Abstract
Numerous in vitro as well as genetic studies have demonstrated that the activities of the E2A proteins are regulated at multiple levels, including modulation of DNA binding by the Id proteins, association with the transcriptional modulators p300 and ETO, and posttranslational modifications. Here, we use affinity purification of tagged E47 combined with mass spectrometry in order to show that E47 interacts with the entire ensemble of Id proteins, namely, Id1, Id2, Id3, and Id4. Furthermore, we find that the lysine-specific histone demethylase 1 (LSD1), the protein arginine N-methyltransferase 5 (PRMT5), the corepressor CoREST, and the chaperones of the 14-3-3 family associate with affinity-purified E47. We also identify a spectrum of amino acid residues in E47 that are phosphorylated, including an AKT substrate site. We did, however, find that mutation of the identified AKT substrate site by itself did not perturb B cell development. In sum, these studies show that the entire ensemble of Id proteins has the ability to interact with E47, identify factors that associate with E47, and reveal a spectrum of phosphorylated residues in E47, including an AKT substrate site.
INTRODUCTION
B-lineage development and specification are tightly regulated by the orchestrated function of critical transcription factors, including E2A, early B cell factor (EBF), Pax5, and Foxo1 (21, 23, 30). The E2A proteins, E12 and E47, arise through differential splicing of the E2A gene. They belong to the class I family of helix-loop-helix (HLH) proteins, also termed E proteins (25). Four E proteins, E12, E47, E2-2, and HEB have been identified in vertebrates. The E2A proteins play particularly important roles during lymphoid development by promoting developmental progression, expansion, and survival of lymphocytes (8, 18, 25). At the hematopoietic stem cell stage, E2A proteins are required to maintain the stem cell pool and to promote the development of lymphoid-primed multipotent progenitors (LMPPs), common lymphoid precursors (CLPs) and pre-pro-B cells (5, 9, 42, 48, 51). B cell development in E2A−/− as well as E2A+/− HEB−/− mice is completely blocked at the all-lymphoid progenitor (ALP) cell stage before the onset of IgH DJ gene rearrangement (2, 11, 46, 52). At this stage E2A and HEB act to induce the expression of FOXO1, which, in concert with E2A, activates EBF expression to promote developmental progression into the B-cell-biased lymphoid progenitor (BLP) compartment (19, 46). During the pro-B cell stage, the E2A proteins induce a B-lineage program of gene expression and promote survival (15). Once a pre-B cell receptor (BCR) has been generated, pre-BCR-mediated signaling leads to a transient decline in E47 protein levels in order to facilitate clonal expansion of large pre-B cells (35). E47 protein levels increase again in proliferating pre-B cells to induce Igκ VJ gene rearrangement by direct binding to the Igκ 3′ and intronic enhancers (10, 12, 26, 35). In the absence of surface expression of a functional and innocuous BCR, E47 levels remain high and together with E12 initiate Igλ light chain rearrangements (3, 35). Once an antigen receptor has been generated that lacks auto-reactivity, tonic BCR signaling leads to a decline in E2A protein levels, and positively selected B cells migrate to the peripheral lymphoid organs (35). In peripheral activated B cells, E47 protein abundance is elevated again to induce Aicda expression and class switch recombination (CSR) (34, 40).
The E2A proteins are transcription factors that contain a basic DNA binding domain just N-terminal of the HLH-dimerization domain. They act as transcriptional regulators by forming either homodimers or heterodimers with other E proteins or other lineage-specific HLH proteins (22, 25, 41). The E proteins contain at least two transactivation domains, named AD1 and AD2. The AD1 domain acts to modulate the transcriptional activities of E proteins by recruitment of p300 or members of the ETO family (6, 33, 49). Recruitment of p300 leads to activation of gene expression, whereas association with members of the ETO family mediates transcriptional repression. Recent genome-wide studies have demonstrated that E2A occupancy is primarily associated with islands that contain mono- and dimethylated lysine 4 of histone 3 (H3K4) (19). However, within the immediate genomic proximity of E2A occupancy, the degree of H3K4 methylation is severely reduced compared to levels in its flanking regions (19).
The DNA binding activities of E2A proteins are regulated by members of the Id gene family, named Id1 to Id4 (4, 37, 39). Since Id proteins lack the basic DNA binding region, they inactivate E protein DNA binding upon heterodimerization. Posttranslational phosphorylation of E2A proteins has also been reported to influence E2A protein stability and activity. Phosphorylation of E47 by the mitogen-activated protein kinase (MAPK) p38 interferes with its transcriptional activity despite normal heterodimerization with MyoD and normal DNA binding (32). In contrast, phosphorylation of E47 by casein kinase II prevents homodimer formation and favors heterodimer formation with myogenic basic HLH (bHLH) proteins, potentially facilitating myogenesis (13). In mature B cells and aged B cell precursors, Notch-induced phosphorylation of E47 by MAPKs leads to enhanced E47 ubiquitination and degradation (14, 28). Additionally, two serine phosphorylation sites in E47 have been shown to be hypophosphorylated in B cell lines, and phosphorylation of these two sites disrupts E47 homodimer formation (43). Functionally, the E47-dependent activation of Rag gene transcription has been shown to depend on phosphorylation of E47 through the extracellular signal-regulated kinase (ERK)/MAPK pathway but not the phosphatidylinositol 3-kinase (PI3K) pathway in a B cell line (29).
