Abstract
CRISPR (clustered regularly interspaced short palindromic repeats) elements and cas (CRISPR-associated) genes are widespread in Bacteria and Archaea. The CRISPR/Cas system operates as a defense mechanism against mobile genetic elements (i.e., viruses or plasmids). Here, we investigate seven CRISPR loci in the genome of the crenarchaeon Thermoproteus tenax that include spacers with significant similarity not only to archaeal viruses but also to T. tenax genes. The analysis of CRISPR RNA (crRNA) transcription reveals transcripts of a length between 50 and 130 nucleotides, demonstrating the processing of larger crRNA precursors. The organization of identified cas genes resembles CRISPR/Cas subtype I-A, and the core cas genes are shown to be arranged on two polycistronic transcripts: cascis (cas4, cas1/2, and csa1) and cascade (csa5, cas7, cas5a, cas3, cas3′, and cas8a2). Changes in the environmental parameters such as UV-light exposure or high ionic strength modulate cas gene transcription. Two reconstitution protocols were established for the production of two discrete multipartite Cas protein complexes that correspond to their operonic gene arrangement. These data provide insights into the specialized mechanisms of an archaeal CRISPR/Cas system and allow selective functional analyses of Cas protein complexes in the future.
INTRODUCTION
Clustered regularly interspaced short palindromic repeats (CRISPR) are a recently discovered type of direct repeats found in prokaryotes. The repeats, ranging in size from 24 to 48 nucleotides (nt), are separated by spacer sequences of similar sizes (18, 38, 57). CRISPR loci are flanked by an AT-rich leader sequence of up to 550 nt in length. In most cases, the repeats are highly conserved within a CRISPR array, whereas spacers are unique within a given locus, even among strains of the same species (59). These remarkable sequence patterns are widely distributed in Bacteria and Archaea, since loci are found in ca. 46% of the bacterial genomes and in 84% of all Archaea, but not in viruses or eukaryotic genome sequences (23, 24). Similarity searches of CRISPR spacers showed that some sequences match viruses and other extrachromosomal elements, such as plasmids, but rarely also chromosomal DNA (7, 15, 56, 76). CRISPR loci are transcribed and processed into a series of smaller CRISPR RNAs (crRNAs), corresponding to spacer units with termini processed within the repeat region (29, 45, 72, 73). Initially, four genes, always located near a CRISPR locus and only found in species containing CRISPR sequences, were identified in numerous prokaryotic genomes and therefore designated as CRISPR-associated (cas) genes (38). Further cas genes were identified and assembled into 41 cas gene families (27, 47, 49) that define a set of 10 CRISPR system subtypes (50).
Recently, it has been demonstrated that in response to phage infection, Streptococcus thermophilus integrates new spacers into its CRISPR arrays, which results in CRISPR-mediated phage resistance. The newly integrated spacers were derived from the genome of the challenging phage, evidenced by 100% identity of spacers and phage sequences (5, 17, 35, 36). In Escherichia coli, the Cascade complex (CRISPR-associated complex for antiviral defense), composed of the Cas proteins Cse1, Cse2, Cas7, Cas5, and Cas6e, cleaves long CRISPR transcripts specifically within the repeat sequence into crRNAs with a length of 61 nt. Subsequently, the Cascade bound crRNA serves as a guiding molecule to target and attack the foreign nucleic acid (10, 40). In addition, the CRISPR/Cas system prevents conjugation and plasmid transformation in Staphylococcus epidermidis and E. coli by targeting plasmid DNA (52, 54, 78). Thus, the CRISPR/Cas system seems not only limited to phage defense but also to play a more general role in the prevention of horizontal gene transfer and the maintenance of genetic integrity in Bacteria and Archaea (41, 53).
Furthermore, CRISPR and cas genes seem to be involved in multiple processes in prokaryotes. E. coli cells with a growth adaptation to 41.5°C for 2,000 generations yield duplicated and recombined CRISPR loci, leading to an increased fitness of the strain (60). Microarray data of Pyrococcus furiosus cells exposed to ionizing radiation revealed a 10-fold increased transcript level of a cas operon (81). Finally, the protein Cas1 has a dual function in E. coli, since it also interacts with key components of DNA repair systems (4).
In Archaea, it remains challenging to pinpoint the different roles of the CRISPR/Cas systems since the molecular mechanisms of only a few CRISPR and Cas functions have been characterized. Details on crRNA maturation and type III interference were obtained for P. furiosus (30, 31). The study of Sulfolobus solfataricus provided insights into the structural basis of an archaeal Cascade-like complex for a type I CRISPR/Cas system (46).
Here, we present genome and transcription analyses of CRISPR loci and cas genes in the crenarchaeon Thermoproteus tenax. T. tenax is the first described hyperthermophilic archaeon, which has an optimal growth temperature of 86°C and was isolated from a solfatare in Iceland (20, 85). Nine T. tenax Cas proteins could be reconstituted as two Cas complexes: a six-protein extended Cascade-like complex and a novel Cas complex that is proposed to mediate the integration of spacers (CRISPR-associated complex for the integration of spacers [Cascis]).
Our results assess the T. tenax CRISPR/Cas-system as a member of the archaeal subtype I-A system, found, e.g., also in Aeropyrum pernix, Pyrobaculum aerophilum, or S. solfataricus. Subtype I-A CRISPR/Cas systems are mainly characterized by the presence of csa1 in the vicinity of cas4, cas1, and cas2 genes. Furthermore, cas3 is split into two discrete genes (cas3 and cas3′) and found next to csa5, cas8a, cas5, cas7, and cas6 genes (50). Finally, we provide further indications that the CRISPR/Cas systems of prokaryotes have a multifunctional response, not only toward phage infection and horizontal gene transfer, including transfection and transformation, but also toward abiotic stress situations (e.g., ionic stress or irradiation).
MATERIALS AND METHODS
Strains and growth conditions.
Mass cultures of T. tenax Kra1 (DSM 2078) (20, 85) were grown heterotrophically in Brock's medium containing 0.2% yeast extract (wt/vol) at 86°C as described previously (66). To induce salt stress, the NaCl concentration of the growth medium was increased up to 50 to 150 mM by adding respective volumes of a sterile 5 M NaCl stock solution to cultures at late exponential phase following further incubation for 3 to 6 h. Temperature variation was achieved by incubating the culture for 3 h in a water bath at 91°C; a reference vessel filled with water served as a control. For UV treatment, cultures concentrated to 50 ml (cell count: 8 × 108 cells/ml) were transferred to petri dishes (12 cm by 12 cm by 3 mm) under anaerobic atmosphere, irradiated with UV light for 30 s to 2 min (5 to 20 J/m2) at 254 nm (6 W), while shaking the culture carefully, and further incubated for 3 h at 86°C in the dark.
E. coli strains DH5α (Invitrogen) and Rosetta (DE3) (Stratagene) for cloning and expression studies were cultured in LB medium under standard conditions according to the manufacturer's instructions.
CRISPR and cas gene analyses.
The complete genome sequence of T. tenax is available (68). Archaeal and bacterial genome sequences were obtained from the National Center for Biotechnology Information. The CRISPR database CRISPRdb (24), the CRISPRFinder software tool (25), and the CRISPI database (61) were used to retrieve and identify CRISPR repeats and spacer sequences. CRISPR orientations were determined by analyzing the AT-content of the 300 bp located up- and downstream region and by identifying conserved BRE site and TATA box motifs. For spacer sequence similarity analyses, comparisons to public sequences were carried out using BLASTn (2).
