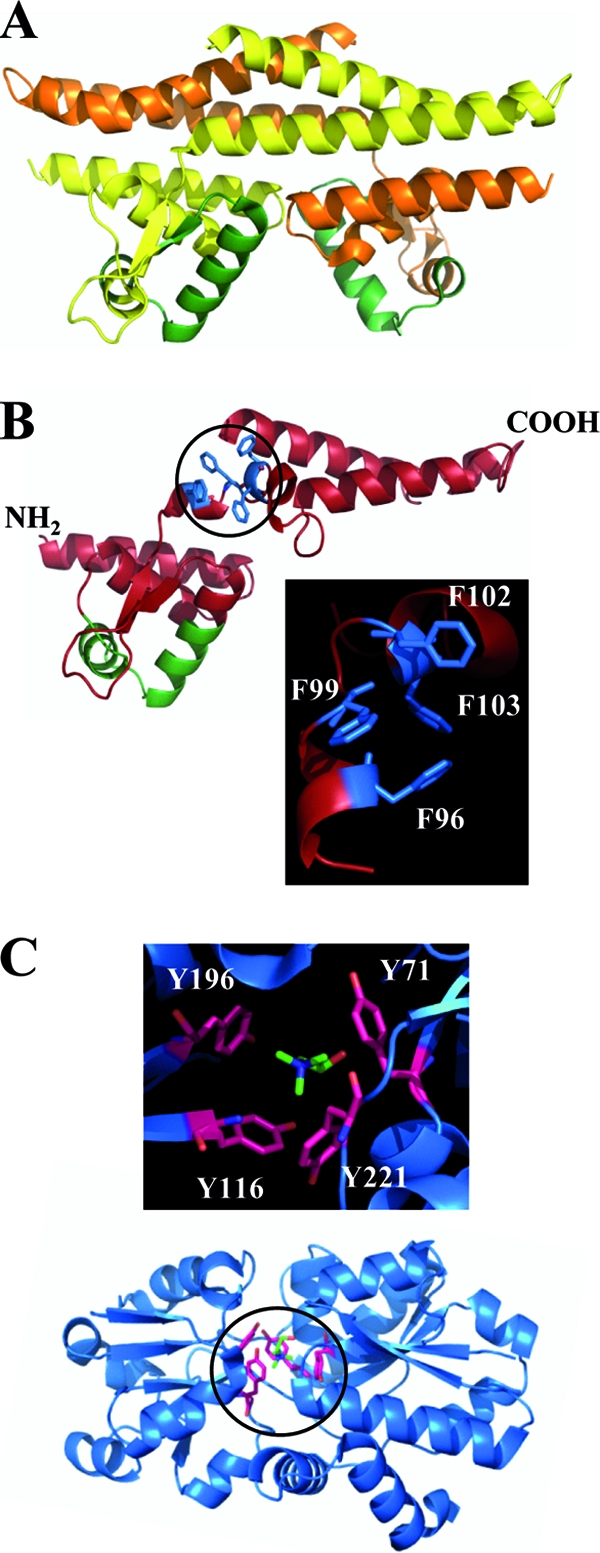
In silico model of the B. subtilis GbsR protein. (A) X-ray structure of the dimer of the M. jannaschii Mj223 protein (PDB code 1ku9) (50). The monomers are shown in either yellow or orange; the winged-helix DNA binding region is shown in green. (B) Monomer of the in silico-derived GbsR structure (red color). The predicted winged-helix DNA binding region is shown in green, and the four Phe residues predicted to form part of the choline-binding box are shown in blue. The insert enlarges a section of the GbsR in silico model and focuses on the spatial arrangement of the aromatic cage predicted to accommodate the trimethlyammonium head group of choline. (C) Overall crystal structure (PDB code 3r6u) (48) of the B. subtilis OpuBC solute binding protein in complex with its ligand choline. The insert enlarges a section of the OpuBC crystal structure and focuses on the spatial arrangement of the aromatic cage entrapping the choline ligand.
Choline transport assays.
Uptake assays with [methyl-14C]choline as the substrate were conducted as described previously (35, 36). For these transport experiments, the B. subtilis cells were grown to early exponential growth phase (optical density at 578 nm [OD578] of about 0.4) in either SMM or SMM containing 0.4 M NaCl. The final substrate concentration of choline used in these uptake assays was 10 μM.
TreA enzyme activity assays.
Aliquots (1.5 ml) from cultures of B. subtilis fusion strains carrying operon fusions to the promoterless treA reporter gene were withdrawn and assayed for TreA reporter enzyme activity as described previously, using para-nitrophenyl-α-d-glucopyranoside as the substrate (22). TreA [phospho-α-(1,1)-glucosidase]-specific activity is expressed in units per mg of protein; protein concentrations were estimated from the optical density of the cell culture (44).
Overproduction and purification of recombinant GbsR protein in E. coli.
To obtain purified GbsR protein, we used the gbsR overexpression plasmid pDH1. Plasmid pDH1 carries the B. subtilis gbsR gene under the control the anhydrotetracycline (AHT)-inducible tet promoter, and its AHT-dependent expression leads to the production of a hybrid GbsR protein with an N-terminal Strep-Tag II affinity peptide. Derivatives of E. coli strain BL21 carrying pDH1 were grown at 37°C in a chemically defined medium (MMA) (44) to avoid binding of possible ligands (e.g., choline or glycine betaine) to the GbsR protein that might be provided through components (e.g., yeast extract) of rich growth media during protein overproduction. Expression of the pDH1-carried gbsR gene was induced by adding AHT (final concentration, 0.2 μg ml−1) to the cells, when the cultures reached an OD578 of about 0.5. The cultures were then grown for an additional 2 h, and the cells were then harvested by centrifugation, resuspended in a lysis buffer (100 mM Tris-HCl [pH 7.5], 2.5% glycerol, 2 mM dithiothreitol, 0.4 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride [PMSF], 0.5 mM benzamidine), and disrupted by passage several times through a French pressure cell. A cleared cell lysate was prepared from these disrupted cells by ultracentrifugation (1 h at 4°C at 140,000 × g), and the soluble proteins were loaded onto a Strep-Tactin column for affinity purification according to procedures described by the manufacturer of the affinity resin (IBA, Göttingen, Germany). Elution fractions containing the purified GbsR protein were pooled, and the proteins were concentrated using VivaSpin columns (VivaSciences Ltd., Stonehouse, United Kingdom) with a simultaneous exchange of the elution buffer to a buffer containing 10 mM Tris-HCl (pH 7). The purity of the isolated GbsR protein was assessed by SDS-polyacrylamide gel electrophoresis.
Determination of the dissociation constants of the GbsR/choline complex.
The dissociation constant of GbsR for choline was determined by intrinsic tyrosine and tryptophan fluorescence spectroscopy as described by Pittelkow et al. (48) for the OpuBC/choline complex using a Carry Eclipse fluorescence spectrometer (Varian, Surry, United Kingdom). Purified GbsR protein (5 μM) (in 10 mM Tris-HCl [pH 7]) was equilibrated for 5 min in the cuvette, and various concentrations (from 25 μM to 1,000 μM) of choline were then titrated to the GbsR protein solution. The excitation wavelength of the fluorescence spectrometer was set to 280 nm, the slit width was 5 nm, and an emission spectrum was recorded (from 290 nm to 400 nm). For these experiments, the photomultiplier tube voltage (PMT) of the fluorescence detector was set to 800 V. Upon choline binding by the purified GbsR protein (see Fig. 6A), the intrinsic fluorescence intensity decreases in response to the choline concentration in the assay (see Fig. 6B). Differences in the fluorescence intensities of the GbsR protein solution observed at 335 nm for the GbsR protein solutions incubated with choline (see Fig. 6B) were used to calculate the apparent KD (equilibrium dissociation constant) value (see Fig. 6C) as previously reported (48).
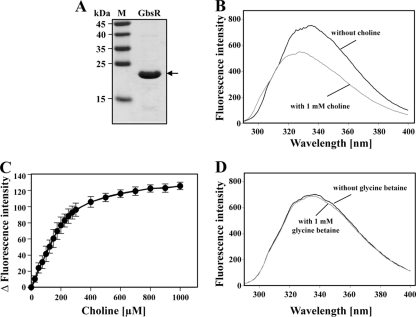
Fig 6.