Here, we have used immunopurification as well as affinity purification of tagged E47 combined with mass spectrometry to identify interacting factors. We find that E47 interacts with the entire ensemble of Id gene products in human embryonic kidney cells (HEK293T), including Id1, Id2, Id3, and Id4, demonstrating that E47 promiscuously associates with all members of the Id protein family. We also find that the lysine-specific histone demethylase 1 (LSD1), the protein arginine N-methyltransferase 5 (PRMT5), and the corepressor CoREST associate with affinity-purified E47. We identify a wide spectrum of E47 phosphorylation sites in pro-B and HEK293T cells, including a highly conserved AKT substrate site that is also present in HEB but not in the splice variant E12 and the isoform E2-2. We mapped the AKT phosphorylation site to a specific serine residue, located just N-terminal to the basic DNA binding region. However, we found that mutation of the identified AKT substrate site did not perturb B cell development, suggesting the presence of redundant pathways that link the PI3K-PTEN-AKT axis with E protein activity. Taken together, our observations verified that the entire ensemble of Id proteins has the ability to interact with E47. Furthermore, we identified novel factors interacting with E47, most notably the histone demethylase LSD1 and CoREST. We also identified a spectrum of posttranslational modifications of E47, including an AKT substrate site. Finally, we propose that redundant pathways connect E47 and the PTEN-PI3K-AKT axis to modulate B cell development.
MATERIALS AND METHODS
Cell culture.
Cells were cultured at 37°C under 5% CO2. HEK293T and EcR293T (Invitrogen) cells were cultured in Dulbecco's modified Eagle medium (DMEM) with 10% fetal bovine serum (FBS). For the generation of pro-B cells, total B cells from the bone marrow of adult C57BL/6 mice were isolated using anti-B220-coupled magnetic beads (Miltenyi Biotec) and cultured in Opti-MEM medium with 10% FBS, 100 U/ml penicillin-streptomycin, 2 mM l-glutamine, and 50 μM 2-mercaptoethanol in the presence of interleukin-7 (IL-7) and stem cell factor (SCF) for 8 days at 37°C in 5% CO2.
Expression of E47 in EcR293T cells.
Sequences encoding Flag and His peptides were fused to the N terminus of E47 and constructed into a vector containing a ponasterone-inducible promoter. A stable cell line was generated in EcR293T cells by isolating a clone which expresses high levels of E47 upon ponasterone stimulation. Cells were cultured in DMEM with 10% FBS, 400 μg/ml Zeocin, and 1 mg/ml Geneticin. E47 expression was induced using 5 mM ponasterone A.
E47 protein purification and mass spectrometry analysis.
In order to prepare human E47 protein for mass spectrometric or Western blot analysis, nuclear extracts were isolated from the E47-expressing EcR293T cell clone after ponasterone induction, and E47 protein complexes were purified using EZview Red anti-Flag M2 affinity gel (Sigma) and eluted twice with 100 mg/ml Flag peptide. The eluted fractions were combined and subjected to a second purification step using Talon metal affinity resins (Upstate). Tagged E47 was eluted using 200 mM imidazole in 300 mM sodium chloride and 50 mM sodium phosphate. The eluted fractions were combined, reduced, and alkylated using 2 mM Tris(2-carboxyethyl) phosphine (AC36383; Fisher) for 30 min at 37°C, followed by another 30-min incubation in the presence of 5 mM iodoacetamide (AC12227; Fisher) at 37°C. For analyzing mouse E47, nuclear extracts were isolated from in vitro pro-B cell cultures, and E47 protein complexes were immunoprecipitated using an anti-E47 antibody (clone 32.1; BD) and purified using protein G-Sepharose beads. The proteins were digested with 1 mg of trypsin (03 708 969 001; Roche) at 37°C overnight.
Automated two-dimensional (2D) nanoflow liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis was performed using a linear trap quadrupole (LTQ) tandem mass spectrometer (Thermo Electron Corporation, San Jose, CA) employing automated data-dependent acquisition. Raw data were extracted and searched using Spectrum Mill (version A.03.02.060b; Agilnet). MS/MS spectra with a sequence tag length of 1 or less were considered poor spectra and discarded. A concatenated forward-reverse data set was used to calculate the in situ identification false-positive rates (FDR). The enzyme parameter was limited to full tryptic peptides with a maximum miscleavage of 1. All other search parameters were set to Spectrum Mill's default settings (carbamidomethylation of cysteines, ± 2.5 Da for precursor ions, ± 0.7 Da for fragment ions, and a minimum matched peak intensity of 50%).
Generation of E2AS528A mice.