To evaluate the significance of BLAST matches, only matches with E-values below 0.005 and above significant cutoffs, estimated as previously described (45), were retained and manually those matches identified, which had identities greater than 50%. cas genes were identified by analyzing protein sequences by BLASTp and comparison to respective Pfam (19), TIGRFam (28), and COG numbers. Multiple sequence alignments and phylogenetic analyses were carried out by using CLUSTAL W2 (43). Secondary structure predictions were obtained by using CDD (51), Jpred 3 (14), and 2Zip (9).
Northern blot analyses of small RNAs.
For preparations of small RNA species (<200 nt) the mirVana miRNA isolation kit (Ambion) was used according to the manufacturer's instructions. Electrophoretic separation of 1 μg of small RNA was achieved by fractionation in 12% polyacrylamide gels (8 M urea, 90 mM Tris, 90 mM boric acid, 2 mM EDTA [pH 8]), together with a 10- to 300-bp Ultra Low Range DNA Ladder (Fermentas). The RNA was blotted by capillary transfer onto nylon membranes (Roti-Nylon Plus; Roth) and immobilized by UV cross-linking. Hybridization was performed at room temperature for 18 h in DIG Easy Hyb buffer (Roche) with 0.5 pmol/ml oligonucleotides (43-mer probes; see Table S1 in the supplemental material) complementary to spacer sequences and end labeled with DIG-11-ddUTP (Roche). The sample was washed three times (2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate] and 0.1% sodium dodecyl sulfate [SDS]) at room temperature for 10 min and subsequently at 42°C in the same buffer. The blot was directly used for immunological detection. The 5S rRNA served as a loading control. The secondary structure of ssRNA was predicted using the RNAfold web server (26).
RT-PCR and Southern blot analyses.
For the detection of cas transcripts a semiquantitative reverse transcription-PCR (RT-PCR)-based assay was combined with Southern hybridization methodology. Total RNA was prepared from T. tenax cells by using TRIzol reagent and the RNeasy minikit according to the instructions of the manufacturers (Invitrogen and Qiagen, respectively). On-column DNase I treatment was performed as described previously (83). Equal amounts of total RNA (1.5 to 3 μg) was reverse transcribed with 5 μM random hexamer primer and Moloney murine leukemia virus (M-MuLV) reverse transcriptase at 45°C for 60 min using a First-Strand cDNA synthesis kit (Fermentas). In a negative control, total RNA was mixed without M-MuLV reverse transcriptase in the cDNA generating process. PCR amplification was performed with 2 μl of synthesized cDNA as a template and 1 μM cas gene-specific primers in a 50-μl reaction volume (primer sequences [see Table S2 in the supplemental material]). PCR products were cleaned with a PCR purification kit (Qiagen). Then, 20-μl portions of secondary cDNAs were fractionated in 1% agarose gels and transferred to nylon membranes by capillary blotting. Subsequently, Southern hybridization was performed at 52°C for 18 h in DIG Easy Hyb buffer with DIG-11-UTP-labeled antisense mRNA probes of cas genes (Roche). After hybridization, the blots were stringently washed up to 68°C in 0.1× SSC and 0.1% SDS and directly used for immunological detection. Different mRNA levels of stressed and control cells were determined with the help of the ImageJ software. Values were normalized to a 16S rRNA internal standard.
Cloning and heterologous expression of cas genes in E. coli.
The single core cas genes organized in operons cascis (cas4, cas1/2, and csa1) and cascade (csa5, cas7, cas5a, cas3, cas3′, and cas8a2) were amplified by PCR using Pfu polymerase (Fermentas), genomic T. tenax DNA as a template, and specific primer sets (PCR primers [see Table S3 in the supplemental material]). Afterward, the cleaned PCR products were cloned via restriction digestion and ligation (Fermentas) into vector pET-15b or pET-24a(+), respectively (Novagen). The sequences of the cloned genes were confirmed by automated sequencing of both strands (LGC Genomics). Expression of the recombinant enzymes in E. coli Rosetta(DE3) was performed by the addition of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) in accordance with the manufacturer's instructions (Stratagene).
Reconstitution of recombinant Cas complexes.
Due to very low amounts of proteins in the soluble fractions, 5 g of recombinant E. coli Rosetta(DE3) Cas4, Cas1/2, Csa1, Cas5a, Cas3, Cas3′, and Cas8a2 cells was used for the purification of inclusion bodies and protein solubilization in 4 M guanidine hydrochloride (GuHCl) as described previously (44, 62). The concentration of solubilized proteins was determined by Bradford protein quantification method. Reconstitution of the protein complex Cascis was carried out by rapid dilution in GuHCl-free buffer. Equal amounts (170 μg) of each solubilized protein Cas4, Cas1/2, and Csa1 were pooled and refolded by adding the solution stepwise to 20 ml of refolding buffer 1 (40 mM Tris-HCl [pH 7], 10 mM β-mercaptoethanol, 10% glycerol, 300 mM NaCl, 500 mM l-arginine) at room temperature. After refolding, the solution was centrifuged (14,000 × g, 15 min, 4°C) to remove precipitated protein and analyzed by SDS-PAGE. The protein complex Cascade was reconstituted by removal of the denaturing agent in a stepwise dialysis against GuHCl-free buffer. Equal amounts (100 μg) of each solubilized protein—Cas5a, Cas3, Cas3′, and Cas8a2—were pooled with 100 μg of the soluble and purified proteins Csa5 and Cas7. For purification, 1 g of recombinant E. coli Rosetta(DE3) Csa5 or the respective Cas7 cells were resuspended in buffer 2 (100 mM HEPES/KOH [pH 7], 10% glycerol, 10 mM β-mercaptoethanol, 10 mM CaCl2, 300 mM NaCl), passed three times through a French pressure cell at 1,100 lb/in2, centrifuged (45,000 × g, 45 min, 4°C), heat precipitated for 30 min at 80°C, and again centrifuged (13,000 × g, 30 min, 4°C). The supernatant was dialyzed overnight at 4°C against buffer 2 without NaCl, and Csa5 was applied on a Q-Sepharose Fast Flow column (Amersham) or Cas7 was applied on a heparin Sepharose 6 Fast Flow column (Amersham), respectively (flow rate, 0.5 ml/min), followed by elution with a linear salt gradient of 0 to 1 M NaCl in a total volume of 100 ml. Fractions containing Csa5 or Cas7, respectively, were pooled, dialyzed, and stored in the presence of 25% glycerol at −20°C. The pooled six proteins of Cascade were mixed with 5 ml of buffer 3 (3 M GuHCl, 2 M urea, 100 mM HEPES/KOH [pH 7], 10% glycerol, 300 mM NaCl, 10 mM CaCl2, 10 mM β-mercaptoethanol) and 30 μg of total T. tenax RNA for supporting the reconstitution process. The Cascade solution was stepwise dialyzed at room temperature to remove denaturing agents against native buffer 2 and after dialysis handled in accordance with the Cascis preparation.
Molecular mass determination.
The native molecular mass was determined by gel filtration on a Superose 6 10/300 preparatory-grade column (Pharmacia, volume 24 ml) as described previously (67).
RESULTS AND DISCUSSION
Characterization of CRISPRs and cas genes in T. tenax.
