Abstract
As a pioneer colonizer of the oral cavity, Actinomyces oris expresses proteinaceous pili (also called fimbriae) to mediate the following two key events in biofilm formation: adherence to saliva deposits on enamel and interbacterial associations. Assembly of type 2 fimbriae that directly facilitate coaggregation with oral streptococci and Actinomyces biofilm development requires the class C sortase SrtC2. Although the general sortase-associated mechanisms have been elucidated, several structural attributes unique to the class C sortases require functional investigation. Mutational studies reported here suggest that the N-terminal transmembrane (TM) region of SrtC2, predicted to contain a signal peptide sequence, is cleaved off the mature protein and that this processing is critical for the proper integration of the enzyme at the cytoplasmic membrane, which is mediated by the extended hydrophobic C terminus containing a TM domain and a cytoplasmic tail. Deletion of this putative TM or the entire cytoplasmic domain abolished the enzyme localization and functionality. Alanine substitution of the conserved catalytic Cys-His dyad abrogated the SrtC2 enzymatic activity. In contrast, mutations designed to alter a “lid” domain that covers the catalytic pocket of a class C sortase showed no effect on enzyme activity. Finally, each of the deleterious mutations that affected SrtC2 activity or membrane localization also eliminated Actinomyces species biofilm development and bacterial coaggregation with streptococci. We conclude that the N terminus of SrtC2, which contains the signal sequence, is required for proper protein translocation and maturation, while the extended C-terminal hydrophobic region serves as a stable membrane anchor for proper enzyme functionality.
INTRODUCTION
Oral biofilms are a complex communities of microbial organisms that dwell on enamel and gingival tissue surfaces. Commonly referred to as dental plaque, this complex microbial community, consisting of over 700 identified species, is associated with root caries, gingivitis, and periodontal disease (11). Actinomyces and oral streptococcal species are the predominant pioneer colonizers of this environment and thus important for establishing favorable conditions for the incorporation of other microbes (11, 24), including Fusobacterium species, bridging bacteria for late colonizers (24). Actinomyces species produce two antigenically and functionally distinct types of fimbriae or pili that are required for the aforementioned interaction of Actinomyces bacteria and oral streptococci and the adherence of Actinomyces cells to the tooth surface (33). Type 1 fimbriae promote bacterial adherence to salivary proline-rich proteins (PRPs) coating the tooth surface (8), while type 2 fimbriae mediate adherence of Actinomyces not only to oral streptococci but also to various host cells, including erythrocytes and epithelial cells (4, 19, 33).
In Actinomyces oris, the most abundant species among various Actinomyces spp. in the human oral cavity (28), the genetic components for type 1 and 2 fimbrial assembly are arranged in two distinct gene clusters (20). Encoded by the fimQ-fimP-srtC1 cluster, a type 1 fimbria is composed of the fimbrial shaft FimP and the tip fimbrillin FimQ, which is the major adhesin interacting with PRPs (32). On the other hand, a type 2 fimbria, encoded by the fimB-fimA-srtC2 cluster, is made of the fimbrial shaft FimA and the tip fimbrillin FimB (20). We showed that FimA is essential for A. oris coaggregation with oral streptococci, adherence to red blood cells (RBCs), and biofilm development (22). Assembly of type 1 fimbrial polymers requires sortase SrtC1 (32), whereas type 2 fimbrial assembly involves sortase SrtC2 (22). An Actinomyces mutant lacking srtC2 fails to coaggregate with oral streptococci, adhere to RBCs, and form biofilms (22).
SrtC1 and SrtC2 are membrane-bound transpeptidase enzymes (16) that belong to class C sortases (5, 7) or fimbria-specific sortases (13, 20). The first sortase enzyme was discovered in Staphylococcus aureus, termed SrtA (18), which is the prototype of class A sortases. Classification of sortases was based solely on their primary sequence and phylogenetic analysis (5, 7). What distinguishes class C sortases from those of class A is the presence of a carboxy-terminal hydrophobic domain after the sortase signature motif TLXTC (7). Sortases of both classes have an amino-terminal transmembrane (TM) helix that harbors a signal peptide sequence (5, 7). In S. aureus, it is thought that the N-terminal signal peptide of SrtA is not cleaved during protein translocation and thus serves as a membrane anchor (17, 18). Our previous studies of C. diphtheriae pilin-specific sortase SrtA (a class C sortase) revealed that the C-terminal hydrophobic domain of SrtA is essential for the enzyme to be inserted into the membrane, hence its polymerization activity (9). Consistently, work in Enterococcus faecalis demonstrated that the requirement of the C-terminal domain of pilin-specific sortase SrtC in efficient pilus polymerization (10). More recently, it was shown in Streptococcus agalactiae that both the N- and C-terminal TM regions of pilin-specific sortase SrtC1 are required for the enzyme activity (6). A key remaining question is whether the N-terminal TM of pilin-specific sortases is cleaved, thus liberating the enzyme N terminus from membrane association.