The linearized targeting vector, containing the E47 S528A mutation was electroporated into 129Sv/Ev embryonic stem (ES) cells, and correctly targeted clones were identified by Southern blot analyses. After injection into blastocysts and generation of chimeric mice, the correctly germ line-targeted mouse was backcrossed to Ella-cre mice in order to delete the neo cassette (16). Mice were subsequently backcrossed to C57BL/6 mice for 10 generations and, if applicable, to E2A-deficient mice. The following primers were used for genotyping: genoS528A_fd, CGGAGACATGGTCTAAGGAC; genoS528A_rev, GCCAATGTGCACAGTGATAG; 3′E2A, TGGTGCAGGATGAGCAGTTT; 5′E2A, GACGAGGACGAGGACGACCTTCT.
All animal experiments were carried out according to the guidelines of the University of California, San Diego (San Diego, CA).
FACS analyses.
Fluorescence-activated cell sorting (FACS) studies were performed on isolated bone marrow and spleens of mice that were between 6 to 12 weeks of age. Single-cell suspensions were generated and stained with fluorochrome-conjugated antibodies purchased from either eBioscience or BD Biosciences. Data were collected either with a FACSCalibur (BD Biosciences) or an LSRII flow cytometer (BD Biosciences) and were analyzed using FlowJo software (Tree Star). The following antibodies were used for flow cytometry: anti-B220 (RA3-6B2); CD19 (1D3); CD117/c-kit (2B8); CD25 (PC61); IgM (II/41); IgD (11-26); Ig(λ1), Ig(λ2), and Ig(λ3) light chain (R26-46); Ter119 (TER-119); Gr1 (RB6-8C5); CD11b/Mac1 (M1/70); CD3ε (145-2C11); and CD11c (N418).
RESULTS
Tandem affinity purification of E47-associating proteins.
To purify and characterize proteins associated with E47, we used a tandem affinity purification strategy. To accomplish this objective, sequences encoding Flag and His peptides were fused to the N terminus of E47 and stably transfected into EcR293T cells. One of the isolated clones (E47-13C2) showed particularly high levels of induction when cells were cultured in the presence of ponasterone A (data not shown). To identify protein components associating with E47, 108 cells were grown in culture, lysed, and salt extracted. E47 was purified from the nuclear extracts using affinity chromatography. Specifically, E47 was purified using anti-Flag affinity beads as well as a Talon affinity resin. Fractions were isolated from the elution and examined by Western blotting for the presence of E47. Fractions containing E47 were combined, reduced, alkylated, and digested with trypsin. Peptides derived from the digestion were subsequently analyzed by mass spectrometry for protein identification. Peptide sequences were then used to infer the identities of the E47-associating protein components (Table 1). As expected, the known suppressors of E2A activity, Id1, Id2, Id3, and Id4, were copurified, indicating that E47 has the ability to interact with all members of the Id protein family (Table 1). Additional bHLH proteins, including Twist and class B bHLH protein 8 were associated with E47 (Table 1). Interestingly, the histone demethylase LSD1, the arginine N-methyltransferase PRMT5, and the transcriptional corepressor CoREST were detected as factors associated with E47 (Table 1).
Table 1.
Proteins associated with E47 in human embryonic kidney cells as determined by mass spectrometric analysis
| Protein | Database accession no. | % amino acid coverage | No. of distinct peptides |
|---|---|---|---|
| E47 of transcription factor E2A | IPI00216911 | 61 | 27 |
| Id4 | IPI00026864 | 72 | 8 |
| Id1 | IPI00012718 | 61 | 7 |
| Id2 | IPI00294210 | 55 | 5 |
| Isoform 2 of LSD1 | IPI00217540 | 8 | 5 |
| PRMT5 | IPI00441473 | 11 | 4 |
| 39S ribosomal protein L37, mitochondrial precursor | IPI00162330 | 11 | 3 |
| 22-kDa protein | IPI00789159 | 17 | 4 |
| Ubiquinol-cytochrome-c reductase complex core protein 2, mitochondrial precursor | IPI00305383 | 10 | 3 |
| Mitochondrial 39S ribosomal protein L49 | IPI00013195 | 15 | 2 |
| Twist-related protein 1 | IPI00018907 | 14 | 2 |
| Isoform 3 of probable ATP-dependent RNA helicase DDX17 | IPI00651653 | 4 | 2 |
| Scaffold attachment factor B (SAF-B) | IPI00646058 | 2 | 2 |