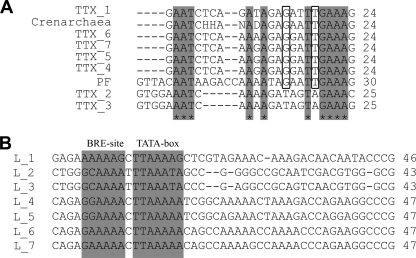
In the genome of T. tenax, seven CRISPR loci with a repeat length of 24 to 25 bp were identified; these were termed TTX_1 to TTX_7 (68). An alignment (Fig. 1A) of the seven repeat elements clearly shows an identical repeat sequence for TTX_4 to TTX_7 (24 bp), slight differences for TTX_1 (24 bp), and a diverging sequence for TTX_2 and TTX_3 (25 bp). BLAST analyses revealed that all seven T. tenax repeats have the highest homology to members of the family Thermoproteaceae (e.g., T. neutrophilus and P. aerophilum), indicating a similar repeat variation among different cluster within these genomes. A comparison of the seven T. tenax repeat elements to the well-studied repeat element of P. furiosus (12, 13, 29, 77) and to the consensus sequence of all crenarchaeal repeat elements (retrieved from reference 61), revealed a highly conserved 7-bp tag and a conserved upstream G residue (5′-GxxTTGAAAG-3′). In TTX_2 and TTX_3, the 7-bp tag is slightly varied (5′-TxxTAGAAAS-3′). A total of 142 unique spacer sequences with lengths ranging from 37 to 57 bp were identified within the seven CRISPR loci, and 73% of all spacers averaged between 41 and 46 bp. The leader sequences that encode the CRISPR promoter and potentially direct the integration of new spacers were allocated for each CRISPR locus by analyzing the AT-rich intergenic CRISPR-flanking regions and within these archaeal BRE sites and TATA box motifs were defined. The differences detected among the seven repeat elements are also reflected in the homology of the leader sequences, since identical sequence blocks were only found between TTX_1 and TTX_4 to TTX_7 (up to 63.5% sequence identity) and between TTX_2 and 3 (up to 77% sequence identity). Homology of all leader sequences (overall only up to 26% sequence identity) could only be detected for the BRE sites and TATA boxes (Fig. 1B).
Fig 1.
Conservation of repeat elements and leader alignment of CRISPR loci. (A) The repeat elements of the seven CRISPR loci were compared to the crenarchaeal consensus repeat and the well-studied repeat element of P. furiosus (PF). The crenarchaeal consensus repeat was defined by performing a multiple sequence alignment of all repeat elements of CRISPR-containing crenarchaeal organism retrieved from the CRISPI database. Identical nucleotides in all repeats were highlighted by gray boxes and identical nucleotides in all repeats except of TTX_2 and TTX_3 were highlighted by black borders. (B) The leader sequences were allocated for each CRISPR locus by analyzing the AT-rich intergenic CRISPR-flanking regions and within these archaeal BRE sites and TATA box motifs were defined. Potential BRE sites and TATA box motifs were highlighted by gray shading.
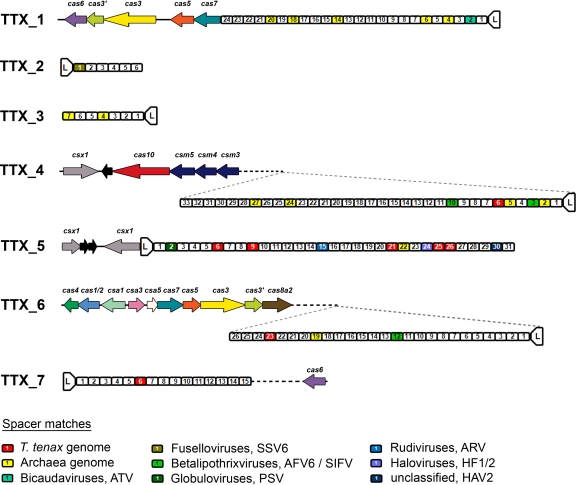
A total of 23 conserved cas genes was identified adjacent to five of the seven CRISPR loci. cas genes were not found in the vicinity of TTX_2 and TTX_3. The core cas genes (cas1 to cas7) and the subtype-specific cas genes (cas8a2, csa1, csa3, and csa5, subtype I-A, Apern) were located between TTX_5 and TTX_6 (Fig. 2). cas6 is not located directly adjacent to this set of genes, but found 17 open reading frames downstream and close to TTX_7. In addition, a csm gene cluster (cas10 and csm3 to csm5, subtype III-A, Mtube) was detected upstream of TTX_4. A subset of core cas genes (cas3, cas3′, and cas5 to cas7) was also identified upstream of TTX_1. It is noteworthy that a unique feature of the T. tenax CRISPR/Cas system is the fusion of the sole cas1 and cas2 genes, which is strong evidence for an interrelated function of the respective proteins. Fusions of these cas genes was also observed in other genomes, e.g., cas1 is fused to cas4 in Geobacter sulfurreducens, and cas2 and cas3 are fused in subtype I-F (75).
Fig 2.
CRISPR loci, spacer similarities and adjacent cas genes in T. tenax. Seven CRISPR loci (TTX_1 to TTX_7) were analyzed from the genome and spacers consecutively numbered (for example, spacer 1.1 is spacer 1 from TTX_1) starting from the localization of the leader sequence (L). All 142 spacer sequences were searched for sequence similarities and spacers with significant homology to archaeal viruses, Archaea, or T. tenax genes are indicated by colors. For every CRISPR loci the adjacent cas genes were identified by analyzing protein sequences by BLASTp and comparison to respective Pfam (19), TIGRFam (28), and COG numbers. CRISPR locus TTX_1 is surrounded by a subset of core cas genes (ORFs, TTX_0232 to TTX_0235). At locus TTX_4 the csm gene cluster (csx1, cas10, and csm3 to csm5, subtype III-A; ORFs, TTX_1228 to TTX_1233) is located and, in addition, at locus TTX_5 (ORFs, TTX_1240 and TTX_1243) two genes with homology to csx1 were identified. The specific genes for subtype I-A can be found at TTX_6 with two clusters (cas4, cas1/2, and csa1 and csa5, cas7, cas5, cas3, cas3′, and cas8a2, subtype I-A; ORFs, TTX_1245 to TTX_1255) and a potential regulator csa3 (ORF, TTX_1249) in between. A second cas6 (ORF, TTX_1272) gene is located near to TTX_7. Identical genes were marked by the same colors.
Abundance of crRNAs.
Different CRISPR loci are usually transcribed into long RNA transcripts, potentially covering the entire cluster and subsequently stepwise processed into small crRNAs (29, 72, 73). In P. furiosus mature crRNAs were characterized by having common 5′-sequence tags (8 nt) and distinct 3′ ends ranging from 0 to 22 nt (31).