We present here a structure-function analysis of the fimbria-specific sortase (class C sortase) SrtC2 of A. oris. Our mutational analysis reveals the importance of the signal peptide sequence and its cleavage in the proper enzyme localization at the membrane and fimbrial polymerization activity. We show further that the C-terminal region, which contains a TM helix and a cytoplasmic domain, is indispensable for the enzyme membrane localization and pilus polymerization. Furthermore, the conserved catalytic residues Cys246 and His184 are essential to the enzymatic activity of SrtC2. Importantly, we show that mutations that disrupt the N- and C-terminal regions of SrtC2 result in the elimination of Actinomyces biofilms and bacterial coaggregation with streptococci. Together, these findings provide a better understanding of the structural features that distinguish the two families of sortases involved in pilus biogenesis critical for pathogenesis of Gram-positive bacteria.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
Bacterial strains and plasmids used in this study are listed in Table 1. Actinomyces bacteria were grown in heart infusion broth (HIB) or on heart infusion agar (HIA) plates. Escherichia coli strains were grown in Luria-Bertani broth (LB). When needed, kanamycin was added at a concentration of 50 μg ml−1. Rabbit-raised polyclonal antibodies against recombinant fimbrial proteins were obtained previously (22). Reagents were purchased from Sigma unless indicated otherwise.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| A. oris strains | ||
| MG1 | Type strain, expressing type 1 and 2 fimbriae | 20 |
| AR1 | ΔsrtC2; an isogenic derivative of MG1 | 20 |
| AR2 | AR1 containing pSrtC2 | 22 |
| WU10 | AR1 containing pSrtC2VV | This study |
| MR1 | AR1 containing pSrtC2TT | This study |
| MR2 | AR1 containing pSrtC2-L45P | This study |
| MR2 | AR1 containing pSrtC2Δsp | This study |
| WU11 | AR1 containing pSrtC2Δ3 | This study |
| WU12 | AR1 containing pSrtC2Δ5 | This study |
| WU13 | AR1 containing pSrtC2Δ59 | This study |
| WU14 | AR1 containing pSrtC2Δ110 | This study |
| WU15 | AR1 containing pSrtC2-H184A | This study |
| WU16 | AR1 containing pSrtC2-C246A | This study |
| Plasmids | ||
| pSrtC2 | pJRD215 derivative expressing wild-type SrtC2 from MG1 | 20 |
| pH 6-SrtA | pMCSG7 derivative expressing SrtA (residues 52 to 253) for antibody production | This study |
| pUC-SrtC2 | The srtC2 fragment from pSrtC2, cut by SalI/HindIII and cloned into pUC19 | This study |
| pSrtC2ΔVV | pJRD215 derivative expressing SrtC2 that carries mutations of SALVTS to VVVVVV at positions 36 to 41 | This study |
| pSrtC2ΔTT | pJRD215 derivative expressing SrtC2 that carries mutations of SIMALVGM to TTTTTTTT at positions 41 to 48 | This study |
| pSrtC2ΔSP | pJRD215 derivative expressing SrtC2 lacking the first 44 residues | This study |
| pSrtC2ΔTM | pJRD215 derivative expressing SrtC2 lacking the residues from 285–302 (transmembrane region) | This study |
| pSrtC2Δ5 | pJRD215 derivative expressing SrtC2 lacking the last 5 residues | This study |
| pSrtC2Δ59 | pJRD215 derivative expressing SrtC2 lacking the last 59 residues | This study |
| pSrtC2Δ110 | pJRD215 derivative expressing SrtC2 lacking the last 59 residues | This study |
| pSrtC2-L45P | pJRD215 derivative expressing SrtC2 with L45P mutation | This study |
| pSrtC2-E104A | pJRD215 derivative expressing SrtC2 with E104A mutation | This study |
| pSrtC2-N106A | pJRD215 derivative expressing SrtC2 with N106A mutation | This study |
| pSrtC2- H184A | pJRD215 derivative expressing SrtC2 with H184A mutation | This study |
| pSrtC2- C246A | pJRD215 derivative expressing SrtC2 with C246A mutation | This study |
| pSrtC1 | pJRD215 derivative expressing wild-type SrtC1 from MG1 | 32 |
| pSrtC1-D129A | pJRD215 derivative expressing SrtC1 with D129A mutation | This study |
| pSrtC1-W131A | pJRD215 derivative expressing SrtC1 with W131A mutation | This study |
Plasmid construction.
SrtC2 truncations and site-directed mutagenesis of recombinant plasmids were based on previous protocols (21, 32), as follows. (i) For SrtC2-truncated mutants, primers (Table 2) were designed to selectively amplify the plasmid pUC-SrtC2 (Table 1), which was obtained by cloning the srtC2 fragment from pSrtC2 (20) into pUC19 at SalI and HindIII sites. (ii) For site-direct mutagenesis of SrtC2, the mutation sites were incorporated into the 5′ end of synthesized primers. Plasmid DNA of pUC-SrtC2 was used as a template for PCR amplification with Pfu DNA polymerase using appropriate primer sets (Table 2). The PCR products were purified by gel extraction and phosphorylated to facilitate religation of the amplicon into circular plasmids, which were then transformed into E. coli DH5α. Mutant plasmids were verified by DNA sequencing, and srtC2 fragments carrying desired mutations were released by SalI and HindIII and subcloned into pJRD215 pretreated with the same enzymes. For site-directed mutagenesis of SrtC1, plasmid DNA of pSrtC1 was used as a template for PCR amplification with Pfu DNA polymerase using appropriate primer sets (Table 2). The PCR product was treated with DpnI and cloned into E. coli as previously described (21). The resulting plasmids (Table 1) were further confirmed by DNA sequencing and transformed into A. oris by electroporation.
Table 2.
Primers used in this study
| Primer | Sequencea |
|---|---|
| LIC-SrtA-5 | TACTTCCAATCCAATGCAATTGACGCCAACGCCAGC |
| LIC-SrtA-3 | TTATCCACTTCCAATGTTAGGTCGTTGAGCACGGACT |
| SrtC2ΔVV-F | GTCGTCATCATGGCGCTGGTGGGCATG |
| SrtC2ΔVV-R | GACCACGACGACGACCGACAGGCGCAGGCGCGGGG |
| SrtC2Δsp-F | CTGGTCACCTCCATCATGGCGCTG |
| SrtC2Δsp-R | CATGGGGCCTCGTACGCTTCCA |
| SrtC2ΔTT-F | ACCACGACCACGACCGGGCTGCTGACCTATCCGACG |
| SrtC2ΔTT-R | GGTGGTGGTGGTGACCAGGGCGGAGACCGA |
| SrtC2-L45P-F | CCGGTGGGCATGGGGGTGGTG |
| SrtC2-L45P-R | CGCCATGATGGAGGTGGAGAC |
| SrtC2ΔTM-F | CGCTCGGGCTACGCGGCGGCGA |
| SrtC2ΔTM-R | GGGGAAGTGGGGAACGTCGGG |
| SrtC2Δ5-F (SalI) | GGCGGTCAGCCTACGTCCTGGTTGAGACC |
| SrtC2Δ5-R (HindIII) | GGCGAAGCTTCTACCCACTGGTCCCCGCGCTCGG |
| SrtC2Δ59-R (HindIII) | GGCGAAGCTTCTAGTCGGTGAACCACCGTTGGGGGCT |
| SrtC2Δ110-R (HindIII) | GGCGAAGCTTCTACCACAGGTACAGCCCGACGAC |
| SrtC2-E104A-F | GCGGCCAACAACCACGTCCCCACC |
| SrtC2-E104A-R | CAGGACCGCCCCGGCCGAGAG |
| SrtC2-N106A-F | GCCAACCACGTCCCCACCGGTGCCG |
| SrtC2-N106A-F | GGCCTCCAGGACCGCCCCGGCCG |
| SrtC2-H184A-F | GCC CGCGGCCTGGCCGAGGCC |
| SrtC2-H184A-R | CCCCGTGATGACCGAGCGGGTC |
| SrtC2-C246A-F | GCCACGCCCCTGGGCATCAACACC |
| SrtC2-C246A-R | GGTCACCAGGGTGAGCAGGTC |
| SrtC1-D129A-F | GAGAGCGCGCCGATCCTCGCCCCCTGGCTCGAGTCGCAG |
| SrtC1-D129A-F | CAGGTTGTTGTTGTAGGTCTCGGCGGAGGCGCGCTCCTT |
| SrtC1-W131A-F | GCGCCGATCCTCGACCCCGCCCTCGAGTCGCAGCGCCCC |
| SrtC1-W131A-R | GCTCTCCAGGTTGTTGTTGTAGGTCTCGGCGGAGGCGCG |
Restriction sites in the primers are underlined.