Binding of choline by the GbsR regulatory protein. (A) SDS-polyacrylamide gel electrophoresis of the purified GbsR protein. (B) Fluorescence spectrum of the purified GbsR protein (5 μM) in the absence or the presence of 1 mM choline. (C) Binding kinetics of choline to the purified GbsR protein (5 μM) as assessed by intrinsic fluorescence spectroscopy. (D) Intrinsic fluorescence spectrum of the purified GbsR protein (5 μM) in the absence or the presence of 1 mM glycine betaine.
Database searches, alignments of amino acid sequences, and modeling of the GbsR structure.
Proteins that are homologous to the B. subtilis GbsR protein were searched for via the Web server of the Department of Energy (DOE) Joint Genome Institute (JGI) (http://www.jgi.doe.gov/) or that of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) using the BLAST algorithm (2). Protein sequences related to the B. subtilis GbsR and the Methanococcus jannaschii Mj223 (50) proteins were aligned and analyzed using ClustalW (57). The genome context of finished and unfinished microbial genomes in the vicinity of gbsR-related and Mj223-type genes was assessed using the gene neighborhood tool provided by the JGI Web server. An in silico model of the B. subtilis GbsR protein was generated with the aid of the SWISS Model server (http://swissmodel.expasy.org) (3) using crystallographic data deposited in the Protein Data Bank (PDB) file 1ku9 for the M. jannaschii Mj223 protein (50) as the template. Figures of the crystal structure of the M. jannaschii Mj223 protein, of the in silico-modeled B. subtilis GbsR protein, and of the OpuBC protein in complex with its ligand choline (PDB code 3r6u) were prepared with PyMol (http://www.pymol.org/).
RESULTS
Induction of gbsAB transcription in response to choline and glycine betaine aldehyde.
To provide quantitative data on the regulation of the gbsAB operon in response to external stimuli, we constructed a gbsA-treA transcriptional fusion and integrated it stably as a single copy into the amyE gene in the B. subtilis chromosome through a homologous double-recombination event. Expression of the gbsA-treA fusion was very low when the resulting reporter strain GNB45 was grown in minimal medium (SMM) with glucose as the carbon source. However, the addition of 1 mM choline or 1 mM glycine betaine aldehyde to the growth medium led to a strong induction of gbsA-treA transcription: 29-fold in response to choline and 22-fold in response to glycine betaine aldehyde (Fig. 2A). In contrast to the inducing effects of the precursor and intermediate for glycine betaine synthesis (Fig. 1), the end product of the glycine betaine biosynthetic route did not trigger enhanced gbsA-treA expression (Fig. 2A). The addition of 25 μM choline to the growth medium of strain GNB45 already resulted in a significant increase in the level of gbsA-treA expression, and the addition of about 100 to 125 μM choline was sufficient to fully induce the expression of the reporter fusion (data not shown).
Fig 2.
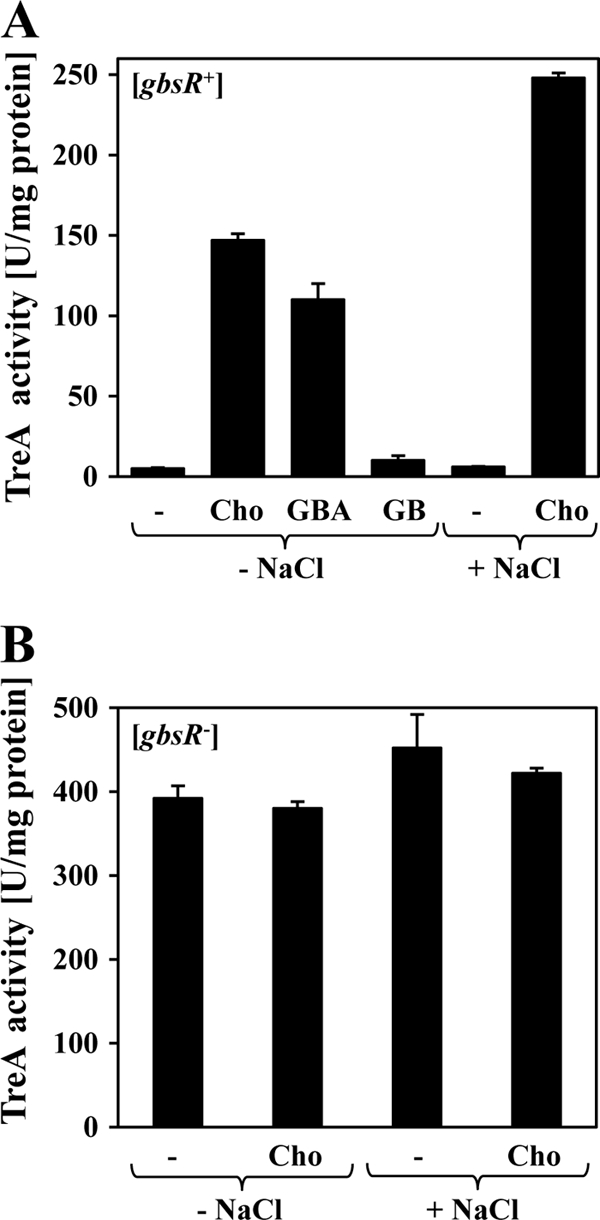
Genetic control of gbsAB expression in response to the presence of external inducers, high salinity, and the GbsR regulator. Cells of the gbsA-treA reporter strains GNB45 (gbsR+) (A) or GNB48 (gbsR::neo) (B) were grown at 37°C either in SMM or in SMM with 0.4 M NaCl to an OD578 of 0.25; 1 mM choline (Cho), glycine betaine aldehyde (GBA), or glycine betaine (GB), respectively, was then added to some of the cultures, and the activity of the TreA reporter enzyme was determined after 90 min of further cultivation.
Sustained growth of the cells of the gbsA-treA reporter strain GNB45 at high osmolarity (SMM with 0.4 M NaCl) did not trigger enhanced gbsA-treA expression. However, the addition of 1 mM choline to the osmotically stressed cells resulted in a strong induction of the reporter gene fusion, and the degree of gbsA-treA expression exceeded that observed in cells grown in the presence of choline alone (Fig. 2A). The enhancing effect of high salinity on the induction of gbsA-treA expression by an exogenous supply of choline can be rationally explained by the fact that the expression of the operons encoding the choline importers OpuB and OpuC is stimulated by high-osmolarity growth conditions (24, 36, 55).
GbsR is a negative regulator of gbsAB expression.
The yuaC gene (locus tag BSU31070) is oriented divergently from the gbsAB operon (Fig. 1) and encodes a 180-amino-acid-comprising protein (21 kDa). Our database searches and computer predictions suggest that it is a regulatory protein carrying a winged-helix DNA-binding motif (20, 33) between amino acids 49 and 73 (see below). YuaC-related proteins are encoded by genes found in close vicinity of functionally characterized glycine betaine biosynthetic genes from Staphylococcus xylosus (51), Halobacillus dabanensis (23), and Halobacillus halophilus (15). We therefore considered the possibility that the B. subtilis YuaC protein might function as a transcriptional regulator for the gbsAB operon. To test this idea, we constructed a gene disruption in the chromosomal yuaC gene and introduced this mutation (yuaC::neo) into the above-described gbsA-treA reporter strain. In the resulting strain, GNB48, transcription of gbsA-treA was strongly derepressed when the cells were grown in the absence of choline, and the availability of choline in the growth medium of cells cultivated in either SMM or SMM with 0.4 M NaCl no longer enhanced gbsA-treA transcription (Fig. 2B). These data therefore show that the yuaC-encoded protein serves as a regulator for gbsAB expression and genetically functions as a repressor. We refer in the following to this protein as the GbsR repressor (glycine betaine synthesis regulator) and redesignate the yuaC locus as the gbsR structural gene (Fig. 1).
Mapping of the gbsR promoter.
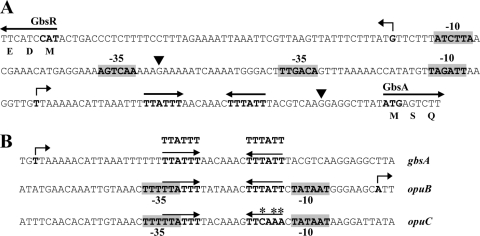
The transcriptional start site for the gbsAB operon has previously been mapped by primer extension analysis by Boch et al. (5). This allowed the identification of the positions of −10 and −35 regions characteristic for SigA-type promoters (28); the gbsAB promoter possesses a canonical spacing of 17 bp between the −10 and −35 sequences (Fig. 3A). We now have mapped the transcriptional start site for the gbsR mRNA by primer extension analysis, and a single transcript was found (see Fig. S1 in the supplemental material). Upstream of this mRNA start site, DNA sequences that resemble −35 and −10 regions of SigA-type promoters are present. The inferred gbsR promoter possesses a spacing of 16 bp (Fig. 3A), a suboptimal spacing for −10 and −35 regions of typical SigA-dependent promoters, suggesting that the gbsR promoter is probably not a particularly strong promoter (28). The promoter regions of the divergently transcribed gbsR and gbsAB genes are compactly organized, but the two promoters do not overlap; their −35 regions are separated by a 22-bp A·T-rich DNA segment (Fig. 3A).