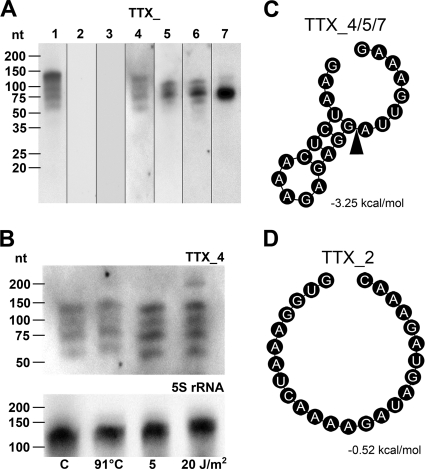
To find out whether all seven CRISPR loci of T. tenax are transcribed, we performed Northern blot analyses of small RNA species (≤200 nt) probed with antisense CRISPR spacers (each 43 nt) of the seven loci. Different crRNA transcripts were identified for five CRISPR loci (TTX_1, TTX_4, and TTX_5 to TTX_7) (Fig. 3A). The detected RNAs corresponded to the theoretical sizes of precursor-crRNAs (pre-crRNA) and mature crRNAs, with lengths of ∼130 nt (pre-crRNA [2× spacer/repeat]: 2× 43 nt + 2× 24 nt), 110 nt (pre-crRNA [2× spacer/repeat/trimmed repeat]: 2× 43 nt + 24 nt + 8 nt), 70 nt (pre-crRNA [spacer/repeat]: 43 nt + 24 nt), and 50 nt (crRNA [spacer/trimmed repeat]: 43 nt + 8 nt). These results suggest that the large pre-crRNAs are stepwise processed within the 24-nt repeats, leading to a crRNA with a full spacer flanked by the hallmark 8-nt-long 5′ handle and a variable 3′ handle. The alignment of crenarchaeal repeats (Fig. 1A) shows the importance of the conserved 3′-terminal repeat bases for the generation of this 8-nt-long 5′ handle of the mature crRNA. This handle was shown to be utilized by the endoribonuclease Cas6 in P. furiosus (12, 13). To analyze whether CRISPR transcription is an ongoing process under different growth conditions (Fig. 3B), we utilized Northern blot methodology to compare small RNAs of T. tenax cultures grown under standard conditions (heterotrophic, 86°C) and of cultures grown under differentiating conditions (irradiated at 5 or 20 J/m2 by UV light or incubated at 91°C). No quantitative differences were detected between the control and stressed cells of T. tenax. For the two CRISPR loci TTX_2 and TTX_3, which display a divergent structure of their repeats, no transcripts could be observed either due to missing transcription or due to a lower stability of the crRNAs. The calculation of the minimum free energy folding of the repeat elements revealed a similar stem-loop structure for all transcribed CRISPR loci (calculated stabilization energy for TTX_1 of 6.38 kcal/mol and for TTX_4 to TTX_7 of 3.25 kcal/mol) (Fig. 3C and D). For the CRISPR loci without detectable crRNAs, however, the formation of a secondary structure seems to be improbable (calculated stabilization energy for TTX_2 and TTX_3 of 0.52 kcal/mol). Since TTX_2 and TTX_3 are not flanked by cas genes, one explanation for different crRNA abundance could be that this repeat sequence or structure is not or only weakly processed by a Cas6 protein that is associated with the other five clusters. The general stability of the RNAs might also be affected. The leader sequences of both active and seemingly inactive CRISPR clusters appear to contain all of the elements necessary for the transcription of the precursor crRNA (Fig. 1B).
Fig 3.
Northern blot analyses of CRISPR loci and secondary structure prediction for repeat sequences. (A) Transcripts of the seven T. tenax CRISPR loci was detected by Northern blot methodology of small RNAs (≤200 nt). Antisense spacers of the respective CRISPR loci served as probes (see sequences in Table S1 in the supplemental material). For CRISPR loci TTX_2 and TTX_3, no distinct signals were detectable. (B) Comparison of CRISPR transcripts TTX_4 from control (lane C, at 86°C), 91°C temperature increase (91°C), and 5- and 20-J/m2 UV-light-stressed T. tenax cells. 5S rRNA served as a loading control. (C and D) Secondary structure prediction of repeat sequences analyzed with RNAfold (17). TTX_4/5/7 clusters share the same repeat sequence (C), and predictions for TTX_1 and TTX_6 revealed comparable secondary structures, as depicted. The black arrow marks the potential cutting site for the generation of the 8-nt long 5′ handle. Repeats of TTX_2/3 clusters share a potential unfolded repeat (D). Depicted are the minimum free energy structures and their free energy values.
Similarity of CRISPR spacers with T. tenax genes and viruses.
CRISPR/Cas systems are described as a prokaryotic immune system against extrachromosomal elements, such as plasmids or viruses. This function requires a significant sequence similarity of the CRISPR spacers with these genetic elements (7, 15, 56, 76). Hence, all T. tenax CRISPR spacers were checked for similarity against the genome sequences of 42 viruses known to infect archaeal organisms (Fig. 2). Surprisingly, CRISPR spacers showed no significant similarity to Thermoproteus tenax virus 1 (TTV1) or Thermoproteus tenax spherical virus 1 (TTSV1) and only one spacer matched Pyrobaculum spherical virus (PSV), which specifically infects T. tenax (34). One spacer matches to Hyperthermophilic archaeal virus 2 (HAV2), whose currently unknown host is among hyperthermophilic neutrophiles (21), six spacers match to viruses known to infect archaeal acidothermophiles, including Acidianus two-tailed virus (ATV), Acidianus filamentous virus 6 (AFV6), or Sulfolobus islandicus filamentous virus (SIFV) and one spacer matches to the mesophilic haloviruses 1 and 2 (HF1/2; see Table S4 in the supplemental material). Nearly all target sites were located within viral reading frames, but partial mismatches suggest that ancient viruses or close relatives of sequenced viruses left their traces in T. tenax CRISPR loci upon previous infections. Interestingly, studies of the interaction between P. aeruginosa and the bacteriophage DMS3 showed that multiple point mutations are tolerated between the spacer and target sequence, which demonstrates that imperfect matches should be taken into account in spacer analysis (11, 84).
In addition, 12 T. tenax spacers were identified to have significant similarity with other prokaryotic genomes, mostly of closely related crenarchaeal organisms (Thermofilum pendens, P. aerophilum, and T. neutrophilus). One remarkable feature of the T. tenax CRISPRs are the matches between spacers and T. tenax reading frames. Complete identity (37 nt) could be detected between spacer 4.6 (spacer number six of CRISPR locus TTX_4) and TTX_0660, a gene which includes a transmembrane-helices pattern and probably encodes an adhesin-like protein. Partial identities were detected for spacer 5.6 targeting a gene encoding an ATPase of the Cdc46/MCM family (TTX_0274), spacer 5.9 with homology to a gene coding for a Fe2+-dependent formamide hydrolase and spacer 6.23 targeting the TATA box motif of a phosphatase of the histone macroH2A1 family (TTX_1457). Spacers 5.21 and 5.26, as well as spacers 5.25 and 7.6 are homologous to each other, indicating their shared origin (see Table S4 in the supplemental material).
The presence of endogenous sequences within CRISPR spacers raises the question of escaping targeting of the host genome, and mechanisms for the discrimination of host DNA and viral targets were described (54, 70). Targets contain a protospacer adjacent motif (PAM) and the differential complementarity of the 5′ tag of a crRNA with a PAM is supposed to discriminate between the CRISPR DNA and the foreign target DNA (40). The relevance of the PAM was shown in S. thermophilus, since phages that had overcome the host immunity were mutated within this short motif (17). In the S. epidermidis Csm-type CRISPR this discrimination is apparently utilized without a specific motif, but only by the lack of complementarity between the 5′ tag of a crRNA and target (54). In T. tenax, these potentially self-destructing spacers are not located within inactive CRISPR loci with mutated repeats, not surrounded by degenerated cas operons and do not have a proviral origin (70). Analysis of the PAM of the T. tenax spacers matching viral and archaeal genomes revealed a low conservation of an NCC motif upstream of the target, which corresponds to the PAM sequence in the genus Sulfolobus (45, 55). The analysis of the PAM motif for the four identified matches within the T. tenax genome did not reveal the NCC motif, probably preventing the function of crRNAs on host DNA. Interestingly, self-targeting was shown for an active spacer against essential tRNA synthetase in Pelobacter carbinolicus (1).