Cell fractionation and Western blotting.
Cell fractionation and Western blotting were followed by using previously published protocols (9, 31), with some modifications. Overnight cultures of various Actinomyces strains were used to inoculate fresh cultures grown to the mid-log phase (1:50 dilution) at 37°C in HIB. All aliquots with equal numbers of cells, as measured by optical density at 600 nm (OD600), were fractionated into medium and cell pellets by centrifugation. Cell pellets were suspended in SMM buffer (0.5 M sucrose, 10 mM MgCl2, and 10 mM maleate, pH 6.8) and treated with mutanolysin for 4 h at 37°C. After mutanolysin treatment, cell wall fractions and protoplasts were obtained by centrifugation. The protoplasts (pellets) were frozen on dry ice for 15 min and resuspended in 0.5 ml of cold buffer H (20 mM HEPES, pH 8.0, 200 nM NaCl, 1 mM dithiothreitol, and protease inhibitor cocktail III). Then, 5 μl 0.1 M MgCl2, 5 μl 0.1 M CaCl2, 2 μl DNase I (BioLab), and 2 μl RNase 20 mg/ml (Sigma) were added, and the mixture was incubated on ice for 1 h. Ultracentrifugation of the resulting lysates was performed at 100,000 × g at 4°C to separate membrane and cytoplasmic fractions. Cell wall and membrane fractions were also collected from cells grown on HIB plates. In this case, cells were scraped from plates and washed in SMM buffer. Cell pellets of equal bacterial numbers were suspended in the same buffer and treated with mutanolysin as described above. All fractions were subjected to trichloroacetic acid precipitation and acetone wash. Samples were then boiled in sample buffer containing sodium dodecyl sulfate (SDS), separated by 4 to 12% Tris-glycine gradient gels, subjected to immunoblotting with rabbit antisera (1:2,000 for α-SrtA, 1:1,333 for α-SrtC2, 1:10,000 for α-FimA, and 1:1,333 for α-FimB), followed by anti-rabbit HRP-linked IgG antibody, and detected by chemiluminescence.
In vitro biofilm formation.
Biofilm assays were performed as previously described, with some modifications (21). Briefly, Actinomyces strains were grown overnight in HIB at 37°C with shaking before being diluted 1:100 in HIB containing 1% sucrose, and 1.5-ml aliquots were dispensed into 24-well polystyrene plates (Corning, NY), followed by incubation at 37°C with 5% CO2 for 48 h. The wells were gently washed three times with 1 ml sterile phosphate-buffered saline (PBS) and air dried for 30 min. Biofilms were stained with 0.5% crystal violet for 30 min, washed extensively to remove any unbound dye, air dried, and photographed by a FluorChem Q Imager (Alpha Innotech).
Bacterial coaggregation.
Coaggregation assays were performed with various strains of Actinomyces and Streptococcus oralis as previously described (21). Briefly, stationary-phase cultures of bacterial strains were grown in a previously described complex medium (22) with 0.2% glucose, harvested by centrifugation, washed in Tris-buffered saline (TBS; pH 7.5) containing 0.1 mM CaCl2, and suspended to an equal cell density of approximately 2 × 109 ml−1 based upon OD600 values. For coaggregation, 0.25-ml aliquots of Actinomyces and streptococcal cell suspensions were mixed in 24-well plates for a few minutes on a rotator shaker and photographed by a FluorChem Q Imager.
Immunoelectron microscopy (IEM).
Actinomyces cells were grown on HIA plates, suspended in 0.1 M NaCl, washed with PBS, and resuspended in PBS. Immunogold labeling was followed as previously described (21). Briefly, a drop of bacterial suspension in PBS was placed on carbon-coated nickel grids, washed three times with PBS containing 2% bovine serum albumin (BSA), and blocked for 1 h in PBS with 0.1% gelatin. Fimbriae were stained with primary antibodies diluted in PBS (1:100 for α-FimA and 1:50 for α-FimB) with 2% BSA for 1 h, followed by washing four times with PBS containing 2% BSA. Samples were then treated with 12 nm gold goat anti-rabbit IgG (Jackson ImmunoResearch) diluted 1:20 in PBS with 2% BSA for 1 h. The samples were washed five times with water before being stained with 1% uranyl acetate and viewed in a JEOL JEM-1400 electron microscope.
RESULTS
Importance of the predicted signal sequence and the N-terminal transmembrane-spanning domain of sortase SrtC2 in type 2 fimbrial assembly.
A bioinformatics analysis of the primary sequence of SrtC2 of A. oris MG1 using the TMHMM server (12) suggested that SrtC2 contains two putative TM helices, one at the amino terminus and another located between the catalytic domain and the positively charged carboxy-terminal tail (Fig. 1A). Sequence analysis by SignalP 3.0 (2) further predicted that the N-terminal TM helix harbors a putative signal peptide with a cleavage site between residues 37 and 38 (Fig. 1A, VSALV). To probe the function of this predicted signal peptide by site-directed mutagenesis, we utilized an expression plasmid and produced an SrtC2 mutant in which the residues at 36 to 38 and 40 to 41 were all replaced by valine (Fig. 1A, VV). We then tested the effect of the mutation on SrtC2 expression and function by introducing this expression plasmid into an A. oris strain that lacks srtC2. For analysis of SrtC2 and type 2 fimbrial production, cultures of MG1 strain and its isogenic derivatives were subjected to cell fractionation as previously described (22), whereby equivalent amounts of early log-phase cells were fractionated into culture medium, cell wall, membrane, and cytoplasmic fractions. The resultant protein samples were then boiled in SDS-containing sample buffer and subjected to SDS-PAGE and Western blotting using antisera specific for SrtC2 (α-SrtC2), SrtA (α-SrtA), or FimA (α-FimA) (see Materials and Methods).