| Isoform 2 of eukaryotic translation initiation factor 5A-1 (EIF5A) | IPI00376005 | 19 | 3 |
| Isoform 1 of myosin-lb | IPI00376344 | 2 | 2 |
| ELAV-like protein 1 | IPI00301936 | 8 | 2 |
| Id3 | IPI00219112 | 16 | 2 |
| Isoform 1 of genetic suppressor element 1 | IPI00215963 | 2 | 2 |
| Hypothetical protein | IPI00053535 | 4 | 2 |
| 39S ribosomal protein L12, mitochondrial precursor | IPI00005537 | 10 | 2 |
| Class B bHLH protein 8 | IPI00219822 | 20 | 2 |
| Prolactin-inducible protein precursor | IPI00022974 | 21 | 2 |
| Isoform long of delta 1-pyrroline-5-carboxylate synthetase | IPI00008982 | 3 | 2 |
| Splicing factor 3A subunit 3 | IPI00029764 | 7 | 2 |
| DnaJ (Hsp40) homolog, subfamily C, member 10 | IPI00293260 | 3 | 2 |
| Similar to large subunit ribosomal protein L36a | IPI00479645 | 13 | 2 |
| Isoform 1 of mitochondrial inner membrane protein | IPI00009960 | 3 | 2 |
| 40S ribosomal protein S13 | IPI00221089 | 16 | 2 |
| Probable ATP-dependent RNA helicase DDX20 | IPI00005904 | 3 | 2 |
| KIAA1546 protein (fragment) | IPI00175151 | 3 | 2 |
| Pre-mRNA processing-splicing factor 8 | IPI00007928 | 1 | 2 |
| REST corepressor 1 | IPI00008531 | 3 | 1 |
| Hypothetical class II bHLH protein (fragment) | IPI00737972 | 12 | 1 |
| RPL14 protein | IPI00555744 | 8 | 1 |
| Similar to serum paraoxonase/arylesterase 2a | IPI00742670 | 4 | 1 |
| Single-stranded DNA-binding protein, mitochondrial precursor | IPI00029744 | 10 | 1 |
| 60S ribosomal protein L31 | IPI00026302 | 11 | 1 |
| 39S ribosomal protein L44, mitochondrial precursor | IPI00009680 | 5 | 1 |
| Protein C14orf166 | IPI00006980 | 6 | 1 |
| Spindlin-1 | IPI00550655 | 6 | 1 |
| T-complex protein 1 subunit beta | IPI00297779 | 2 | 1 |
| Myosin light polypeptide 6B | IPI00027255 | 6 | 1 |
| Coiled-coil-helix-coiled-coil-helix domain-containing protein 3 | IPI00015833 | 6 | 1 |
| Sarco/endoplasmic reticulum Ca2+-ATPase isoform e | IPI00303760 | 1 | 1 |
| LanC-like protein 2 | IPI00032995 | 3 | 1 |
| Ezrin-radixin-moesin-binding phosphoprotein 50 | IPI00003527 | 3 | 1 |
| Isoform 2 of Import inner membrane translocase subunit TIM50, mitochondrial precursor | IPI00418497 | 3 | 1 |
| SPFH2 protein (fragment) | IPI00026942 | 4 | 1 |
| Isoform 2 of mitochondrial 39S ribosomal proteinL39 | IPI00084571 | 4 | 1 |
| Atonal homolog 8 | IPI00045865 | 6 | 1 |
| Serpin H1 precursor | IPI00032140 | 3 | 1 |
| Similar to protein SET | IPI00735319 | 4 | 1 |
| Isoform 1 of SON protein | IPI00218624 | 0 | 1 |
| rRNA 2′-O-methyltransferase fibrillarin | IPI00025039 | 5 | 1 |
| ADP/ATP translocase 1 | IPI00022891 | 3 | 1 |
| Elongation factor 1-α2 | IPI00014424 | 2 | 1 |
| Galectin-7 | IPI00219221 | 8 | 1 |
| Complement component 1 Q subcomponent-binding protein, mitochondrial precursor | IPI00014230 | 7 | 1 |
| Carboxypeptidase D precursor | IPI00027078 | 1 | 1 |
| 19-kDa protein | IPI00658024 | 9 | 1 |
| 9-kDa protein | IPI00397963 | 13 | 1 |
| 35-kDa protein | IPI00170779 | 6 | 1 |
| HLH protein 2 | IPI00026246 | 14 | 1 |
| 60S ribosomal protein L13 | IPI00465361 | 3 | 1 |
| Cytochrome c oxidase polypeptide Vlla-liver/heart, mitochondrial precursor | IPI00026570 | 11 | 1 |
| Cystatin A | IPI00032325 | 18 | 1 |
| Poly(rC)-binding protein 1 | IPI00016610 | 4 | 1 |
| NADH dehydrogenase (ubiquinone) 1 beta subcomplex subunit 4 | IPI00220059 | 14 | 1 |
| Trefoil factor 2 precursor | IPI00018909 | 7 | 1 |
| Similar to 40S ribosomal protein S26 isoform 1 | IPI00477047 | 13 | 1 |
| Lfa psoriasin | IPI00397801 | 0 | 1 |
| HNRPR protein | IPI00644055 | 2 | 1 |
| Isoform HERA-A of GTP-binding protein ra homolog | IPI00026512 | 3 | 1 |
| Dolichyl-diphosphooligosaccharide-protein glycosyltransferase 63-kDa subunit precursor | IPI00028635 | 2 | 1 |
| Isoform 1 of RNA-binding protein 39 | IPI00163505 | 3 | 1 |
| 19-kDa protein | IPI00793696 | 4 | 1 |
| Lamin-B receptor | IPI00292135 | 3 | 1 |
| Caspase-14 precursor | IPI00013885 | 4 | 1 |
| Serine-threonine protein kinase 38 | IPI00027251 | 2 | 1 |
| Calnexin precursor | IPI00020984 | 2 | 1 |
Synonyms: EC 3.1.1.2, EC 3.1.8.1, PON2; serum aryldialkylphosphatase 2/A-esterase 2/aromatic esterase 2.