It remains unknown why and how T. tenax integrated such host spacers into its CRISPR. So what can be the function of potentially self-targeting spacers in the CRISPR/Cas system? The interfering reaction is utilized via the complementary binding of the crRNA to its target DNA or RNA, which lead to its nucleolytic degradation (39, 74, 75). It is possible that partially mismatching crRNAs might bind a host target, which could result in the recruitment of Cas proteins without subsequent DNA degradation for the regulation of the encoded target.
cas genes are organized in operons, and the transcription is regulated by abiotic stress conditions.
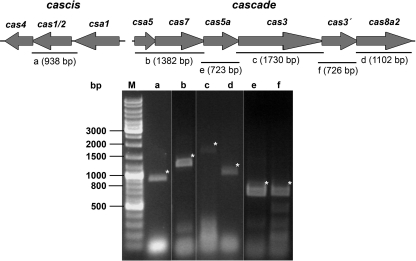
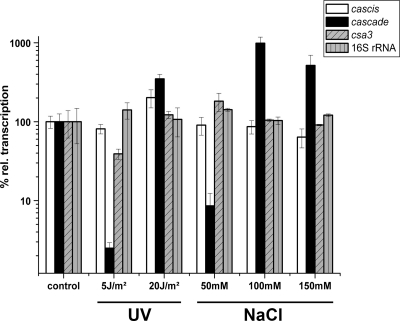
Because of clustering, gene orientations, TATA box motifs, and BRE sites in front and overlapping start/stop codons of open reading frames (ORFs), two core cas gene operons—cascis and cascade—could be identified. A separate putative transcriptional regulator csa3 with a typical HTH-motif (TTX_1249) is located between these operons (73 bp downstream of the cascis gene csa1 and 36 bp upstream of the first cascade gene csa5). RT-PCR results confirmed that both genomic units are organized as operons, since every overlapping part of the polycistronic transcripts could be detected by a specific PCR product (Fig. 4). The transcript of cascade is most likely leaderless, as the csa5 gene lacks a consensus Shine-Dalgarno (SD) motif and the “internal” genes possess well-defined SD motifs in front of the initiation codons (GGAG or GGGG, at nucleotide positions −7 to −4 of the start codon). For the cascis operon, consensus SD motifs were located at nt −12 to −15 upstream of the start codon. To assess the transcription levels of the operons cascis and cascade in response to applied abiotic stress, a semiquantitative RT-PCR-based assay with gene specific primers for cas4 (representing cascis), cas3 (representing cascade), and csa3 was combined with Southern hybridization for identification of the products. The results indicate that the transcription level of cascade was affected by the varied parameter (Fig. 5). The cas3 gene showed a >3-fold-increased transcript level in cells treated with UV light at 20 J/m2 in comparison to the control cells. Furthermore, the mRNA levels of the cas3 gene were >10-fold increased for cells grown with 100 mM NaCl and 5-fold increased in the presence of at 150 mM NaCl. It is noteworthy that, in the presence of 50 mM NaCl, the transcript level was decreased 7-fold, indicating a strong regulation of cas3 depending on the environmental input. Also, the transcript level of the cas4 gene was slightly increased 2-fold in UV light-treated cells (20 J/m2). The csa3 gene and the 16S rRNA were not significantly affected. We conclude that cascade is more sensitive toward our tested abiotic stress conditions and therefore potentially also for environmental changes than cascis or the regulator csa3.
Fig 4.
Core cas genes are organized in two operon structures. Polycistronic mRNAs cascis (TTX_1245 to TTX_1248) and cascade (TTX_1250 to TTX_1255) were identified by amplification of cDNA and the products were separated on agarose gels. PCR products (lanes a to f, marked by asterisks) of overlapping gene regions correlated to the calculated length (primer sets see Table S2 in the supplemental material). M, DNA ladder. As a negative control, primer sets binding outside of the polycistronic mRNA were used (data not shown).
Fig 5.
Transcript levels of cascis, cascade, and csa3 in control and stressed T. tenax cells. The mRNA levels of cascis, cascade, and csa3 in control cells and stressed cells (UV light for 5 and 20 J/m2, as well as 50, 100, and 150 mM NaCl) were analyzed by electrophoretic separation of amplified cDNA and subsequent Southern blot identification. Values were normalized to a 16S rRNA internal standard and negative controls were performed without the RT step. The data quantification of signals from three independent experiments was conducted with ImageJ. Black bars show the standard deviation.
Our results complement previous studies on the influence of abiotic stress on the transcription of cas genes. Further examples for induction effects of cascade by abiotic stress factors are Methanocaldococcus jannaschii cells that were heat shocked from 88 to 98°C, which showed a remarkable upregulation of heat shock genes and cas7 (8). Microarray data of P. furiosus exposed to ionizing radiation (2,500 Gy) revealed up to 10-fold increased level for the polycistronic transcript of a cas operon, resembling cascade of T. tenax, whereas a second cas operon, similar to cascis showed no significant changes (81). The addition of 0.5 mM H2O2 to a growing P. furiosus culture increased the transcription of some genes encoded in the above-mentioned cas operon up to 6-fold (71). UV-light stress (200 J/m2) studied in S. solfataricus revealed that cas genes were slightly affected and, e.g., 1.5 to 2 h of UV-light exposition resulted in the 2-fold upregulation of cas8a2 (22).
In summary, the Cascade complex appears to be the main target for differentiated transcriptional regulation under stress conditions. Potentially, such stress conditions mimic an attack of mobile genetic elements, in which it is the role of the Cascade complex to target the invading nucleic acid for destruction.
Recombinant Cas proteins are strongly interacting.
The core cas genes proposed to be involved in the adaptation of spacers and the interference against invading nucleic acids are arranged on polycistronic transcripts (Fig. 4). It has been shown for few CRISPR/Cas subtypes that at least some Cas proteins form complexes, termed Cascade, to attack viral DNA (10, 40, 46, 65). In E. coli, Cascade is a 405-kDa complex comprised of five essential proteins (CasA1B2C6D1E1) and a 61-nt crRNA (40).
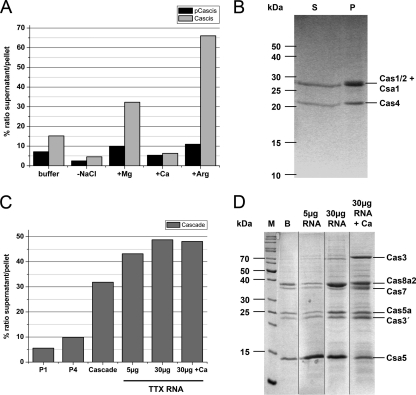
To understand functionality and molecular interaction of the proteins, we sought to express the individual proteins heterologously in E. coli. Various proteins could not be detected in soluble form in crude extracts, but only as inclusion bodies, suggesting that the single proteins were unable to adopt their correct structure during expression and might require interactions with other Cas proteins in vivo. To obtain Cas proteins for biochemical studies, a synergistic reconstitution by mixing equal amounts of the three unfolded proteins of operon cascis (Cas4, Cas1/2, and Csa1) in 4 M GuHCl, followed by rapid dilution in GuHCl-free refolding buffer, was applied. The highest recovery of soluble protein was obtained in the presence of 500 mM l-arginine and 10 mM Mg2+. Surprisingly, the recovery of soluble protein was improved significantly in the presence of all three proteins (Fig. 6A) in comparison to parallel assays with only one or two components of the proposed complex. Overall, ∼1 mg of recombinant Cascis complex was obtained from 1 g of cells by this methodology.