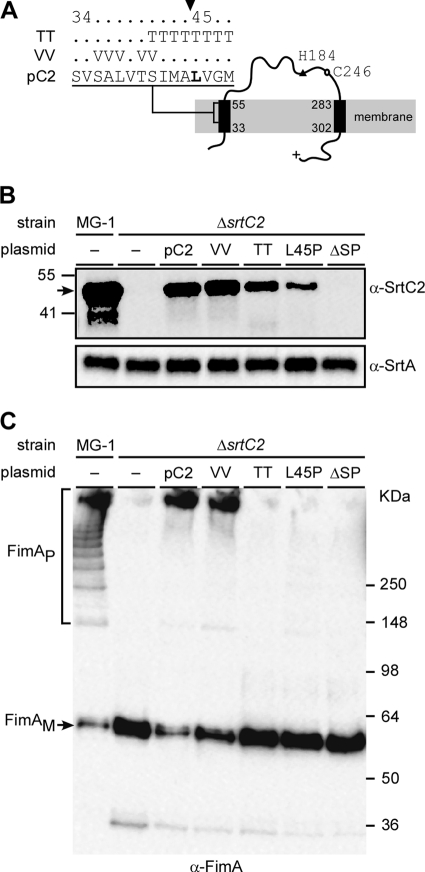
Fig 1.
Requirements for fimbrial polymerization of the SrtC2 N-terminal signal peptide sequence. Predicted membrane topology of the type 2 fimbria-specific sortase SrtC2 is shown in panel A, with the N terminus possessing a signal peptide sequence. The filled arrowhead specifies a cleavage site. Threonin (TT) and valine (VV) substitutions of this cleavage site are indicated. Catalytic residues H184 and C246 are located upstream of the C-terminal transmembrane helix. (B and C) Cell wall and membrane fractions were harvested from Actinomyces cells of the wild-type MG-1 strain and its isogenic derivative strains. Equivalent protein samples were separated on 4 to 12% Tris-glycine gradient gel and detected by immunoblotting with antibodies against SrtC2 and SrtA (α-SrtC2 and α-SrtA; membrane fractions) (B) or FimA (α-FimA; cell wall fractions) (C). The positions of high-molecular-mass polymers (FimAP) and monomers (FimAM) and molecular mass markers are indicated.
As shown in Fig. 1B, SrtC2 (predicted mass of 45 kDa) of wild-type MG1 cells migrated around the 41 and 55 kDa markers as detected in the membrane fractions; the Western blot data also revealed some degradation products. In the isogenic srtC2-deleted strain, however, the major SrtC2-reactive bands were eliminated as expected, and the introduction of the plasmid expressing wild-type SrtC2 into this mutant strain restored these SrtC2-reactive bands (Fig. 1B). As for SrtC2 function in type 2 fimbrial assembly, the cell wall fraction of the srtC2-deleted strain showed no FimA polymers, while that of the MG1 parent showed an abundant amount of these polymers (Fig. 1C, first 2 lanes). Finally, the wild-type SrtC2-expression plasmid restored FimA polymerization in the ΔsrtC2 mutant (Fig. 1C, third lane from left), in agreement with results we reported previously (20). Surprisingly, the plasmid expressing SrtC2 with mutations of the putative signal peptide also resulted in fimbrial polymerization in the ΔsrtC2 strain, similar to that of the mutant expressing WT SrtC2 (Fig. 1C, compare lane pC2 with lane VV), as the expression of SrtC2 was comparable to that of the wild-type complemented strain (Fig. 1C, lane VV). We interpret this result to indicate that the signal peptide is not cleaved between Ala37 and Leu38, as predicted by the bioinformatics analysis.
We therefore turned to another potential cleavage site between Ala44 and Leu45 (Fig. 1A), based on the annotation of A. oris MG1 SrtC2 (A. oris MG1, ANA_0025 [http://www.oralgen.lanl.gov]). If this prediction is correct, mutations that change the +1 residue to proline would prevent the signal peptide from getting processed (1, 26). Consequently, we generated recombinant plasmids expressing an SrtC2 mutant that had residues 41 to 48 all replaced by threonine (Fig. 1B, lane TT), an SrtC2 mutant with Leu45 changed to proline (Fig. 1B, lane L45P), and another SrtC2 mutant lacking the first 44 amino acids (Fig. 1, lane ΔSP). All recombinant plasmids were introduced into the ΔsrtC2 mutant, and SrtC2 expression and polymerization of type 2 fimbriae were analyzed as described above. Significantly, while membrane expression of SrtC2 was detected in the threonine and proline mutants, albeit at a reduced level (Fig. 1B, lanes TT and L45P, respectively), FimA polymerization was not observed in these mutants (Fig. 1C, lanes TT and L45P). As expected, when the first 44 amino acids of SrtC2 were deleted, SrtC2 expression was absent (Fig. 1B, last lane), concomitant with the loss of FimA polymerization (Fig. 1C, last lane). Note that the SrtC2 signal was not observed in the culture medium, cell wall, and cytoplasmic fractions of all SrtC2 mutants (data not shown). The results indicate that the first 44 residues of SrtC2 contain a signal peptide, which is potentially cleaved between residues Ala44 and Leu45 (see Discussion).
C-terminal TM domain and cytoplasmic tail are essential for SrtC2-mediated fimbrial assembly.
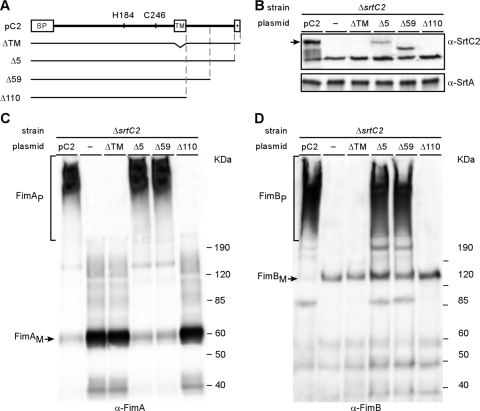
If the N-terminal TM domain is cleaved off by processing of the signal sequence, then the membrane insertion of the mature SrtC2 protein might involve the second predicted TM domain of the protein. To determine whether this C-terminal transmembrane helix is required for SrtC2 function, we generated a series of C-terminally truncated mutant SrtC2 proteins expressed from a recombinant plasmid (Fig. 2A) and tested their abilities to complement the ΔsrtC2 mutant by performing cell fractionation studies similar to those described above. Strikingly, SrtC2 was not detected in the membrane fraction of mutants that lack the C-terminal TM domain (Fig. 2B, lane ΔTM) or the last 110 amino acids immediately downstream of this TM (Fig. 2B, lane Δ110); consistent with loss of SrtC2, no FimA and FimB polymers were detected in these mutants (Fig. 2C and D, lanes ΔTM and Δ110). By comparison, deletion of either the basic patch or 59 residues of the SrtC2 carboxy terminus reduced membrane association of SrtC2 proteins (Fig. 2B, lanes Δ5 and Δ59, respectively), but these mutations did not affect the overall polymerization of FimA and FimB fimbrillins (Fig. 2C and D, lanes Δ5 and Δ59).