Fig 3.
DNA sequence of the intergenic gbsR-gbsAB regulatory region. (A) The −10 and −35 sequences of the SigA-type promoters for the gbsR gene and the gbsAB operon are indicated, and the mapped transcriptional start sites for these genes are shown by bent arrows. The transcriptional start site for the gbsAB operon was mapped by Boch et al. (5) and that of the gbsR gene was determined in this study (see Fig. S1 in the supplemental material). The putative GbsR binding site positioned downstream of the gbsAB transcription initiation site is indicated by a pair of inverted arrows. The beginning of the coding regions for the GbsR and GbsA proteins is marked. The two black triangles indicate the extent of the inferred minimal DNA fragment (Fig. 5) that permits GbsR-mediated induction of gbsAB transcription in response to choline. (B) Putative GbsR binding sites in the gbsAB and opuC regulatory regions. The −10 and −35 sequences of the SigA-type promoters for the opuB and opuC operons are indicated; the transcriptional start site for the opuB mRNA has been experimentally determined (36) (indicated by a bent arrow), and the promoter for the opuC operon was predicted by DNA sequence gazing. The putative GbsR binding sites are indicated by pairs of inverted arrows; the asterisks (*) mark differences in the DNA sequences of the putative GbsR binding site in the opuB and opuC regulatory regions.
GbsR controls the expression of the opuB operon but not that of the opuC operon.
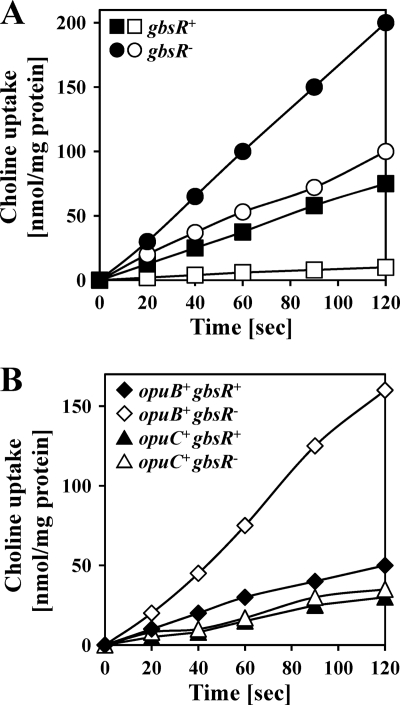
Synthesis of glycine betaine by B. subtilis depends on the GbsAB enzymes but requires the prior import of the precursor choline through the OpuB or OpuC ABC transport systems (4, 5, 36) (Fig. 1), since B. subtilis cannot synthesize choline. To test whether the gbsR::neo gene disruption mutation would influence choline uptake, we introduced this allele into strain JH642 (opuB+ opuC+ gbsR+) and compared the transport activity of the parent strain JH642 with the resulting gbsR::neo mutant derivative, strain JBB8. As reported previously (36), uptake of [methyl-14C]choline was stimulated in cells that were grown at high salinity (SMM with 0.4 M NaCl) in comparison to those cultivated in SMM without additional salt (Fig. 4A). However, the levels of choline uptake by the gbsR::neo mutant strain JBB8 with or without osmotic induction well exceeded the choline import activity exhibited by the gbsR+ strain JH642 (Fig. 4A). This finding suggested that GbsR is involved in controlling the expression of either the opuB or opuC operons or that of both loci.
Fig 4.
Influence of GbsR on choline import via the OpuB and OpuC ABC transporters. Cells were cultivated at 37°C in SMM or in SMM containing 0.4 M NaCl until they had reached an OD578 of about 0.4. Subsequently, uptake of [14C]choline was assayed at a final substrate concentration of 10 μM. (A) Uptake of [14C]choline in the wild-type strain JH642 (opuB+ opuC+ gbsR+) (□, ■) and strain JBB8 (opuB+ opuC+ gbsR::neo) (○, ●) grown in SMM (□, ○) or SMM with 0.4 M NaCl (■, ●). (B) Uptake of [14C]choline in strain RMKB25 [opuB+ opuC::Tn10 (spc) gbsR+] (♦), GNB25 [opuB+ opuC::Tn10 (spc) gbsR::neo] (♢), strain RMKB26 [opuB::Tn10 (spc) opuC+ gbsR+] (▲) and GNB26 [opuB::Tn10 (spc) opuC+ gbsR::neo] (▵); cells were grown at 37°C in SMM containing 0.4 M NaCl.
Since both the OpuB and OpuC systems can mediate high-affinity choline transport, we genetically constructed an isogenic set of strains in which either only the OpuB transporter or only the OpuC transporter was functional and which differed in the genetic status of their gbsR loci. Transport assays with radiolabeled [methyl-14C]choline revealed that the uptake activity via the choline-specific OpuB transporter (36, 48) was strongly enhanced in a gbsR::neo mutant strain (Fig. 4B), whereas choline import via the OpuC transporter, an uptake system with a broad substrate specificity (19, 30, 36) (Fig. 1), was not affected by the gbsR::neo gene disruption (Fig. 4B).
The different effects of a gbsR::neo mutation on the choline import activity of the OpuB and OpuC transporters was further corroborated by testing the effect of this mutation on the expression level of opuB-treA and opuC-treA reporter gene fusions. As documented in Table 2, and fully consistent with the [methyl-14C]choline transport assays shown in Fig. 4A, loss of GbsR affected only opuB expression. Hence, the entire choline-to-glycine betaine biosynthetic route of B. subtilis, i.e., the gbsAB operon encoding the glycine betaine biosynthetic enzymes (5) and the opuB operon encoding the choline-specific import system OpuB (36) (Fig. 1), is under the control of the GbsR repressor.
Table 2.
Influence of GbsR on the transcriptional activity of the opuB and the opuC genesa
| Strain | tre fusion | gbsRb | TreA activity (U/mg protein) |
|
|---|---|---|---|---|
| Without NaCl | 1.2 M NaCl | |||
| GNB74 | opuB-treA | + | 38 ± 5 | 104 ± 2 |
| GNB76 | opuB-treA | − | 308 ± 5 | 724 ± 16 |
| GNB67 | opuC-treA | + | 34 ± 3 | NDc |
| GNB69 | opuC-treA | − | 36 ± 5 | ND |
Cells carrying chromosomal copies of the indicated opuB-treA and opuC-treA operon fusions were cultivated in SMM or SMM with 1.2 M NaCl to mid-log phase and were then harvested for TreA reporter enzyme activity.
Strains carrying a gbsR mutation (denoted with −) harbored the gbsR::neo allele.
ND, not determined.
High salinity increases the expression of the opuB operon (24, 36, 55). We therefore wondered whether a gbsR gene disruption would impinge on the osmotic induction of opuB transcription. The data summarized in Table 2 show that this is not the case. Under the conditions tested, a gbsR mutant increased opuB-treA expression about 8-fold in cells grown in SMM, and a similar level of opuB-treA derepression (about 7-fold) was observed in cells grown in SMM that contained 1.2 M NaCl. Likewise, osmotic induction of opuB-treA expression was not significantly affected by the presence of a gbsR mutation: transcription of the reporter fusion was increased about 2.6-fold in the gbsR+ strain GNB74 and about 2.3-fold in the gbsR::neo strain GNB76 (Table 2). In contrast to the opuB operon (24, 36, 55), expression of the gbsAB gene cluster is not enhanced in cells cultivated under sustained high-salinity conditions (Fig. 2A) (5), but it responds to an osmotic stimulus in cells subjected to a severe and growth-restricting sudden osmotic upshift with 1.7 M NaCl (24).
Delineation of the GbsR-binding site within the gbsAB regulatory region by deletion analysis.