Fig 6.
Reconstitution assays for recombinant Cas complexes Cascis and Cascade. (A) Denatured proteins Cas4, Cas1/2, and Csa1 were combined and refolded into the Cascis complex by rapid dilution. The ratio (%) between complete protein content of supernatant and pellet in different refolding buffers is shown: buffer (40 mM Tris-HCl [pH 7], 10 mM β-mercaptoethanol, 10% glycerol, 300 mM NaCl), −NaCl (buffer without NaCl), +Mg (buffer plus 10 mM MgCl2), +Ca (buffer plus 10 mM CaCl2), and +Arg (buffer plus 500 mM l-arginine). Cascis, complete complex; pCascis, partial complex (Cas4, Csa1). (B) SDS gel electropherogram of the reconstitution assay (buffer plus 500 mM l-arginine). Lane S, supernatant fraction (refolded complex); lane P, pellet fraction (aggregated protein). (C) Denatured proteins Cas5a, Cas3, Cas3′, and Cas8a2 and soluble purified proteins Csa5 and Cas7 were reconstituted to Cascade by stepwise dialysis. The ratio (%) between the complete protein content of the supernatant and pellet in refolding buffer (100 mM HEPES/KOH [pH 7], 10% glycerol, 300 mM NaCl, 10 mM β-mercaptoethanol; for refolding of p1CasA2, p4CasA2, and CasA2) and the addition of 5 or 30 μg of total T. tenax RNA or 30 μg of total T. tenax RNA and 10 mM CaCl2 is shown. Partial complexes: P1 (single Cas proteins) and P4 (four proteins [Cas5a, Cas3, Cas3′, and Cas8a2]). (D) SDS-gel electropherogram for the identification of the constituents of Cascade in refolding buffer under the conditions described above. Depicted are the supernatant fractions (M, protein marker; B, refolding of Cascade without addition of RNA).
For the complex Cascis, the Cas1 protein is a universal marker of the CRISPR/Cas system, represented nearly in all genomes (48–50). Cas1 of Pseudomonas aeruginosa showed DNA-endonuclease activity, generating fragments of ∼80 bp (80). In contrast, Cas1 of S. solfataricus bound DNA and RNA with high affinity in a nonspecific manner, but no nuclease activity was detected (33). Cas2 of S. solfataricus was characterized as an Mg-dependent endoribonuclease with a ferredoxin-like fold, specific for single-stranded RNA, but not for crRNAs with a preferential cleavage site within U-rich regions (6, 63). Cas4 and Csa1 are not yet enzymatically characterized, but both proteins belong to a restriction endonuclease-like superfamily, defined by a conserved PD-(D/E)XK-motif and are functional similar to RecB exonucleases (42). These proteins are generally thought to be involved in the adaptation and integration of new spacers (50); however, complex formation of these Cas proteins has not yet been detected in vivo and the described activities only partly correspond to this proposed role. Our in vitro results indicate for the first time that these Cas proteins might not act individually in the cell but would require closely coordinated activities within the proposed Cascis interactions.
A similar strategy for refolding was applied for proteins of the Cascade complex, since four of the six individual proteins were found to be insoluble (Cas5a, Cas3, Cas3′, and Cas8a2). The soluble proteins Csa5 and Cas7 could be directly purified and enriched by ion-exchange chromatography. Equal amounts (100 μg) of the solubilized and purified proteins were refolded by mixing them in a GuHCl-containing buffer and stepwise decreasing the GuHCl concentration in the buffer. Refolding assays with one (P1, refolding of Cas3 or Cas3′) or four proteins (P4, refolding of Cas5a, Cas3, Cas3′, and Cas8a2) resulted in minimal amounts of refolded protein in the supernatant (5 to 10%). The refolding experiments with all proteins encoded in the cascade operon revealed maximal recovery of a complex in the presence of up to 30 μg of T. tenax total RNA and of 10 mM CaCl2 exhibiting a 1:1 ratio of the six proteins (Fig. 6D) and an approximate molecular mass of 300 kDa, which differs from the uneven stoichiometry of Cas7 found in E. coli Cascade (40).
Some proteins of the analyzed Cascade complex were previously biochemically characterized. The E. coli Cascade complex has a minimal protein core that is composed of a backbone of six CasC (Cas7) subunits, in conjunction with CasD (Cas5e) and CasE (Cas6) (40, 79). In S. solfataricus, a putative arCascade complex was analyzed in which Cas7 and Cas5a play a central role in binding crRNA and complementary ssDNA (46). The Cas3 protein is a member of the DEAD/DEAH-box helicases with typical motifs for substrate binding (Mg2+, ATP and RNA) and helicase activity (37, 58, 69). The recombinant Cas3′ protein from S. solfataricus showed unspecific degradation of double-stranded DNA and double-stranded RNA (32) and belongs to a superfamily of metal-dependent phosphohydrolases (82). In T. tenax, we see further indications for a Cas protein complex that fulfills the interference function and comprises the proteins described for the minimal core of Cascade and of arCascade. However, our in vitro studies indicate that proper refolding of the Cascade complex requires the presence of six proteins, which suggests that the archaeal Cascade complex might be larger than previously thought. Interaction of other Cas proteins within the arCascade complex has been indicated before (46) but has not yet been biochemically verified.
The intermolecular stability of these heterologously produced complexes (Cascis and Cascade) and their operon organization support that this might represent their native structure in vivo. The established reconstitution assay for Cascis and Cascade offers the advantage of studying the function of the whole complexes and enabling defined mutagenesis approaches gaining insight into the roles played by individual proteins.
Conclusion.
Our results provide further indications that CRISPR/Cas systems have complex roles in prokaryotes. They function as a defense system that is able to protect a host cell against invading foreign nucleic acid. In T. tenax spacer sequences similar to archaeal viruses underline this role. However, since spacers are similar not only to foreign genetic sequences but also to chromosomal sequences of the T. tenax host genome, it is possible that CRISPR/Cas systems similarly fulfill a regulatory or interfering function. Our data also show that the transcription of cas genes is modulated by environmental factors. Previously, it was hypothesized that the CRISPR/Cas system is involved in multiple cellular information processes, such as chromosomal segregation, homologous recombination, or DNA repair (4, 16, 57, 60). Indeed, a stabilizing function of the genome, of plasmids or other cell components is plausible for the CRISPR/Cas system, since the presence of CRISPR loci within the chromosome is proposed to have a stabilizing effect on the DNA structure (18, 38, 64). This can be an explanation for the observed correlation of the number of CRISPR loci and cas genes with an increased optimal growth temperature (3). Taken together, the diverse impact on the archaeal cell, besides its defense against mobile genetic elements, is only beginning to be fully realized. The established reconstitution of two Cas protein complexes is an important step for future in vitro studies to characterize their involvement in diverse CRISPR/Cas functions within Archaea.
Supplementary Material
ACKNOWLEDGMENTS
We thank Melanie Zaparty (University of Regensburg) for providing T. tenax 16S rRNA and hexokinase primer.
This study was supported by the Deutsche Forschungsgemeinschaft (GRK1431) and the Max-Planck Society.