Fig 2.
Requirements for membrane localization of the C-terminal transmembrane domain of SrtC2. (A) Various C-terminal truncations of SrtC2 were generated. Plasmids expressing wild-type SrtC2 (pC2) and mutants were transformed into an Actinomyces sp. strain lacking srtC2. (B to D) Protein samples from cell wall and membrane fractions of Actinomyces sp. cells were obtained as described in the legend to Fig. 1 and analyzed by Western blotting with α-SrtC2 and α-SrtA (membrane fractions) (B), α-FimA (cell wall fractions) (C), and α-FimB (cell wall fractions) (D). The positions of high-molecular-mass polymers (P) and monomers (M) and molecular mass markers are indicated.
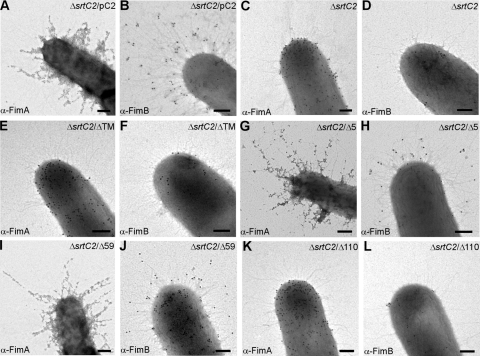
We next extended the studies described above by immunoelectron microscopy (IEM) to determine how surface assembly of the type 2 fimbriae and/or fimbrillins is affected by the mutations described above affecting SrtC2 processing. Using a previously developed protocol (22), we labeled A. oris cells of various strains with specific antisera α-FimA and α-FimB, followed by staining with 12 nm gold particles conjugated with IgG, and cells were viewed by an electron microscope. With cells expressing wild-type SrtC2, FimA and FimB signals were observed abundantly along the fimbrial structures as well as on the bacterial surface (Fig. 3A and B). Consistent with the role of SrtC2 in fimbrial polymerization (22), no fimbrial structures labeled with α-FimA and α-FimB were observed in the ΔsrtC2 mutant, whereas surface display of non-polymeric FimA and FimB proteins was not abrogated (Fig. 3C and D). In agreement with the Western blot analysis described above, deletion of the TM domain or the last 110 residues of SrtC2 abrogated fimbrial assembly (Fig. 3E to H), while deletion of the last 5 or 59 residues of SrtC2 did not (Fig. 3I to L). Altogether, the data indicate that the TM domain and part of the cytoplasmic tail are critical for SrtC2 membrane localization and function.
Fig 3.
Requirements of the SrtC2 C-terminal transmembrane domain for surface assembly of type 2 fimbriae. Bacterial cells were immobilized on nickel grids, stained with α-FimA (A, C, E, G, I, and K) or α-FimB (B, D, F, H, J, and L) and goat anti-rabbit IgG conjugated to 12-nm (for anti-FimA) or 18-nm (for anti-FimB) gold particles. Samples were viewed by transmission electron microscopy after being stained with 1% uranyl acetate. Bars, 0.2 μm.
Characterization of the catalytic center of SrtC2: Cys246 and His184 are essential for SrtC2 activity.
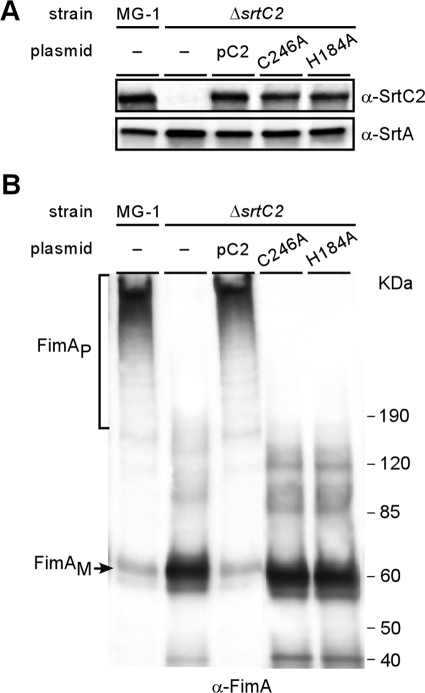
Sortase acts by cleaving a LPXTG-containing substrate between threonine and glycine, thereby forming an acyl-enzyme intermediate with the substrate via a thioester bond generated by the reactive cysteine residue of sortase and the threonine residue of the substrate (29). Alanine substitution of Cys184 or His120 of staphylococcal SrtA completely abrogates the enzymatic activity (30). To determine whether the corresponding residues Cys246 and His184 are essential for A. oris SrtC2 activity, we expressed in the ΔsrtC2 mutant recombinant SrtC2 with indicated alanine substitution mutations and analyzed these mutants by using Western blots. Importantly, mutations of the conserved Cys246 and His184 did not alter the expression and localization of SrtC2 in the cytoplasmic membrane compared to that of the wild-type enzymes (Fig. 4B; compare last MG1 and pC2 lanes with the last two lanes), indicating the unaltered export, processing, folding, membrane insertion, and stability of the mutant proteins. Yet, mutations of the conserved Cys246 and His184 abolished fimbrial polymerization (Fig. 4B, last two lanes) as well as surface assembly of type 2 fimbriae (data not shown). Hence, the conserved Cys246 and His184 are the catalytic residues of pilin-specific sortase SrtC2.
Fig 4.
Role of catalytic residues C246 and H184 in SrtC2-mediated FimA polymerization. Membrane (A) and cell wall (B) fractions were collected from wild-type MG-1 and its isogenic derivative strains carrying a deletion of srtC2 or the derivative-expressing SrtC2 mutants. Equivalent protein samples were separated on a 4 to 12% Tris-glycine gradient gel and detected by immunoblotting with α-SrtC2 and α-SrtA (A) and α-FimA (B). The positions of FimA monomers (FimAM), FimA high-molecular-weight polymers (FimAP), and molecular mass markers are indicated.
Dispensability of the catalytic site lid domain in SrtC2 activity.