Mass spectrometry identifies 14-3-3 proteins as interaction partners of E47 in pro-B cells.
To identify proteins that interact with E47 in primary B cells, mouse pro-B cells were generated by purification from bone marrow and cultured for 1 week in the presence of IL-7 and SCF. Nuclear lysates were prepared from the expanded pro-B cell population and E47-protein complexes were isolated using a monoclonal anti-E47 antibody. Associating proteins were eluted, digested with trypsin, and identified by mass spectrometry. The peptide sequences were used to infer the identities of the E47-associating proteins (Table 2). Among the identified proteins was the known E47-interacting protein Id3, the predominantly expressed Id protein in pro-B cells. Furthermore, among other novel putative E47-interacting proteins, 14-3-3 proteins were also identified to be associated with E47 in pro-B cells (Table 2). 14-3-3 scaffolding proteins interact preferentially with proteins containing phosphorylated serine or threonine residues (45). The original 14-3-3 binding motif has the consensus sequence RSX(pS)XP (where pS is phosphoserine); however, it seems that there is significant diversity in 14-3-3 binding motifs, resulting in a minimal consensus of RXX(pS/pT) or RXXX(pS/pT) (27, 50). Screening of the E47 protein sequence for 14-3-3 binding motifs using the PhosphoMotif Finder (http://www.hprd.org/PhosphoMotif_finder) revealed the presence of five putative 14-3-3 binding motifs (data not shown).
Table 2.
Proteins associated with E47 in mouse pro-B cells as determined by mass spectrometric analysis
| Protein name (mouse) | % amino acid coverage |
|---|---|
| Nucleolin | 22 |
| Ribosomal protein S3 | 48 |
| Ribosomal protein l27a | 28 |
| Ribosomal protein L4 | 19 |
| Nucleophosmin 1 | 36 |
| Replication factor C (activator 1) 3 | 17 |
| Laminin receptor 1 (ribosomal protein SA) | 27 |
| Stratifin (14-3-3 protein sigma) | 6 |
| 14-3-3 protein theta | 24 |
| Eukaryotic translation initiation factor 3 | 25 |
| Eukaryotic translation initiation factor 4A1 | 38 |
| Eukaryotic translation initiation factor 4A2 | 14 |
| Eukaryotic translation elongation factor 1 | 43 |
| DEAD (Asp-Glu-Ala-Asp) box polypeptide 5 | 16 |
| DNA methyltransferase (cytosine-5) 1 | 6 |
| Non-POU-domain, octamer binding protein | 23 |
| Histone 1, h4h | 52 |
| Histone 4, H4 | 52 |
| Histone 1, h4c | 52 |
| Histone 2, h2ab | 49 |
| Histone 3, h2a | 49 |
| Histone 1, h4k | 52 |
| TAF15 RNA polymerase II, TBP-associated | 7 |
| Retinoblastoma binding protein 4 | 16 |
| Retinoblastoma binding protein 7 | 7 |
| Proteasome 26S non-ATPase subunit 11 | 22 |
| Proteasome 26S non-ATPase subunit 13 | 17 |
| Proteasome 26S non-ATPaase subunit 2 | 13 |
| Proteasome activator subunit 3 | 16 |
| Ubiquitin-conjugating enzyme E2N | 23 |
| Ubiquitin-conjugating enzyme E2I | 8 |
| Ubiquitin B | 23 |
| Pre-mRNA processing factor 8 | 2 |
| PRP31 | 5 |
| Splicing factor (ASF/SF2) | 18 |
| Splicing factor 3b, subunit 3 | 8 |
| snRNP core protein SMX5 | 14 |
| Inhibitor of DNA binding 3 | 10 |
| Stress-induced phosphoprotein 1 | 10 |
| Proliferating cell nuclear antigen | 37 |
| SWI/SNF related regulator of chromatin | 8 |
| Rpl7a protein | 17 |
| Programmed cell death 6 | 29 |
| p21-activated kinase 2 | 14 |
| Chromosome condensation protein G | 5 |
| Chromodomain helicase DNA binding 4 | 6 |
| S-phase kinase-associated protein 1A | 33 |
| Chromosome condensation-related SMC 1 | 4 |
| DJ-1 protein | 22 |
| RNA polymerase II transcriptional coactivator | 15 |
Mass spectrometry identifies a spectrum of phosphorylated residues across E47.