Footnotes
Published ahead of print 9 March 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Aklujkar M, Lovley DR. 2010. Interference with histidyl-tRNA synthetase by a CRISPR spacer sequence as a factor in the evolution of Pelobacter carbinolicus. BMC Evol. Biol. 10:230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 3. Anderson RE, Brazelton WJ, Baross JA. 2011. Using CRISPRs as a metagenomic tool to identify microbial hosts of a diffuse flow hydrothermal vent viral assemblage. FEMS Microbiol. Ecol. 77:120–133 [DOI] [PubMed] [Google Scholar]
- 4. Babu M, et al. 2011. A dual function of the CRISPR-Cas system in bacterial antivirus immunity and DNA repair. Mol. Microbiol. 79:484–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barrangou R, et al. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712 [DOI] [PubMed] [Google Scholar]
- 6. Beloglazova N, et al. 2008. A novel family of sequence-specific endoribonucleases associated with the clustered regularly interspaced short palindromic repeats. J. Biol. Chem. 283:20361–20371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. 2005. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 151:2551–2561 [DOI] [PubMed] [Google Scholar]
- 8. Boonyaratanakornkit BB, Miao LY, Clark DS. 2007. Transcriptional responses of the deep-sea hyperthermophile Methanocaldococcus jannaschii under shifting extremes of temperature and pressure. Extremophiles 11:495–503 [DOI] [PubMed] [Google Scholar]
- 9. Bornberg-Bauer E, Rivals E, Vingron M. 1998. Computational approaches to identify leucine zippers. Nucleic Acids Res. 26:2740–2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brouns SJ, et al. 2008. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321:960–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cady KC, O'Toole GA. 2011. Non-identity-mediated CRISPR-bacteriophage interaction mediated via the Csy and Cas3 proteins. J. Bacteriol. 193:3433–3445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Carte J, Pfister NT, Compton MM, Terns RM, Terns MP. 2010. Binding and cleavage of CRISPR RNA by Cas6. RNA 16:2181–2188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Carte J, Wang R, Li H, Terns RM, Terns MP. 2008. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev. 22:3489–3496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cole C, Barber JD, Barton GJ. 2008. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 36:W197–W201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cui Y, et al. 2008. Insight into microevolution of Yersinia pestis by clustered regularly interspaced short palindromic repeats. PLoS One 3:e2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. DeBoy RT, Mongodin EF, Emerson JB, Nelson KE. 2006. Chromosome evolution in the Thermotogales: large-scale inversions and strain diversification of CRISPR sequences. J. Bacteriol. 188:2364–2374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Deveau H, et al. 2008. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J. Bacteriol. 190:1390–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fadiel A, Lithwick S, Ganji G, Scherer SW. 2003. Remarkable sequence signatures in archaeal genomes. Archaea 1:185–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Finn RD, et al. 2008. The Pfam protein families database. Nucleic Acids Res. 36:D281–D288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fischer F, Zillig W, Stetter KO, Schreiber G. 1983. Chemolithoautotrophic metabolism of anaerobic extremely thermophilic archaebacteria. Nature 301:511–513 [DOI] [PubMed] [Google Scholar]
- 21. Garrett RA, et al. 2010. Metagenomic analyses of novel viruses and plasmids from a cultured environmental sample of hyperthermophilic neutrophiles. Environ. Microbiol. 12:2918–2930 [DOI] [PubMed] [Google Scholar]
- 22. Gotz D, et al. 2007. Responses of hyperthermophilic crenarchaea to UV irradiation. Genome Biol. 8:R220.1–R220.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Grissa I, Vergnaud G, Pourcel C. 2008. CRISPRcompar: a website to compare clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 36:W145–W148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Grissa I, Vergnaud G, Pourcel C. 2007. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinform. 8:172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Grissa I, Vergnaud G, Pourcel C. 2007. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 35:W52–W57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gruber AR, Lorenz R, Bernhart SH, Neuboock R, Hofacker IL. 2008. The Vienna RNA Websuite. Nucleic Acids Res. 36:W70–W74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Haft DH, Selengut J, Mongodin EF, Nelson KE. 2005. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput. Biol. 1:e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Haft DH, Selengut JD, White O. 2003. The TIGRFAMs database of protein families. Nucleic Acids Res. 31:371–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hale C, Kleppe K, Terns RM, Terns MP. 2008. Prokaryotic silencing (psi)RNAs in Pyrococcus furiosus. RNA 14:2572–2579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hale CR, et al. 2012. Essential features and rational design of CRISPR RNAs that function with the Cas RAMP module complex to cleave RNAs. Mol. Cell, 45:292–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hale CR, et al. 2009. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell 139:945–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Han D, Krauss G. 2009. Characterization of the endonuclease SSO2001 from Sulfolobus solfataricus P2. FEBS Lett. 583:771–776 [DOI] [PubMed] [Google Scholar]
- 33. Han D, Lehmann K, Krauss G. 2009. SSO1450: a CAS1 protein from Sulfolobus solfataricus P2 with high affinity for RNA and DNA. FEBS Lett. 583:1928–1932 [DOI] [PubMed] [Google Scholar]
- 34. Haring M, et al. 2004. Morphology and genome organization of the virus PSV of the hyperthermophilic archaeal genera Pyrobaculum and Thermoproteus: a novel virus family, the Globuloviridae. Virology 323:233–242 [DOI] [PubMed] [Google Scholar]
- 35. Horvath P, Barrangou R. 2010. CRISPR/Cas, the immune system of bacteria and archaea. Science 327:167–170 [DOI] [PubMed] [Google Scholar]
- 36. Horvath P, et al. 2008. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J. Bacteriol. 190:1401–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Howard JA, Delmas S, Ivancic-Bace I, Bolt EL. 2011. Helicase dissociation and annealing of RNA-DNA hybrids by Escherichia coli Cas3 protein. Biochem. J. 439:85–95 [DOI] [PubMed] [Google Scholar]
- 38. Jansen R, Embden JD, Gaastra W, Schouls LM. 2002. Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol. 43:1565–1575 [DOI] [PubMed] [Google Scholar]
- 39. Jore MM, Brouns SJ, van der Oost J. 2 February 2011. RNA in defense: CRISPRs protect prokaryotes against mobile genetic elements. Cold Spring Harbor Perspect. Biol. doi:10.1101/cshperspect.a003657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jore MM, et al. 2011. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat. Struct. Mol. Biol. 18:529–536 [DOI] [PubMed] [Google Scholar]
- 41. Karginov FV, Hannon GJ. 2010. The CRISPR system: small RNA-guided defense in bacteria and archaea. Mol. Cell 37:7–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kinch LN, Ginalski K, Rychlewski L, Grishin NV. 2005. Identification of novel restriction endonuclease-like fold families among hypothetical proteins. Nucleic Acids Res. 33:3598–3605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Larkin MA, et al. 2007. CLUSTAL W and CLUSTAL X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 44. Lilie H, Schwarz E, Rudolph R. 1998. Advances in refolding of proteins produced in Escherichia coli. Curr. Opin. Biotechnol. 9:497–501 [DOI] [PubMed] [Google Scholar]
- 45. Lillestol RK, et al. 2009. CRISPR families of the crenarchaeal genus Sulfolobus: bidirectional transcription and dynamic properties. Mol. Microbiol. 72:259–272 [DOI] [PubMed] [Google Scholar]