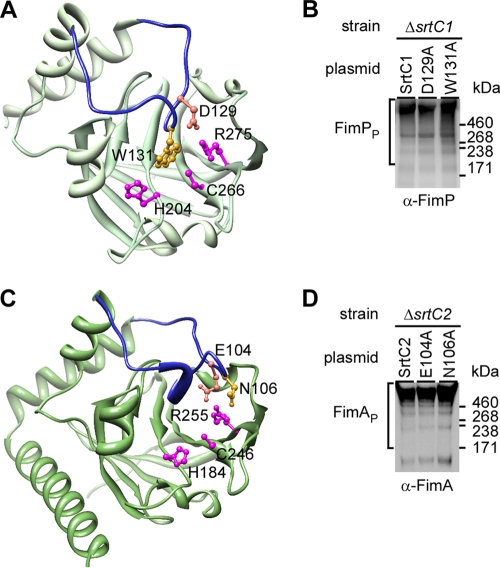
A distinguishing feature found in class C sortases is the presence of a flexible lid covering the catalytic site that was first described in Streptococcus pneumoniae pilin-specific sortase SrtC1 (14). This unique feature is also present in the fimbria-specific sortase SrtC1 in A. oris (23). Similar to S. pneumoniae SrtC1, this flexible lid contains the conserved lid anchor motif DPW (Fig. 5A, D129 and W131). Intriguingly, the flexible lid has been shown to be required for pilus polymerization activity of streptococcal SrtC1 in an E. coli system (15). However, in S. agalactiae, alanine substitution of the aspartate and tryptophan residues in the lid anchor motif of pilin-specific sortase SrtC1 does not affect pilus polymerization (6). Thus, the functional significance of the lid domain in class C sortase activity remains unresolved. Using the I-TASSER analysis that is based on the sequence-to-structure-to-function paradigm (25, 34), the 3-dimentional structure of A. oris SrtC2 was predicted with a confidence (C) score of 0.93 and a structural similarity TM score of 0.84 ± 0.08 (Fig. 5C). Interestingly, although SrtC2 does not contain the conserved DPW motif, the modeled SrtC2 structure appears to have a lid region with Glu104 and Asn106 as part of the lid anchor residues (Fig. 5C).
Fig 5.
Analysis of a flexible lid in Actinomyces sp. fimbria-specific sortases. (A) A lid (blue) with anchor residues D129 and W131 covering catalytic residues H204, C266, and R275 is shown in the three-dimensional (3D) crystal structure of Actinomyces sp. fimbria-specific sortase SrtC1. Shown in panel C is a 3D structure of Actinomyces fimbria-specific sortase SrtC2, as modeled after the SrtC2 structure. (B and D) Western blotting for fimbrial polymerization of Actinomyces sp. cells expressing wild-type SrtC1 and its lid mutants (B) or wild-type SrtC2 and its lid mutants (D) was carried out as described in the legend to Fig. 1.
To further probe the potential function of the lid, we generated recombinant plasmids expressing A. oris SrtC1 with an alanine substitution at the canonical anchor residues D129 and W131, and these recombinant plasmids were introduced into the A. oris ΔsrtC1 mutant. Polymerization of the type 1 fimbriae was analyzed by using Western blots with α-FimP as described above. As was reported for S. agalactiae SrtC1, no significant defect in fimbrial polymerization was detected in strains expressing SrtC1 with D129A or W131A mutations compared to the wild-type SrtC1-expressing strain (Fig. 5B).
To address whether lid function may vary among members of the class C sortases, we went on to analyze fimbrial polymerization of A. oris strains expressing SrtC2 with an alanine substitution at E104 or N106. Compared to the strain expressing wild-type SrtC2 (Fig. 5D, lane SrtC2), no apparent defect in fimbrial polymerization was observed in strains expressing SrtC2 with E104A or N106A mutations (Fig. 5C, last two lanes). With the high confidence levels of the predicted SrtC2 structure, altogether the results support the notion that the lid of A. oris fimbria-specific sortases is dispensable for fimbrial polymerization, like the lid of S. agalactiae pilin-specific sortase SrtC1 (6).
Phenotypic correlation of SrtC2 structural determinants in Actinomyces coaggregation with Streptococcus oralis and biofilm formation.
Our previous work established that the fimbrial shaft FimA of type 2 fimbriae is the major fimbrial adhesin that is required for Actinomyces coaggregation with S. oralis and formation of Actinomyces biofilm (22). As shown above and elsewhere (22), polymerization of FimA on the bacterial surface requires functional sortase SrtC2. To determine whether improper localization of the enzyme affects biological functions of type 2 fimbriae, we subjected the above-described SrtC2 mutants to coaggregation and biofilm formation assays as reported previously (22).
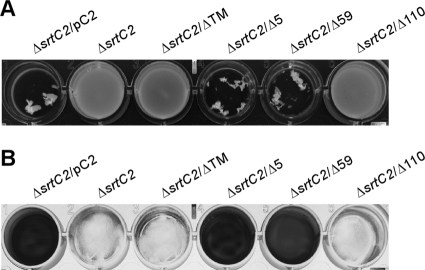
For coaggregation assays, equal numbers of Actinomyces and streptococcal cells were mixed together in microtiter plates, and coaggregation was evaluated by photography. In agreement with previous studies (22), Actinomyces cells lacking srtC2 failed to coaggregate with S. oralis strain 34, unlike cells expressing SrtC2 (Fig. 6A, first two lanes). As expected, coaggregation with S. oralis was not observed in Actinomyces cells which expressed mutant SrtC2 lacking the carboxy-terminal TM domain or the cytoplasmic tail (Fig. 6A, lanes ΔTM and Δ110). No apparent defect in coaggregation was detected in Actinomyces cells expressing SrtC2 mutants that lack the last 5 or 59 residues, consistent with the biochemical evidence for normal sortase activity (Fig. 6A, lanes Δ5 and Δ59).
Fig 6.
Role of the SrtC2 transmembrane domain in coaggregation of Actinomyces species with oral streptococci and biofilm formation. (A) Stationary-phase cultures of the ΔsrtC2 strain and its derivatives expressing wild-type SrtC2 or various mutants were examined for coaggregation with oral streptococci (S. oralis strain 34). (B) The same set of bacterial strains described in panel A was analyzed for biofilm formation, and the generated biofilms were stained with crystal violet.
Finally, we subjected the mutants described above to biofilm assays, whereby Actinomyces cells were grown in the presence of sucrose for 48 h at 37°C with 5% CO2. Biofilm production was quantified by optical density at 450 nm after biofilms were stained with crystal violet. Consistent with the results described above, no biofilms were observed in strains lacking srtC2 or expressing SrtC2 with truncation of the TM domain or the cytoplasmic tail, whereas abundant biofilms were produced in cells expressing wild-type SrtC2 or SrtC2 missing the last 5 or 59 residues (Fig. 6B). Of note, neither coaggregation with S. oralis nor biofilm formation was detected with A. oris cells expressing nonfunctional enzymes, i.e., C246A or H184A mutants, or signal peptide defective enzymes, i.e., TT, L54P, or ΔSP mutants (data not shown).