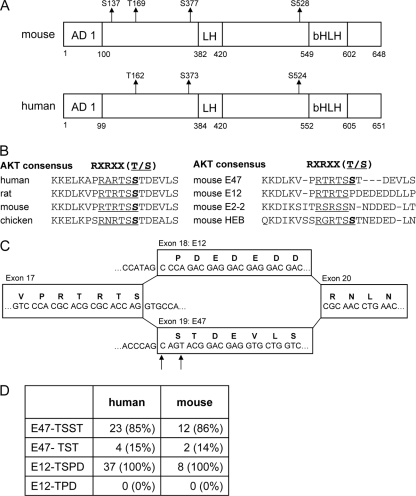
In order to determine whether E47 contains a phosphorylated 14-3-3 binding motif, we purified E47 as described above from murine pro-B cells in the presence of phosphatase inhibitors, digested it with trypsin, and subjected the peptides to mass spectrometry. Furthermore, Flag- and His-tagged human E47 was expressed in EcR293T cells, purified twice by affinity chromatography, trypsin digested, and analyzed by mass spectrometry. This analysis identified three serine residues (S137, S377, and S528) and one threonine residue (T169) that were phosphorylated in mouse pro-B cells (Fig. 1A, top). Three of these sites were also phosphorylated in human E47 expressed in EcR293T cells (Fig. 1A, bottom). One of these residues, murine serine residue 528, mapped to a putative 14-3-3 binding site. Further analysis of the amino acid sequence spanning the S528 residue using the PhosphoMotif Finder identified a potential AKT substrate recognition site (RARTSS). Comparison of this putative AKT phosphorylation site in different species showed a remarkable degree of conservation (Fig. 1B). Notably, whereas S528 is present in E47, it is absent in E12 (Fig. 1B and C). Serine residue 528 is also present in HEB but not in E2-2 (Fig. 1B). Interestingly, there are two splice acceptor sites (consensus sequence, NAG) located at the 5′ boundary of the E47 exon (Fig. 1C) (7). We note that, in principle, alternative splice acceptor usage of exon 19 may lead to the exclusion of residue S528. To further evaluate this possibility, we analyzed expressed sequence tags (ESTs) from the NCBI database, which matched 21 nucleotides spanning the alternatively spliced region (1). We observed that the majority of cDNAs encoding E47 contained two serine residues, whereby the second serine residue matches the potential AKT site (Fig. 1D). However, a minor fraction of ESTs that were analyzed contained only a single serine residue. Thus, differential splicing of the E2A gene results in the formation of two distinct E47 proteins, one of which contains a putative AKT phosphorylation site.
Fig 1.
Phosphoproteomic analysis of E47. (A) Murine and human E47 are phosphorylated at multiple residues. Murine E47 was purified from in vitro pro-B cell cultures using an anti-E47 antibody. His- and Flag-tagged human E47 was expressed in EcR293T cells and purified by affinity chromatography. Purified E47 was subjected to trypsin digestion and analyzed for posttranslational modifications by mass spectrometry. Indicated are the positions of phosphorylated serine and threonine residues. The transactivation domains AD1 and LH (loop-helix) as well as the bHLH region are depicted in the diagram. Numbers indicate amino acid positions. (B) Sequence homology of the E47 protein from human, rat, mouse, and chicken spanning the putative AKT substrate site (RXRXXT/S) (left side) and sequence homology of this RXRXXT/S site in the related E proteins E12, E2-2, and HEB (right side). The phosphorylated serine residue is indicated in bold. (C) Diagram indicating the organization of exons 17 to 20 of the mouse E2A gene. Due to the presence of a tandem acceptor splice sequence (NAGNAG, indicated by arrows), splicing to E47 will result in the protein sequence RTRTSSTDE or RTRTSTDE. (D) Table showing the results of a sequence comparison of ESTs spanning the alternative spliced region. Depicted are the absolute numbers and percentages of ESTs containing the described sequences.
AKT phosphorylation at serine residue S524.
In order to verify that E47 contains a phosphorylated AKT motif as well as a phosphorylated potential 14-3-3 binding site, nuclear extracts were generated from EcR293T cells expressing the Flag- and His-tagged E47. E47 was affinity purified, and the different elution fractions containing purified E47 were analyzed by Western blotting for reactivity to antibodies recognizing the phosphorylated 14-3-3 and AKT substrate sites (Fig. 2A). The data showed that human E47 expressed in EcR293T cells contains putative 14-3-3 [RXX(pS)] and AKT [RXRXX(pS/pT)] substrate sites.
Fig 2.