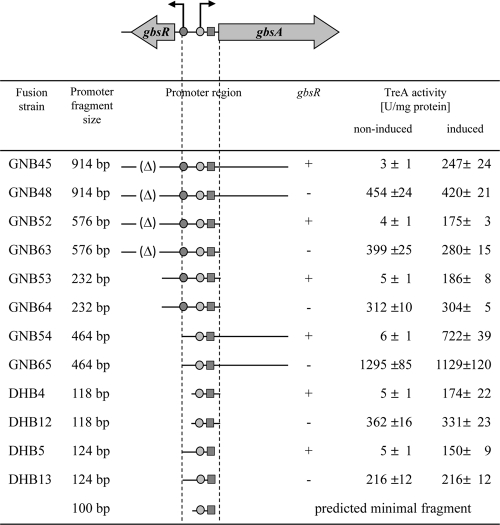
To narrow down the DNA segment required for the GbsR-mediated and choline-responsive control of gbsAB expression, we performed a deletion analysis of the gbsAB regulatory region. For this purpose, we constructed a set of gbsA-treA reporter gene fusions carrying the gbsAB promoter and flanking DNA sequences of various lengths. These transcriptional fusions were integrated in a single copy into the genome of a strain with an intact gbsR gene and also into that of a gbsR::neo mutant. We then determined the expression level of the various gbsA-treA reporter fusions in response to the presence of choline in the growth medium and the functionality of the GbsR regulator. Each of the reporter fusions tested responded to choline availability and was controlled by GbsR (Fig. 5). By viewing these data together, we infer that a DNA fragment with a length of just 100 bp is sufficient to confer GbsR-mediated and choline-responsive control of gbsAB expression (Fig. 5). This regulatory fragment comprises the gbsAB promoter, a 51-bp-long region downstream of the −10 region and a 19-bp-long region upstream of the −35 region. This DNA fragment, whose sequence is highlighted in Fig. 3A, does not carry any part of the gbsR promoter. Hence, GbsR binding is expected to occur at or very close to the gbsAB promoter sequence.
Fig 5.
Choline-responsive and GbsR-dependent regulation of gbsA-treA expression. Strains with either an intact gbsR gene (+) or a gbsR::neo mutation (−) carrying chromosomal gbsA-treA fusions of various lengths were pregrown in SMM to early log phase (OD578 of 0.25), and aliquots were assayed for TreA activity (noninduced condition). At this time point, 1 mM choline (final concentration) and 0.4 M NaCl (final concentration) were added to the cultures; after further growth for 90 min, the cells were harvested for assays of the TreA reporter enzyme (induced condition). NaCl was added to these cultures to elicit enhanced uptake of the inducer choline via the osmotically stimulated OpuB and OpuC transport systems (36). The symbol (Δ) indicates that the corresponding segment of the gbsR gene was deleted from the DNA fragment fused to the treA reporter gene. All fusion strains are derivatives of the B. subtilis strain JH642 (trp phe).
Since GbsR also controls the expression of opuB, but not that of opuC (Table 2), we inspected the regulatory regions of these gene clusters for sets of common DNA sequences that might serve as operator sequences for the GbsR repressor. We found an identical A·T-rich inverted repeat (5′-TTATTT-N7-TTTATT-3′) that is present downstream of the transcription initiation site of the gbsAB operon (Fig. 3B) and which overlaps the −35 and spacer region of the previously mapped (36) opuB promoter (Fig. 3B). This DNA element is not intact in the presumed promoter region of the opuC operon (Fig. 3B). While this type of DNA sequence gazing is certainly suggestive, DNA-binding assays and footprinting studies with the purified GbsR protein have to be conducted in the future to assess the functionality of the indicated inverted repeat sequence for GbsR binding. If the DNA sequences highlighted in Fig. 3B are indeed the operator sites for GbsR, GbsR would likely repress the transcription of gbsAB through a roadblock mechanism impeding progress of the RNA polymerase and that of opuB through a mechanism relying on competition with RNA polymerase for access to the promoter region.
GbsR is a choline-sensing protein and binds choline directly.
Since the addition of choline to the growth medium induces gbsAB expression (Fig. 2A), it seemed to us that the GbsR repressor might serve as an intracellular choline-sensing protein. To investigate this further genetically, we constructed a gbsA-treA reporter strain (GNB46) that is defective in the gbsAB operon. In such a strain, choline accumulated via the OpuB and OpuC transporters cannot be removed by the cell via oxidation to glycine betaine (Fig. 1) and is thus predicted to accumulate to high intracellular levels (5). The addition of choline to the growth medium triggered gbsA-treA expression in both the gbsAB wild-type strain and the gbsAB mutant strain, but the level of gbsA-treA expression was considerably higher (about 5-fold) in strain GNB46 (gbsAB::neo) lacking the glycine betaine biosynthetic enzymes (Table 3). As a matter of fact, the level of gbsA-treA expression in strain GNB46 was, within experimental error, practically identical with that measured in gbsR::neo mutant strain GNB48, in which the gbsA-treA reporter fusion is completely derepressed and no longer answers to choline availability in the growth medium (Table 3). This experiment therefore suggests that choline serves as the inducer of GbsR-controlled gene expression and that the B. subtilis cell continuously removes this inducer from the cytoplasm by oxidizing choline to glycine betaine via the action of the GbsAB enzymes (Table 3). This conclusion can be inferred by comparing the level of gbsA-treA promoter activity in strain GNB45, which is proficient in the GbsAB enzymes, with that of strain GNB46, which lacks these enzymes (Table 3).
Table 3.
Influence of choline and glycine betaine on the activity of the gbsA promotera
| Strain | gbsABb | gbsR | Compound present | TreA activity (U/mg protein) |
|---|---|---|---|---|
| GNB45 | + | + | − | 6 ± 1 |
| + | + | Choline | 106 ± 3 | |
| + | + | Choline + glycine betaine | 5 ± 1 | |
| GNB46 | − | + | − | 6 ± 1 |
| − | + | Choline | 534 ± 7 | |
| − | + | Choline + glycine betaine | 5 ± 1 | |
| GNB48 | + | − | − | 478 ± 16 |
| + | − | Choline | 569 ± 19 | |
| + | − | Choline + glycine betaine | 515 ± 27 |
Cells of the various gbsA-treA reporter fusion strains were cultivated in SMM without (−) or with 1 mM choline or in the presence of 1 mM choline and 1 mM glycine betaine until the cultures had reached on OD578 of 1.5; samples were then assayed for TreA reporter enzyme activity.
The strains carrying mutations in the gbsAB operon [Δ(gbsAB::neo)] or in the gbsR gene [gbsR::neo] are indicated by −.
If choline is the true inducer for the GbsR repressor, then one might be able to biochemically demonstrate the binding of this ligand to the GbsR protein. To test this, we produced the B. subtilis GbsR repressor in E. coli as a fusion protein with an NH2-terminal Strep-Tag and purified the recombinant protein by affinity chromatography to apparent homogeneity (Fig. 6A). GbsR carries two Trp and six Tyr residues, and we reasoned that one could exploit changes in intrinsic fluorescence of this protein to monitor conformational changes that might occur upon ligand binding. We have previously successfully used this technique to determine the binding constant for choline and glycine betaine of ligand-binding proteins associated with bacterial ABC transport systems (32, 47, 48, 53, 58). The addition of 1 mM choline to the purified GbsR protein resulted in a strong quenching of the intrinsic Trp/Tyr fluorescence of GbsR with a simultaneous shift in the wavelength of the emission maximum (Fig. 6B). Titration experiments with choline were then used to assess the binding constant for choline (Fig. 6C), and a KD value of 165 ± 15 μM was calculated. This value is 1 to 2 orders of magnitude higher than that measured for choline binding by ligand-binding proteins associated with high-affinity ABC transport systems: ChoX (KD, 2 μM) from Sinorhizobium meliloti (47), OpuBC (KD, 30 μM) from B. subtilis (48), and ChoX (KD, 2 μM) from Agrobacterium tumefaciens (1).
Glycine betaine tunes down choline-mediated and GbsR-dependent induction of gbsAB expression.
The level to which glycine betaine is amassed by the cell from exogenous sources (30, 31) is linearly dependent on the prevalent osmolarity of the growth medium (T. Hoffmann and E. Bremer, unpublished data). This observation suggests the existence of a not-yet-understood physiological and/or genetic regulatory circuit that links the intracellular glycine betaine pool with the osmolarity of the particular ecological niche that B. subtilis happens to colonize. It therefore follows that the osmoadaptive glycine betaine synthesis pathway needs to be shut down once the cell has balanced the osmotic gradient across its cytoplasmic membrane through synthesis of the compatible solute glycine betaine.