- 46. Lintner NG, et al. 2011. Structural and functional characterization of an archaeal clustered regularly interspaced short palindromic repeat (CRISPR)-associated complex for antiviral defense (CASCADE). J. Biol. Chem. 286:21643–21656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Makarova KS, Aravind L, Grishin NV, Rogozin IB, Koonin EV. 2002. A DNA repair system specific for thermophilic Archaea and Bacteria predicted by genomic context analysis. Nucleic Acids Res. 30:482–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Makarova KS, Aravind L, Wolf YI, Koonin EV. 2011. Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR-Cas systems. Biol. Direct 6:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. 2006. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol. Direct 1:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Makarova KS, et al. 2011. Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 9:467–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Marchler-Bauer A, et al. 2009. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 37:D205–D210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Marraffini LA, Sontheimer EJ. 2008. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322:1843–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Marraffini LA, Sontheimer EJ. 2010. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat. Rev. Genet. 11:181–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Marraffini LA, Sontheimer EJ. 2010. Self versus non-self-discrimination during CRISPR RNA-directed immunity. Nature 463:568–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Almendros C. 2009. Short motif sequences determine the targets of the prokaryotic CRISPR defense system. Microbiology 155:733–740 [DOI] [PubMed] [Google Scholar]
- 56. Mojica FJM, Diez-Villasenor C, Garcia-Martinez J, Soria E. 2005. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 60:174–182 [DOI] [PubMed] [Google Scholar]
- 57. Mojica FJM, Ferrer C, Juez G, Rodriguezvalera F. 1995. Long stretches of short tandem repeats are present in the largest replicons of the archaea Haloferax mediterranei and Haloferax volcanii and could be involved in replicon partitioning. Mol. Microbiol. 17:85–93 [DOI] [PubMed] [Google Scholar]
- 58. Mulepati S, Bailey S. 2011. Structural and biochemical analysis of nuclease domain of clustered regularly interspaced short palindromic repeat (CRISPR)-associated protein 3 (Cas3). J. Biol. Chem. 286:31896–31903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Pourcel C, Salvignol G, Vergnaud G. 2005. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology 151:653–663 [DOI] [PubMed] [Google Scholar]
- 60. Riehle MM, Bennett AF, Long AD. 2001. Genetic architecture of thermal adaptation in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 98:525–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rousseau C, Gonnet M, Le Romancer M, Nicolas J. 2009. CRISPI: a CRISPR interactive database. Bioinformatics 25:3317–3318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Rudolph R, Lilie H. 1996. In vitro folding of inclusion body proteins. FASEB J. 10:49–56 [PubMed] [Google Scholar]
- 63. Samai P, Smith P, Shuman S. 2010. Structure of a CRISPR-associated protein Cas2 from Desulfovibrio vulgaris. Acta Crystallogr. Sect F Struct. Biol. Crystallogr. Commun. 66:1552–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Schleper C, Holz I, Janekovic D, Murphy J, Zillig W. 1995. A multicopy plasmid of the extremely thermophilic archaeon sulfolobus effects its transfer to recipients by mating. J. Bacteriol. 177:4417–4426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Semenova E, et al. 2011. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc. Natl. Acad. Sci. U. S. A. 108:10098–10103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Siebers B, Hensel R. 1993. Glucose catabolism of the hyperthermophilic archaeum Thermoproteus tenax. FEMS Microbiol. Lett. 111:1–8 [Google Scholar]
- 67. Siebers B, Wendisch VF, Hensel R. 1997. Carbohydrate metabolism in Thermoproteus tenax: in vivo utilization of the non-phosphorylative Entner-Doudoroff pathway and characterization of its first enzyme, glucose dehydrogenase. Arch. Microbiol. 168:120–127 [DOI] [PubMed] [Google Scholar]
- 68. Siebers B, et al. 2011. The complete genome sequence of Thermoproteus tenax: a physiologically versatile member of the crenarchaeota. PLoS One 6:e24222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sinkunas T, et al. 2011. Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in the CRISPR/Cas immune system. EMBO J. 30:1335–1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Stern A, Keren L, Wurtzel O, Amitai G, Sorek R. 2010. Self-targeting by CRISPR: gene regulation or autoimmunity? Trends Genet. 26:335–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Strand KR, et al. 2010. Oxidative stress protection and the repair response to hydrogen peroxide in the hyperthermophilic archaeon Pyrococcus furiosus and in related species. Arch. Microbiol. 192:447–459 [DOI] [PubMed] [Google Scholar]
- 72. Tang TH, et al. 2002. Identification of 86 candidates for small non-messenger RNAs from the archaeon Archaeoglobus fulgidus. Proc. Natl. Acad. Sci. U. S. A. 99:7536–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Tang TH, et al. 2005. Identification of novel non-coding RNAs as potential antisense regulators in the archaeon Sulfolobus solfataricus. Mol. Microbiol. 55:469–481 [DOI] [PubMed] [Google Scholar]
- 74. Terns MP, Terns RM. 2011. CRISPR-based adaptive immune systems. Curr. Opin. Microbiol. 14:321–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. van der Oost J, Jore MM, Westra ER, Lundgren M, Brouns SJ. 2009. CRISPR-based adaptive and heritable immunity in prokaryotes. Trends Biochem. Sci. 34:401–407 [DOI] [PubMed] [Google Scholar]
- 76. Vergnaud G, et al. 2007. Analysis of the three Yersinia pestis CRISPR loci provides new tools for phylogenetic studies and possibly for the investigation of ancient DNA. Adv. Exp. Med. Biol. 603:327–338 [DOI] [PubMed] [Google Scholar]
- 77. Wang R, Preamplume G, Terns MP, Terns RM, Li H. 2011. Interaction of the Cas6 riboendonuclease with CRISPR RNAs: recognition and cleavage. Structure 19:257–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Westra ER, et al. 2010. H-NS-mediated repression of CRISPR-based immunity in Escherichia coli K-12 can be relieved by the transcription activator LeuO. Mol. Microbiol. 77:1380–1393 [DOI] [PubMed] [Google Scholar]
- 79. Wiedenheft B, et al. 2011. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature 477:486–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wiedenheft B, et al. 2009. Structural basis for DNase activity of a conserved protein implicated in CRISPR-mediated genome defense. Structure 17:904–912 [DOI] [PubMed] [Google Scholar]
- 81. Williams E, Lowe TM, Savas J, DiRuggiero J. 2007. Microarray analysis of the hyperthermophilic archaeon Pyrococcus furiosus exposed to gamma irradiation. Extremophiles 11:19–29 [DOI] [PubMed] [Google Scholar]
- 82. Yakunin AF, et al. 2004. The HD domain of the Escherichia coli tRNA nucleotidyltransferase has 2′,3′-cyclic phosphodiesterase, 2′-nucleotidase, and phosphatase activities. J. Biol. Chem. 279:36819–36827 [DOI] [PubMed] [Google Scholar]
- 83. Zaparty M, et al. 2008. DNA microarray analysis of central carbohydrate metabolism: glycolytic/gluconeogenic carbon switch in the hyperthermophilic crenarchaeum Thermoproteus tenax. J. Bacteriol. 190:2231–2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Zegans ME, et al. 2009. Interaction between bacteriophage DMS3 and host CRISPR region inhibits group behaviors of Pseudomonas aeruginosa. J. Bacteriol. 191:210–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Zillig W, et al. 1981. Thermoproteales: a novel type of extremely thermoacidophilic anaerobic archaebacteria isolated from Icelandic solfataras. Zentralbl. Bakteriol. Mikrobiol. Hyg. I Abt. Orig. C-Allgemeine Angew. Okol. Mikrobiol. 2:205–227 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.