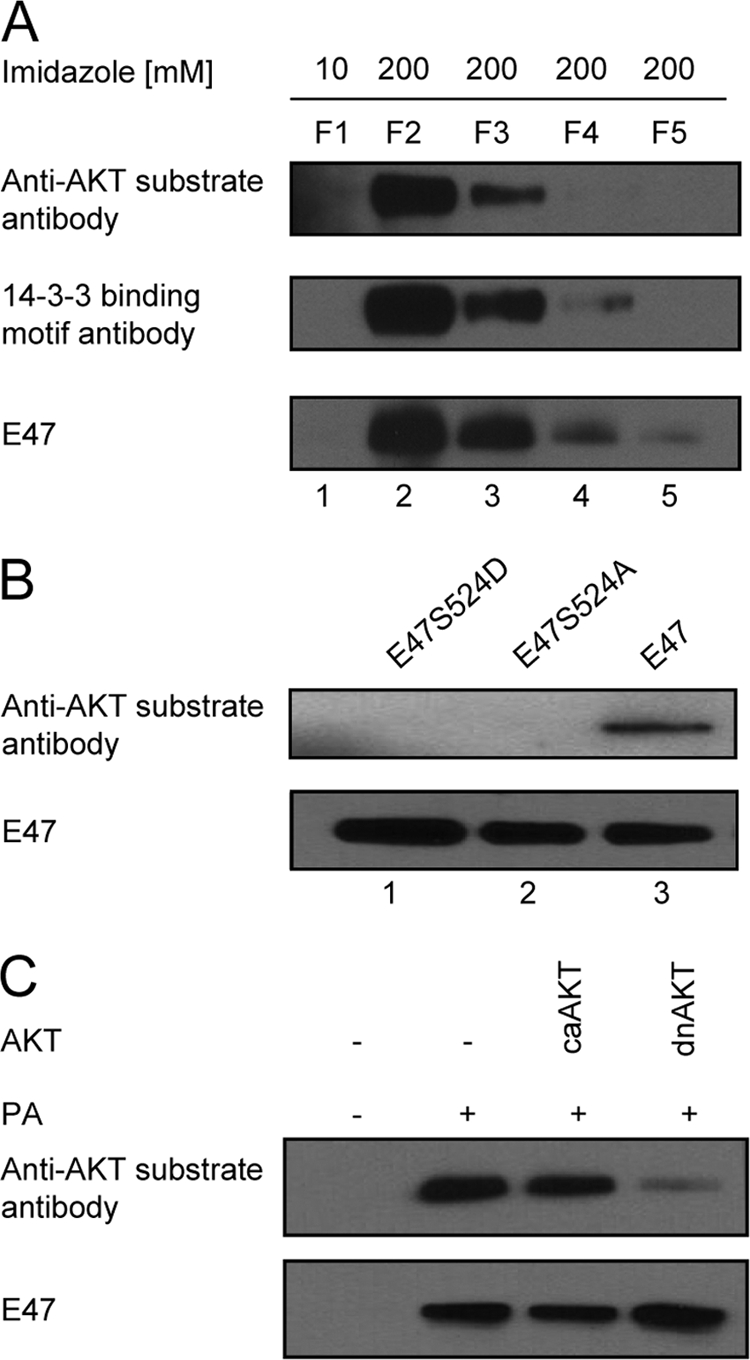
E47 is a substrate for AKT. (A) Epitope-tagged E47 was affinity purified from EcR293T cells. Purified E47 was analyzed by Western blotting using antibodies specific for 14-3-3 and AKT substrate sites as well as E47. F1 indicates the flowthrough fraction eluted with 10 mM imidazole, representing the washing. F2 to F5 are fractions eluted with 200 mM imidazole containing purified E47. (B) EcR293T cells were transfected with expression vectors expressing either wild-type E47, E47-S524A, or E47-S524D. Nuclear extracts were derived from transfected cells, immunoprecipitated using an antibody recognizing E47, and immunoblotted using antibodies recognizing the AKT substrate site and E47. (C) EcR293T cells, which express E47 upon ponasterone (PA) stimulation were transfected with vectors encoding a constitutively active form of AKT1 (caAKT) or a dominant negative form of AKT1 (dnAKT). Upon PA stimulation, E47 protein was purified, and immunoblotting was performed with antibodies recognizing the phosphorylated AKT substrate site and E47.
To further examine if the human E47 S524 is phosphorylated and responsible for the immunoreactivity to the AKT substrate antibody, residue S524 was replaced by either an aspartate (E47-S524D) or alanine (E47-S524A) residue. Expression vectors encoding wild-type (WT) E47, E47-S524D, and E47-S524A were transfected into HEK293T cells. Subsequently, E47 was immunoprecipitated and analyzed by Western blotting using an AKT substrate antibody. Replacing S524 with either an aspartate or alanine residue interfered with the ability of the AKT substrate antibody to detect purified E47, confirming that S524 is the only phosphorylated serine residue in E47 recognized by the AKT substrate antibody (Fig. 2B).
To determine whether AKT activity is required to phosphorylate serine residue 524, EcR293T cells expressing E47 upon ponasterone stimulation were transfected with vectors expressing either a constitutively active AKT (caAKT) or a dominant negative (dnAKT) form of AKT. Expressing the constitutively active form of AKT did not enhance the fraction of E47 recognized by the anti-AKT substrate antibody, probably due to high levels of endogenous AKT activity in the EcR293T cell line (Fig. 2C). On the other hand, the proportion of phosphorylated E47 was substantially reduced upon enforced expression of a dominant negative form of AKT (Fig. 2C). Collectively, these data indicate that E47 is phosphorylated by AKT at serine residue 524, resulting in a putative 14-3-3 binding site.