DISCUSSION
Assembly of covalently linked pilus polymers in Gram-positive bacteria on their cell wall peptidoglycan is achieved by sequential activities of two transpeptidase enzymes named sortases. A pilin-specific sortase or class C sortase catalyzes the covalent linkage between pilin subunits, which are ultimately anchored to the bacterial peptidoglycan also by a covalent linkage. The latter step is typically catalyzed by the housekeeping sortase or class A sortase. In the current work, we have characterized A. oris SrtC2, the fimbria-specific sortase (class C sortase) that is required for the type 2 fimbrial polymerization.
Sortase is a membrane-bound transpeptidase enzyme, the activity of which depends on the proper localization of the mature protein in the membrane. A. oris SrtC2 is predicted to possess two TM helices, which are typical of class C sortases (5, 7). We presented here mutational studies that suggest that the N-terminal TM helix of SrtC2, which contains a signal peptide sequence, is processed by the general secretion machinery (Sec) for the protein precursor to be translocated across the cytoplasmic membrane. Our genetic evidence supports the bioinformatics prediction that the first 44 residues of SrtC2 constitute a TM helix and a signal peptide sequence with a potential cleavage site between Ala44 and Leu45 (Fig. 1). Conventionally, Leu45 represents the first residue of the mature SrtC2 at the +1 position. In E. coli, the signal peptide of the M13 procoat protein with Leu at the +1 position is cleavable, whereas that of the precursor with proline at the same position is not (26). Similarly, a maltose-binding protein precursor with proline at +1 is not processed (1). Consistent with the notion that A. oris SrtC2 is cleaved at its N-terminal signal peptide sequence between Ala44 and Leu45 for proper processing, although mutant SrtC2 with a proline substitution at Leu45 was detected in the membrane, albeit at a reduced level, fimbrial polymerization in the mutant cell was abrogated (Fig. 1). Furthermore, the SrtC2 mutant lacking the first 44 residues displays the same fimbrial polymerization defect (Fig. 1).
The proposed cleavage of the signal peptide sequence of SrtC2 would liberate the N terminus of the mature enzyme from the amino-terminal membrane anchor, unlike the situation established for the S. aureus class A sortase SrtA, in which case the signal peptide is not cleaved and the N-terminal TM domain serves as the essential membrane anchor (17, 18). Processing of the signal sequence in a class C sortase requires that they contain a distinct domain for membrane association. Indeed, the A. oris SrtC2 contains a C-terminal TM helix that may function as the membrane anchor. If so, this TM helix should be essential for the proper localization of the enzyme in the membrane. Our genetic data established that this is the case: an SrtC2 mutant that lacks this C-terminal TM domain was not detected in the membrane fraction (Fig. 2). It is interesting that an SrtC2 mutant lacking the TM-proximal cytosolic domain displays the same defect, whereas mutants missing TM distal parts of the cytosolic domain, including a basic patch, behave like the wild-type enzyme (Fig. 2). Perhaps the essential cytosolic domain helps to retain the mature protein within the cytoplasmic membrane. Alternatively, it may interact with cytosolic factors that control and coordinate pilus assembly. Further biochemical and genetic studies will be needed to illuminate the function of the cytosolic domain of a class C sortase.
Apart from having the C-terminal membrane anchor and a cytoplasmic tail, SrtC2 potentially possesses a flexible lid over the catalytic pocket (Fig. 5), which is a typical feature found in class C sortases, such as A. oris SrtC1 and S. agalactiae SrtC1 (6, 23). While mutations of the lid anchor residues of S. pneumoniae SrtC1 abrogated pilus polymerization (15), similar mutations in S. agalactiae SrtC1 (6) and A. oris fimbria-specific sortases SrtC1 and SrtC2 do not affect pilus polymerization (Fig. 5). It is noteworthy that although S. pneumoniae SrtC1 mutants were expressed and evaluated for their roles in pilus polymerization in E. coli (15), they have not been reexamined in S. pneumoniae. Thus, it is possible that the observed defect of these mutations in the polymerization activity of S. pneumoniae SrtC1 might be due to the instability of the mutant S. pneumoniae SrtC1 enzymes in the heterologous E. coli system. What might then be the role of this flexible lid in pilus assembly? It has been speculated that the lid may have a regulatory role in pilus assembly by restraining access of LPXTG-containing substrates until other ligands or regulatory factors displace them (6). This remains a challenging but important problem to be investigated.
While the lid anchor residues are dispensable for the activity of A. oris sortases, the catalytic residues C246 and H184 of SrtC2 were shown here to be essential for the enzymatic activity, hence the assembly of type 2 fimbriae (Fig. 5). Consequently, the ability of A. oris cells to form a biofilm and to coaggregate with S. oralis was abrogated with mutations of the two residues (data not shown). Importantly, three-dimensional structural studies of sortases of different classes reveal a common fold, with the catalytic cysteine and histidine residues forming a catalytic pocket (27). This has become a signature of sortase families (6, 16, 27), and our finding here validates their essential role. The key puzzle that remains unsolved is how substrate specificity is determined, given that all sortase enzymes display a similar catalytic core. Our previous studies hinted that the substrate specificity is partly endowed by the exact sequence of the LPXTG motif (3). Based on this study and available evidence, it is tempting to speculate that accessory elements such as the lid region may contribute to substrate specificity by gating sortase substrates for pilus polymerization and cell wall anchoring of the resulting pilus polymers. Future work should address this basic and fascinating aspect of sortase-mediated pilus biogenesis in Gram-positive bacteria.
ACKNOWLEDGMENTS
We thank the members of our laboratory for their critical inputs.
This work was supported by the National Institute of Dental and Craniofacial Research (NIDCR) NIH grant DE017382 to H.T.-T.