We therefore asked if an exogenous supply of glycine betaine would repress gbsA-treA expression and found that this was indeed the case (Table 3). The presence of 1 mM glycine betaine in the growth medium repressed even the very high level of transcription of this reporter fusion in a gbsAB mutant strain that cannot remove the inducer choline from the cytoplasm by oxidizing it to glycine betaine (Table 3). Strikingly, repression of gbsA-treA transcription in response to the presence of glycine betaine was abolished in a gbsR mutant strain (Table 3), showing that the GbsR repressor protein is required for the inhibitory effect of glycine betaine on gbsAB expression.
We then employed the intrinsic fluorescence-based ligand-binding assay used to assess the binding of choline to GbsR (Fig. 6B and C) to test whether glycine betaine would bind to the B. subtilis GbsR protein as well. However, we did not detect any effects of glycine betaine (1 mM) on the intrinsic fluorescence of the purified GbsR protein (Fig. 6D) that could be interpreted as an indication for binding of glycine betaine to GbsR. We conducted a further series of competition experiments where we preincubated the GbsR protein in the presence of 1 mM, 10 mM, and 100 mM glycine betaine, subsequently added 1 mM the inducer choline to the reaction mixture, and then recorded the intrinsic fluorescence of GbsR. No noteworthy influence of glycine betaine on the choline-induced emission spectrum of the GbsR protein was observed (see Fig. S2 in the supplemental material). This observation indicates that there is no significant degree of competition between the inducer choline and the “counterinducer” glycine betaine for occupancy of the choline-binding site in GbsR (see Fig. S2) in a ligand-binding assay that relies on changes in the intrinsic fluorescence of the purified GbsR protein (Fig. 6B). However, since this assay provides only an indirect readout of ligand binding, some caution in the interpretation of the data is warranted. Mechanisms other than competition of choline and glycine betaine for the same ligand-binding site come to mind: (i) binding of glycine betaine to an allosteric effector site in GbsR influencing the occupancy of the choline-binding site and (ii) opposing effects of choline and glycine betaine on dimer formation of GbsR. Alternatively, glycine betaine might not bind directly to GbsR at all and might instead exert its regulatory effects indirectly through its “chemical chaperone” function (16, 17) that can influence protein folding and conformation (7, 8, 56).
Osmoprotectants other than glycine betaine can tune down choline-mediated GbsR-dependent induction of gbsAB expression.
Since glycine betaine is the end product of the choline-dependent biosynthetic route for this compatible solute (Fig. 1), we wondered whether osmoprotectants other than glycine betaine would exert a repressing effect on gbsA-treA transcription as well. From the six additional compounds tested that function as osmoprotectants for B. subtilis (9, 30, 31), proline betaine and dimethylsulfoniopropionate (DMSP) repressed gbsA-treA transcription to an extent similar to that of glycine betaine (Fig. 7A). On the other hand, carnitine repressed the expression of the reporter gene fusion only modestly (Fig. 7A), whereas choline-O-sulfate, proline, and ectoine did not significantly influence gbsA-treA expression (Fig. 7A). As observed for glycine betaine (Table 3), repression of gbsA-treA expression by proline betaine, by DMSP, and to a minor degree by carnitine was dependent on an intact GbsR protein (Fig. 7B). Hence, compatible solutes used by B. subtilis that are not associated with glycine betaine production (Fig. 1) can reduce the level of expression of the glycine betaine gbsAB biosynthetic genes either very effectively (e.g., proline betaine), modestly (e.g., carnitine), or not at all (e.g., ectoine) (Fig. 7A). However, the interpretation of the data shown in Fig. 7A is not straightforward, since the effects of these compounds on gene expression cannot be directly correlated with the osmoprotective potential that these compatible solutes possess for B. subtilis. For instance, glycine betaine, carnitine, proline betaine, and choline-O-sulfate confer a similar degree of osmostress resistance to high-salinity-challenged B. subtilis cells (34, 35, 45, 53), yet they have quite disparate effects on gbsA-treA transcription (Fig. 7A).
Fig 7.
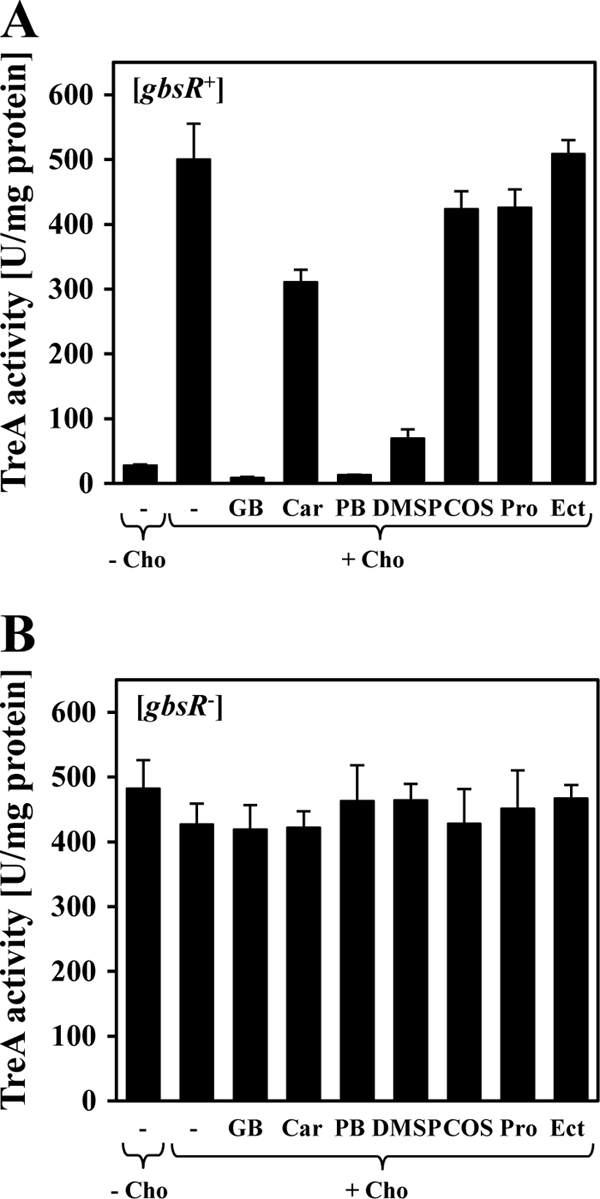
Cells of the gbsA-treA fusion strain GNB46 (gbsAB::neo gbsR+) (A) and of strain GNB48 (gbsAB+ gbsR::neo) (B) were grown either in SMM without a compatible solute (−) or in SMM containing 1 mM choline (Cho) or 1 mM choline in combination with various compatible solutes: glycine betaine (GB), carnitine (Car), proline betaine (PB), dimethylsulfoniopropionate (DMSP), choline-O-sulfate (COS), l-proline (Pro), or ectoine (Ect). Cells were harvested for TreA reporter assays when they reached an OD578 of 1 to 1.5.
A structure-guided in silico model for the GbsR repressor and inference of a putative binding site for the inducer choline.
Database searches with the amino acid sequence of the B. subtilis GbsR protein as the search template alerted us to the crystal structure of the Mj223 protein (50) from the extreme thermophilic methanogenic archaeon Methanococcus jannaschii (14). Using the crystallographic data set deposited for the Mj223 protein (PDB code 1ku9) (50) (Fig. 8A) showing a graphic representation of the Mj223 protein) and by relying on the Web-based SWISS Model server (3), we were able to derive an in silico model of the GbsR protein from B. subtilis (Fig. 8B).
The M. jannaschii Mj223 protein is a DNA-binding protein that possesses a winged-helix DNA binding motif (20, 33), and it is a dimer both in solution and in the crystal structure (50) (Fig. 8A). No ligand that could give a firm clue with respect to the physiological process in which Mj223 might be involved is present in the crystal structure of the Mj223 protein. However, based on the structural similarity of the M. jannaschii Mj223 protein to the B. subtilis BmrR and E. coli MarR transcriptional regulators, Ray et al. (50) speculated that it could be involved in controlling the expression of genes involved in multidrug resistance, but no concrete target gene of the Mj223 DNA-binding protein in the M. jannaschii genome is known. We inspected the genetic context of the structural gene for the Mj223 protein in the genome of M. jannaschii (14), but no genes whose products could be associated with glycine betaine synthesis or the uptake of choline or with any other type of osmoprotectant were identifiable.
Mj223 is a two-domain protein, with the N-terminal module containing the DNA reading head with a winged-helix DNA-binding motif (20, 33) and an extended C-terminal α-helical module that supports the dimerization of the protein (50) (Fig. 8A). In their analysis of the X-ray structure of the M. jannaschii Mj223 protein, Ray et al. (50) noticed that the determined crystal structure (Fig. 8A) would not be conductive for DNA binding of the Mj223 protein to a standard duplex B-form of DNA. Major spatial rearrangements of both the dimerization domain and the DNA-reading head relative to each other have to occur in order to properly fit the two winged helices of the Mj223 dimer into the major groove of the DNA. These movements pivot around the flexible linker region of the Mj223 protein connecting the DNA-binding and dimerization domains (50).
According to our GbsR in silico model, the N-terminal DNA reading head extends from amino acids 1 to 73 and contains the predicted winged-helix DNA-binding motif (20, 33) between amino acids 49 and 73; the predicted C-terminal dimerization domain of GbsR comprises amino acids 95 to 161. The predicted flexible linker region connecting these two domains extends from amino acids 74 to 94 (Fig. 8B).
Since GbsR can directly bind the inducer choline (Fig. 6B and C), we manually inspected our GbsR in silico-derived model for a possible ligand-binding site. This search was based on three high-resolution crystal structures of solute-receptor proteins associated with microbial ABC transporters in complex with their choline ligand: the choline-binding protein ChoX protein from S. meliloti (47) and the OpuBC (48) and OpuCC (19) proteins from B. subtilis. The precise geometries of the choline-binding site vary somewhat among these ligand-binding proteins, but four aromatic residues that are spatially arranged in the form of an aromatic cage (10) are always part of the substrate-binding sites. The bulky head group of choline is wedged into this aromatic cage and is stabilized by cation-pi interactions (18) between the positively charged trimethylammonium group of choline and the electronegative surface potential of side chains of the aromatic residues. The alcohol function of the choline molecule protrudes out of this aromatic cage and is stabilized by additional contacts (19, 47, 48).
We detected a region in our GbsR in silico model that could possibly correspond to an aromatic cage for the coordination of the trimethylammonium head group of the choline ligand. It is formed by four Phe residues that occur in close proximity in the amino acid sequence of the GbsR protein: Phe-96, Phe-99, Phe-102, and Phe-103 (Fig. 8B). The spatial arrangement of an aromatic cage used for the entrapment of choline by the ligand-binding protein, OpuBC, of the B. subtilis OpuB ABC transporter (48) is shown in Fig. 8C for comparison to the putative choline-binding site suggested here for the GbsR protein (Fig. 8B).
Genome context of gbsR-related genes.
We used the amino acid sequence of the B. subtilis GbsR protein as the search template in a BLAST analysis (2) of unfinished and finished microbial genomes and inspected the genome context of structural genes for the recovered GbsR-related proteins. GbsR-related proteins are found ubiquitously in members of the Bacillus and Staphylococcus genera, where they are typically found in the vicinity of either glycine betaine biosynthetic gene clusters or potential uptake systems for compatible solutes.
Rosenstein et al. (51) previously characterized a glycine betaine synthesis gene cluster (cudTCAB) in Staphylococcus xylosus comprising the glycine betaine biosynthetic CudAB enzymes and the choline transporter CudT, a member of the BCCT family of transporters (65). CudC is a regulatory protein exhibiting 54% identity to the GbsR protein from B. subtilis (51). Our database searches showed that the cudTCAB gene cluster is present in every sequenced Staphylococcus genome, suggesting that CudC/GbsR-type proteins function widely as the regulatory protein for glycine betaine synthesis in staphylococci. In the functionally characterized glycine betaine biosynthetic gene clusters of Halobacillus dabanensis (23) and Halobacillus halophilus (15), gbsR-type genes are present as well and are divergently orientated from the gbsAB genes and cotranscribed with a gene (referred to as gbsT and gbsU, respectively) encoding an orphan ligand-binding protein that bears the hallmarks of a substrate-binding protein associated with ABC transport system possessing a specificity for glycine betaine or other osmoprotectants with a trimethylammonium or a dimethlysulfonium head group (47, 52, 53, 58). Burkhardt et al. (15) demonstrated the induction of the transcription of the H. halophilus gbsAB and gbsRU gene clusters in response to choline availability and their repression by glycine betaine but did not experimentally correlate these transcriptional responses with the gbsR-encoded protein.
We detected two types of genome contexts of gbsR-related genes in members of the Bacillus genus. One group of these genes is associated with gbsAB-type glycine betaine biosynthetic genes in a genetic configuration identical with that found in B. subtilis (Fig. 1). The second group of gbsR-related genes is associated with gene clusters annotated as ABC-type uptake systems for compatible solutes. Some bacilli possess more than one gbsR-type gene, and these are found in both mentioned genomic contexts. One example is B. subtilis itself. In addition to the gbsR gene found next to the gbsAB gene cluster (Fig. 1), gbsR-related genes (yvbF and yvaV) are also present upstream of the opuB and opuC operons, and the encoded proteins possess amino acid sequence identities to GbsR of 34% (similarity of 58%) and 35% (similarity of 57%), respectively. Nothing is currently known about a possible function of these putative regulatory proteins, but we noted that both the YvbF and YvaV proteins each possess three out of the four aromatic residues forming the predicted choline-binding site in GbsR (Fig. 8B). We therefore tested a possible involvement of YvbF and YvaV on gbsAB expression, but none was found regardless of whether the yvbF and yvaV genes were disrupted individually, tested as part of a double mutant, or tested in various combinations with the gbsR::neo mutation (Table 4).
DISCUSSION
The data presented here identify a regulatory protein (GbsR) controlling the osmoadaptive choline-to-glycine betaine synthesis pathway of B. subtilis (Fig. 1). Coregulation of the opuB and gbsAB operons by GbsR in response to choline availability is physiologically fitting, since glycine betaine production by B. subtilis relies on the import of the precursor molecule choline (4, 5). It is equally physiologically fitting that GbsR does not control the expression of the opuC operon (Table 2), since the OpuC ABC transporter does not only function for the import of choline (36) but mediates the uptake of a wide spectrum of other osmoprotectants (9, 30) (Fig. 1). Regulation of opuC expression in response to choline availability would therefore be counterproductive for osmotically stressed B. subtilis cells.
The precursor and inducer for glycine betaine production is present in the soil habitat of Bacillus subtilis in physiologically relevant concentrations (41), primarily as a result of the degradation of phosphatidylcholine, a major component of membranes of eukaryotic cells. Glycine betaine aldehyde, the intermediate in glycine betaine synthesis (Fig. 1), is also an inducer of gbsAB expression (Fig. 2A) but is unlikely to be present in significant amounts in natural ecosystems due to its high chemical reactivity. Its function as an inducer might therefore serve as a fail-safe mechanism to ensure the synthesis of the glycine betaine aldehyde dehydrogenase GbsA, an enzyme that efficiently converts the noxious glycine betaine aldehyde (5, 36) into the physiologically well-tolerated glycine betaine molecule (6).
Our genetic and biochemical data show that GbsR acts as a repressor monitoring the presence of choline inside the cell (Table 3) and that the GbsR protein binds this ligand directly (Fig. 6B and C). We took advantage of an in silico model for GbsR to search for a possible choline-binding site (Fig. 8B) that resembles those found in several choline solute receptors of known crystal structure (19, 47, 48), e.g., the ligand-binding protein (OpuBC) of the OpuB transporter (Fig. 8C). These types of ligand-binding proteins accommodate the choline ligand via cation-pi interactions (18) between the bulky and positively charged head group of choline with the electronegative surface potential of the side chains of aromatic amino acids spatially arranged in the form of an open cage (10). The predicted aromatic choline-binding cage in GbsR is strategically positioned close to the flexible linker region connecting the N-terminal DNA-reading head with its winged-helix DNA-binding motif and the extended C-terminal dimerization domain (Fig. 8B). Ligand-induced conformational changes, as we have detected by fluorescence spectroscopy of the purified GbsR protein in response to choline (Fig. 6B), can therefore readily be envisioned as altering the spatial orientation of the N-terminal DNA-binding domain relative to the C-terminal dimerization domain and thereby relieve the repressing effects of GbsR on gbsAB and opuB expression. Indeed, the modeling studies conducted by Ray et al. (50) for the M. jannaschii Mj223 protein strongly suggest that the conformations of the DNA-bound and the DNA-free forms (Fig. 8A) of the Mj223 protein differ greatly. These conformational changes in Mj223 pivot around the flexible linker region (50) (Fig. 8A), in whose vicinity we envision the ligand-binding site for the inducer choline in our GbsR in silico model (Fig. 8B).