Footnotes
Published ahead of print 23 March 2012
REFERENCES
- 1. Barkocy-Gallagher GA, Bassford PJ., Jr 1992. Synthesis of precursor maltose-binding protein with proline in the +1 position of the cleavage site interferes with the activity of Escherichia coli signal peptidase I in vivo. J. Biol. Chem. 267:1231–1238 [PubMed] [Google Scholar]
- 2. Bendtsen JD, Nielsen H, von Heijne G, Brunak S. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783–795 [DOI] [PubMed] [Google Scholar]
- 3. Chang C, Mandlik A, Das A, Ton-That H. 2011. Cell surface display of minor pilin adhesins in the form of a simple heterodimeric assembly in Corynebacterium diphtheriae. Mol. Microbiol. 79:1236–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cisar JO, Takahashi Y, Ruhl S, Donkersloot JA, Sandberg AL. 1997. Specific inhibitors of bacterial adhesion: observations from the study of gram-positive bacteria that initiate biofilm formation on the tooth surface. Adv. Dent. Res. 11:168–175 [DOI] [PubMed] [Google Scholar]
- 5. Comfort D, Clubb RT. 2004. A comparative genome analysis identifies distinct sorting pathways in gram-positive bacteria. Infect. Immun. 72:2710–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cozzi R, et al. 2011. Structure analysis and site-directed mutagenesis of defined key residues and motives for pilus-related sortase C1 in group B Streptococcus. FASEB J. 25:1874–1886 [DOI] [PubMed] [Google Scholar]
- 7. Dramsi S, Trieu-Cuot P, Bierne H. 2005. Sorting sortases: a nomenclature proposal for the various sortases of Gram-positive bacteria. Res. Microbiol. 156:289–297 [DOI] [PubMed] [Google Scholar]
- 8. Gibbons RJ, Hay DI, Cisar JO, Clark WB. 1988. Adsorbed salivary proline-rich protein 1 and statherin: receptors for type 1 fimbriae of Actinomyces viscosus T14V-J1 on apatitic surfaces. Infect. Immun. 56:2990–2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Guttilla IK, et al. 2009. Acyl enzyme intermediates in sortase-catalyzed pilus morphogenesis in gram-positive bacteria. J. Bacteriol. 191:5603–5612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kline KA, et al. 2009. Mechanism for sortase localization and the role of sortase localization in efficient pilus assembly in Enterococcus faecalis. J. Bacteriol. 191:3237–3247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kolenbrander PE, et al. 2006. Bacterial interactions and successions during plaque development. Periodontol. 2000 42:47–79 [DOI] [PubMed] [Google Scholar]
- 12. Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567–580 [DOI] [PubMed] [Google Scholar]
- 13. Mandlik A, Swierczynski A, Das A, Ton-That H. 2008. Pili in Gram-positive bacteria: assembly, involvement in colonization and biofilm development. Trends Microbiol. 16:33–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Manzano C, et al. 2008. Sortase-mediated pilus fiber biogenesis in Streptococcus pneumoniae. Structure 16:1838–1848 [DOI] [PubMed] [Google Scholar]
- 15. Manzano C, Izore T, Job V, Di Guilmi AM, Dessen A. 2009. Sortase activity is controlled by a flexible lid in the pilus biogenesis mechanism of gram-positive pathogens. Biochemistry 48:10549–10557 [DOI] [PubMed] [Google Scholar]
- 16. Marraffini LA, Dedent AC, Schneewind O. 2006. Sortases and the art of anchoring proteins to the envelopes of gram-positive bacteria. Microbiol. Mol. Biol. Rev. 70:192–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mazmanian SK, Liu G, Jensen ER, Lenoy E, Schneewind O. 2000. Staphylococcus aureus sortase mutants defective in the display of surface proteins and in the pathogenesis of animal infections. Proc. Natl. Acad. Sci. U. S. A. 97:5510–5515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mazmanian SK, Liu G, Ton-That H, Schneewind O. 1999. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science 285:760–763 [DOI] [PubMed] [Google Scholar]
- 19. McIntire FC, Vatter AE, Baros J, Arnold J. 1978. Mechanism of coaggregation between Actinomyces viscosus T14V and Streptococcus sanguis 34. Infect. Immun. 21:978–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mishra A, Das A, Cisar JO, Ton-That H. 2007. Sortase-catalyzed assembly of distinct heteromeric fimbriae in Actinomyces naeslundii. J. Bacteriol. 189:3156–3165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mishra A, et al. 2011. Two autonomous structural modules in the fimbrial shaft adhesin FimA mediate Actinomyces interactions with streptococci and host cells during oral biofilm development. Mol. Microbiol. 81:1205–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mishra A, et al. 2010. The Actinomyces oris type 2 fimbrial shaft FimA mediates co-aggregation with oral streptococci, adherence to red blood cells and biofilm development. Mol. Microbiol. 77:841–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Persson K. 2011. Structure of the sortase AcSrtC-1 from Actinomyces oris. Acta Crystallogr. D Biol. Crystallogr. 67:212–217 [DOI] [PubMed] [Google Scholar]
- 24. Rickard AH, Gilbert P, High NJ, Kolenbrander PE, Handley PS. 2003. Bacterial coaggregation: an integral process in the development of multi-species biofilms. Trends Microbiol. 11:94–100 [DOI] [PubMed] [Google Scholar]
- 25. Roy A, Kucukural A, Zhang Y. 2010. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5:725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shen LM, et al. 1991. Use of site-directed mutagenesis to define the limits of sequence variation tolerated for processing of the M13 procoat protein by the Escherichia coli leader peptidase. Biochemistry 30:11775–11781 [DOI] [PubMed] [Google Scholar]
- 27. Spirig T, Weiner EM, Clubb RT. 2011. Sortase enzymes in Gram-positive bacteria. Mol. Microbiol. 82:1044–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ton-That H, Das A, Mishra A. 2011. Actinomyces oris fimbriae: an adhesive principle in bacterial biofilms and tissue tropism, p 63–77 In Kolenbrander PE. (ed), Oral microbial communities: genomic inquiry and interspecies communication. ASM Press, Washington, DC [Google Scholar]
- 29. Ton-That H, Marraffini LA, Schneewind O. 2004. Protein sorting to the cell wall envelope of Gram-positive bacteria. Biochim. Biophys. Acta 1694:269–278 [DOI] [PubMed] [Google Scholar]
- 30. Ton-That H, Mazmanian SK, Alksne L, Schneewind O. 2002. Anchoring of surface proteins to the cell wall of Staphylococcus aureus. Cysteine 184 and histidine 120 of sortase form a thiolate-imidazolium ion pair for catalysis. J. Biol. Chem. 277:7447–7452 [DOI] [PubMed] [Google Scholar]
- 31. Wayne KJ, et al. 2010. Localization and cellular amounts of the WalRKJ (VicRKX) two-component regulatory system proteins in serotype 2 Streptococcus pneumoniae. J. Bacteriol. 192:4388–4394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wu C, et al. 2011. Dual function of a tip fimbrillin of Actinomyces in fimbrial assembly and receptor binding. J. Bacteriol. 193:3197–3206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yeung MK. 1999. Molecular and genetic analyses of Actinomyces spp. Crit. Rev. Oral Biol. Med. 10:120–138 [DOI] [PubMed] [Google Scholar]
- 34. Zhang Y. 2007. Template-based modeling and free modeling by I-TASSER in CASP7. Proteins 69(Suppl 8):108–117 [DOI] [PubMed] [Google Scholar]