One issue that needs to be reckoned with in the physiological context of the osmoadaptive glycine betaine synthesis route of B. subtilis (Fig. 1) is the cellular and genetic events that lead to a repression of glycine betaine production once the osmotically challenged cell has accomplished a physiologically sufficient degree of cellular hydration and raised turgor again. The presence of choline in the soil habitat of B. subtilis (41) will inevitably lead to choline uptake caused by the basal activity of the OpuB and OpuC transporters (36) (Fig. 4A and Table 2). Import of the inducer choline will then relieve GbsR-mediated repression of the opuB and gbsAB operons and will thereby trigger enhanced choline uptake and continued synthesis of glycine betaine, regardless of whether the B. subtilis cell is osmotically stressed or not. If glycine betaine synthesis would remain unchecked, an undue rise in turgor would ensue, and this would force the B. subtilis cell eventually to jettison part of the newly synthesized glycine betaine either through the transient opening of mechanosensitive channels (27, 29, 60) or through specialized excretion systems (21, 40, 43). The genetic data obtained by us suggest that B. subtilis has found an elegant way to avoid such a wasteful behavior, since the properties of the GbsR repressor are responsive to the presence of glycine betaine, which can counteract the inducing effects of choline (Table 3 and Fig. 7).
Whatever might be the precise molecular mechanism that allows glycine betaine to promote the repressor function of the GbsR protein in the presence of the inducer choline, the data presented here show that the GbsR regulator integrates signals required both for the onset of glycine betaine synthesis from the precursor choline and for tuning down glycine betaine production when the B. subtilis cell has relieved itself from osmotic stress through the accumulation of this compatible solute.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jutta Gade, Maritha Lippmann, and Jochen Sohn for their dedicated and expert technical assistance and greatly value the help of Vickie Koogle in the language editing of the manuscript. We greatly appreciate the discussions with our colleague Lutz Schmitt and Sander H. Smits (University of Düsseldorf, Düsseldorf, Germany) on the choline-binding properties of the GbsR protein and thank Gregor Lentzen (Bitop AG) for a kind gift of ectoine.
Our work on the GbsR protein was supported by the LOEWE program of the state of Hessen (via the Centre for Synthetic Microbiology, SYNMIKRO, Marburg) and the Fonds der Chemischen Industrie.
Footnotes
Published ahead of print 9 March 2012
G.N.-W., D.O., and A.R. contributed equally to this article.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Aktas M, Jost KA, Fritz C, Narberhaus F. 2011. Choline uptake in Agrobacterium tumefaciens by the high-affinity ChoXWV transporter. J. Bacteriol. 193:5119–5129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 3. Arnold K, Bordoli L, Kopp J, Schwede T. 2006. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22:195–201 [DOI] [PubMed] [Google Scholar]
- 4. Boch J, Kempf B, Bremer E. 1994. Osmoregulation in Bacillus subtilis: synthesis of the osmoprotectant glycine betaine from exogenously provided choline. J. Bacteriol. 176:5364–5371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boch J, Kempf B, Schmid R, Bremer E. 1996. Synthesis of the osmoprotectant glycine betaine in Bacillus subtilis: characterization of the gbsAB genes. J. Bacteriol. 178:5121–5129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Boch J, Nau-Wagner G, Kneip S, Bremer E. 1997. Glycine betaine aldehyde dehydrogenase from Bacillus subtilis: characterization of an enzyme required for the synthesis of the osmoprotectant glycine betaine. Arch. Microbiol. 168:282–289 [DOI] [PubMed] [Google Scholar]
- 7. Borwankar T, et al. 2011. Natural osmolytes remodel the aggregation pathway of mutant huntingtin exon 1. Biochemistry 50:2048–2060 [DOI] [PubMed] [Google Scholar]
- 8. Bourot S, et al. 2000. Glycine betaine-assisted protein folding in a lysA mutant of Escherichia coli. J. Biol. Chem. 275:1050–1056 [DOI] [PubMed] [Google Scholar]
- 9. Bremer E. 2002. Adaptation to changing osmolarity, p 385–391 In Sonenshein AL, Hoch JA, Losick R. (ed), Bacillus subtilis and its closest relatives. ASM Press, Washington, DC [Google Scholar]
- 10. Bremer E. 2011. A look into the aromatic cage. Environ. Microbiol. Rep. 3:1–5 [Google Scholar]
- 11. Bremer E, Krämer R. 2000. Coping with osmotic challenges: osmoregulation through accumulation and release of compatible solutes, p 79–97 In Storz G, Hengge-Aronis R. (ed), Bacterial stress responses. ASM Press, Washington, DC [Google Scholar]
- 12. Brigulla M, et al. 2003. Chill induction of the SigB-dependent general stress response in Bacillus subtilis and its contribution to low-temperature adaptation. J. Bacteriol. 185:4305–4514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Brill J, Hoffmann T, Bleisteiner M, Bremer E. 2011. Osmotically controlled synthesis of the compatible solute proline is critical for cellular defense of Bacillus subtilis against high osmolarity. J. Bacteriol. 193:5335–5346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bult CJ, et al. 1996. Complete genome sequence of the methanogenic archaeon Methanococcus jannaschii. Science 273:1058–1073 [DOI] [PubMed] [Google Scholar]
- 15. Burkhardt J, Sewald X, Bauer B, Saum SH, Müller V. 2009. Synthesis of glycine betaine from choline in the moderate halophile Halobacillus halophilus: co-regulation of two divergent, polycistronic operons. Environ. Microbiol. Rep. 1:38–43 [DOI] [PubMed] [Google Scholar]
- 16. Chattopadhyay MK, et al. 2004. The chemical chaperone proline relieves the thermosensitivity of a dnaK deletion mutant at 42°C. J. Bacteriol. 186:8149–8152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Diamant S, Eliahu N, Rosenthal D, Goloubinoff P. 2001. Chemical chaperones regulate molecular chaperones in vitro and in cells under combined salt and heat stresses. J. Biol. Chem. 276:39586–39591 [DOI] [PubMed] [Google Scholar]
- 18. Dougherty DA. 1996. Cation-pi interactions in chemistry and biology: a new view of benzene, Phe, Tyr, and Trp. Science 271:163–168 [DOI] [PubMed] [Google Scholar]
- 19. Du Y, et al. 2011. Structures of the substrate-binding protein provide insights into the multiple compatible solute binding specificities of the Bacillus subtilis ABC transporter OpuC. Biochem. J. 436:283–289 [DOI] [PubMed] [Google Scholar]
- 20. Gajiwala KS, Burley SK. 2000. Winged helix proteins. Curr. Opin. Struct. Biol. 10:110–116 [DOI] [PubMed] [Google Scholar]
- 21. Glaasker E, Konings WN, Poolman B. 1996. Glycine betaine fluxes in Lactobacillus plantarum during osmostasis and hyper- and hypo-osmotic shock. J. Biol. Chem. 271:10060–10065 [DOI] [PubMed] [Google Scholar]
- 22. Gotsche S, Dahl MK. 1995. Purification and characterization of the phospho-α-(1,1)glucosidase (TreA) of Bacillus subtilis 168. J. Bacteriol. 177:2721–2726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gu ZJ, Wang L, Le Rudulier D, Zhang B, Yang SS. 2008. Characterization of the glycine betaine biosynthetic genes in the moderately halophilic bacterium Halobacillus dabanensis D-8T. Curr. Microbiol. 57:306–311 [DOI] [PubMed] [Google Scholar]
- 24. Hahne H, et al. 2010. A comprehensive proteomics and transcriptomics analysis of Bacillus subtilis salt stress adaptation. J. Bacteriol. 192:870–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Harwood CR, Archibald AR. 1990. Growth, maintenance and general techniques, p 1–26 In Harwood CR, Cutting SM. (ed), Molecular biological methods for Bacillus. John Wiley & Sons, Chichester, United Kingdom [Google Scholar]
- 26. Harwood CR, Cutting SM. 1990. Molecular biological methods for Bacillus. John Wiley & Sons, Chichester, United Kingdom [Google Scholar]
- 27. Haswell ES, Phillips R, Rees DC. 2011. Mechanosensitive channels: what can they do and how do they do it? Structure 19:1356–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Helmann JD. 1995. Compilation and analysis of Bacillus subtilis sigma A-dependent promoter sequences: evidence for extended contact between RNA polymerase and upstream promoter DNA. Nucleic Acids Res. 23:2351–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hoffmann T, Boiangiu C, Moses S, Bremer E. 2008. Responses of Bacillus subtilis to hypotonic challenges: physiological contributions of mechanosensitive channels to cellular survival. Appl. Environ. Microbiol. 74:2454–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hoffmann T, Bremer E. 2011. Protection of Bacillus subtilis against cold stress via compatible-solute acquisition. J. Bacteriol. 193:1552–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Holtmann G, Bremer E. 2004. Thermoprotection of Bacillus subtilis by exogenously provided glycine betaine and structurally related compatible solutes: involvement of Opu transporters. J. Bacteriol. 186:1683–1693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Horn C, et al. 2006. Molecular determinants for substrate specificity of the ligand-binding protein OpuAC from Bacillus subtilis for the compatible solutes glycine betaine and proline betaine. J. Mol. Biol. 357:592–606 [DOI] [PubMed] [Google Scholar]
- 33. Huffman JL, Brennan RG. 2002. Prokaryotic transcription regulators: more than just the helix-turn-helix motif. Curr. Opin. Struct. Biol. 12:98–106 [DOI] [PubMed] [Google Scholar]
- 34. Kappes RM, Bremer E. 1998. Response of Bacillus subtilis to high osmolarity: uptake of carnitine, crotonobetaine and butyrobetaine via the ABC transport system OpuC. Microbiology 144:83–90 [DOI] [PubMed] [Google Scholar]
- 35. Kappes RM, Kempf B, Bremer E. 1996. Three transport systems for the osmoprotectant glycine betaine operate in Bacillus subtilis: characterization of OpuD. J. Bacteriol. 178:5071–5079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kappes RM, et al. 1999. Two evolutionarily closely related ABC transporters mediate the uptake of choline for synthesis of the osmoprotectant glycine betaine in Bacillus subtilis. Mol. Microbiol. 32:203–216 [DOI] [PubMed] [Google Scholar]
- 37. Kempf B, Bremer E. 1995. OpuA, an osmotically regulated binding protein-dependent transport system for the osmoprotectant glycine betaine in Bacillus subtilis. J. Biol. Chem. 270:16701–16713 [DOI] [PubMed] [Google Scholar]
- 38. Kempf B, Bremer E. 1998. Uptake and synthesis of compatible solutes as microbial stress responses to high osmolality environments. Arch. Microbiol. 170:319–330 [DOI] [PubMed] [Google Scholar]
- 39. Kimura Y, Kawasaki S, Yoshimoto H, Takegawa K. 2010. Glycine betaine biosynthesized from glycine provides an osmolyte for cell growth and spore germination during osmotic stress in Myxococcus xanthus. J. Bacteriol. 192:1467–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Koo SP, Higgins CF, Booth IR. 1991. Regulation of compatible solute accumulation in Salmonella typhimurium: evidence for a glycine betaine efflux system. J. Gen. Microbiol. 137:2617–2625 [DOI] [PubMed] [Google Scholar]
- 41. Kortstee GJ. 1970. The aerobic decomposition of choline by microorganisms. Arch. Microbiol. 71:235–244 [PubMed] [Google Scholar]
- 42. Lamark T, et al. 1991. DNA sequence and analysis of the bet genes encoding the osmoregulatory choline-glycine betaine pathway of Escherichia coli. Mol. Microbiol. 5:1049–1064 [DOI] [PubMed] [Google Scholar]
- 43. Lamark T, Styrvold OB, Strom AR. 1992. Efflux of choline and glycine betaine from osmoregulating cells of Escherichia coli. FEMS Microbiol. Lett. 75:149–154 [DOI] [PubMed] [Google Scholar]
- 44. Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 45. Nau-Wagner G, Boch J, Le Good JA, Bremer E. 1999. High-affinity transport of choline-O-sulfate and its use as a compatible solute in Bacillus subtilis. Appl. Environ. Microbiol. 65:560–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nyyssöl̈ A, Kerovuo J, Kaukinen P, von Weymarn N, Reinikainen T. 2000. Extreme halophiles synthesize betaine from glycine by methylation. J. Biol. Chem. 275:22196–22201 [DOI] [PubMed] [Google Scholar]
- 47. Oswald C, et al. 2008. Crystal structures of the choline/acetylcholine substrate-binding protein ChoX from Sinorhizobium meliloti in the liganded and unliganded-closed states. J. Biol. Chem. 283:32848–32859 [DOI] [PubMed] [Google Scholar]
- 48. Pittelkow M, Tschapek B, Smits SH, Schmitt L, Bremer E. 2011. The crystal structure of the substrate-binding protein OpuBC from Bacillus subtilis in complex with choline. J. Mol. Biol. 411:53–67 [DOI] [PubMed] [Google Scholar]
- 49. Quaye O, Cowins S, Gadda G. 2009. Contribution of flavin covalent linkage with histidine 99 to the reaction catalyzed by choline oxidase. J. Biol. Chem. 284:16990–16997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ray SS, et al. 2003. X-ray structure of an M. jannaschii DNA-binding protein: implications for antibiotic resistance in S. aureus. Proteins 50:170–173 [DOI] [PubMed] [Google Scholar]
- 51. Rosenstein R, Futter-Bryniok D, Götz F. 1999. The choline-converting pathway in Staphylococcus xylosus C2A: genetic and physiological characterization. J. Bacteriol. 181:2273–2278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Schiefner A, et al. 2004. Cation-pi interactions as determinants for binding of the compatible solutes glycine betaine and proline betaine by the periplasmic ligand-binding protein ProX from Escherichia coli. J. Biol. Chem. 279:5588–5596 [DOI] [PubMed] [Google Scholar]
- 53. Smits SH, et al. 2008. The compatible-solute-binding protein OpuAC from Bacillus subtilis: ligand binding, site-directed mutagenesis, and crystallographic studies. J. Bacteriol. 190:5663–5671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Srivatsan A, et al. 2008. High-precision, whole-genome sequencing of laboratory strains facilitates genetic studies. PLoS Genet. 4:e1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Steil L, Hoffmann T, Budde I, Völker U, Bremer E. 2003. Genome-wide transcriptional profiling analysis of adaptation of Bacillus subtilis to high salinity. J. Bacteriol. 185:6358–6370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Street TO, Krukenberg KA, Rosgen J, Bolen DW, Agard DA. 2010. Osmolyte-induced conformational changes in the Hsp90 molecular chaperone. Protein Sci. 19:57–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Thompson JD, Plewniak F, Thierry JC, Poch O. 2000. DbClustal: rapid and reliable global multiple alignments of protein sequences detected by database searches. Nucleic Acids Res. 28:2919–2926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tschapek B, et al. 2011. Arg149 is involved in switching the low affinity, open state of the binding protein AfProX into its high affinity, closed state. J. Mol. Biol. 411:36–52 [DOI] [PubMed] [Google Scholar]
- 59. Waditee R, et al. 2005. Genes for direct methylation of glycine provide high levels of glycinebetaine and abiotic-stress tolerance in Synechococcus and Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 102:1318–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wahome PG, Cowan AE, Setlow B, Setlow P. 2009. Levels and localization of mechanosensitive channel proteins in Bacillus subtilis. Arch. Microbiol. 191:403–414 [DOI] [PubMed] [Google Scholar]
- 61. Whatmore AM, Chudek JA, Reed RH. 1990. The effects of osmotic upshock on the intracellular solute pools of Bacillus subtilis. J. Gen. Microbiol. 136:2527–2535 [DOI] [PubMed] [Google Scholar]
- 62. Whatmore AM, Reed RH. 1990. Determination of turgor pressure in Bacillus subtilis: a possible role for K+ in turgor regulation. J. Gen. Microbiol. 136:2521–2526 [DOI] [PubMed] [Google Scholar]
- 63. Wood JM, et al. 2001. Osmosensing and osmoregulatory compatible solute accumulation by bacteria. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 130:437–460 [DOI] [PubMed] [Google Scholar]
- 64. Yancey PH. 2005. Organic osmolytes as compatible, metabolic and counteracting cytoprotectants in high osmolarity and other stresses. J. Exp. Biol. 208:2819–2830 [DOI] [PubMed] [Google Scholar]
- 65. Ziegler C, Bremer E, Krämer R. 2010. The BCCT family of carriers: from physiology to crystal structure. Mol. Microbiol. 78:13–34 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.