Generation of E2AS528A/S528A mutant mice.
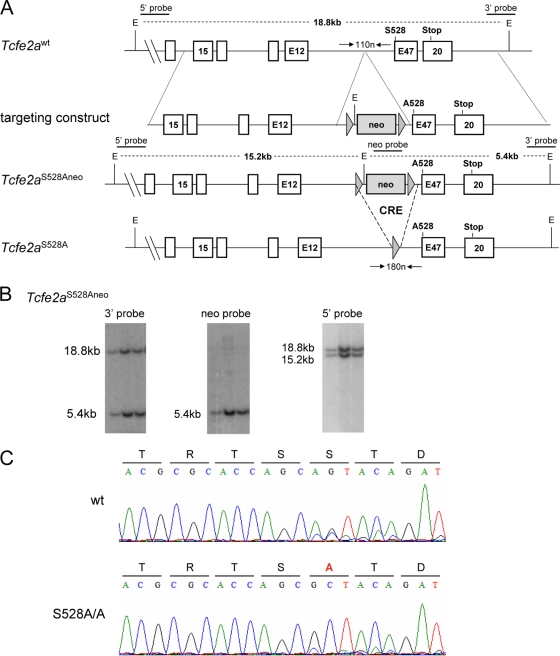
To address the role of this AKT phosphorylation site in vivo, we generated mice in which S528 of E47 was replaced by an alanine residue using insertional mutagenesis (Fig. 3A). ES cells were targeted for replacement using a vector in which the conserved serine residue was replaced by alanine. Genomic DNA was isolated from the targeted ES cells and examined by Southern blotting using the appropriate probes for the presence of the mutation (Fig. 3B). Mutant mice were generated from ES cells carrying the appropriate E2A allele. Subsequently, mice were crossed with Ella-cre transgenic mice in order to excise the neo gene and bred to homozygosity. To verify that the alanine mutation was properly inserted, mRNA was isolated from splenic B cells isolated from wild-type (WT) and homozygous E2AS528A/S528A mice, reverse transcribed, and sequenced (Fig. 3C). As expected, the triplet encoding the serine residue (AGT) was replaced by a triplet encoding an alanine amino acid (GCT) (Fig. 3C). Mice were then backcrossed onto the C57BL/6 background. They were viable, bred with normal Mendelian frequencies, and did not exhibit gross abnormalities (data not shown).
Fig 3.
Generation of E2AS528A knock-in mice by homologous recombination in ES cells. (A) The top bar represents a map of the mouse Tcfe2a gene encompassing the area around the E47 exon (white boxes represent exons), which encodes the targeted amino acid serine 528 (S528). The second bar shows the targeting construct, which contains two arms of homology, as well as the neomycin resistance gene (gray box) flanked by two loxP sites (gray triangles). The 3′ arm contained the substitution of serine 528 to alanine (A528). The third bar depicts the correctly targeted Tcfe2a allele (Tcfe2aS528Aneo), and the bottom bar depicts the correctly targeted Tcfe2a allele upon Cre recombinase-mediated excision of the neomycin resistance gene (Tcfe2aS528A). On top of the wild-type locus and the targeted allele, the positions of the external 5′ and 3′ probes and the internal neo probe for Southern blot hybridization, EcoRI (E) restriction sites, and the appropriate DNA sizes are indicated. Underneath the wild-type locus and the targeted allele, horizontal arrowheads indicate the positions of primers used for genotyping and the size of the expected bands. (B) Southern blot analysis of EcoRI-digested genomic DNA from three correctly targeted ES cell clones, each containing one wild-type and one targeted Tcfe2a allele. The position of the indicated probes is shown in panel A. The sizes of the obtained bands match the expected fragment sizes for the wild-type allele (18.8 kb) and the targeted allele (5.4 and 15.2 kb, respectively) as shown in panel A. (C) mRNA was isolated from splenic B cells from the indicated mice, cDNA was generated, and a PCR to amplify the E47 transcript was performed using specific primers (the forward primer spans exon 16/17, and the reverse primer aligns to exon 20). Sequencing was performed on the PCR product using the forward primer, and the appropriate amino acid translation is depicted above the obtained sequence.
E2AS528A/S528A mice exhibit normal B cell development.
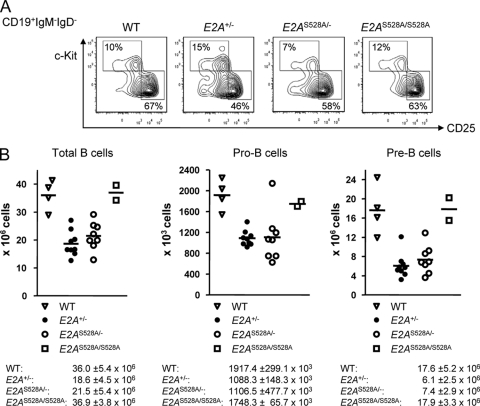
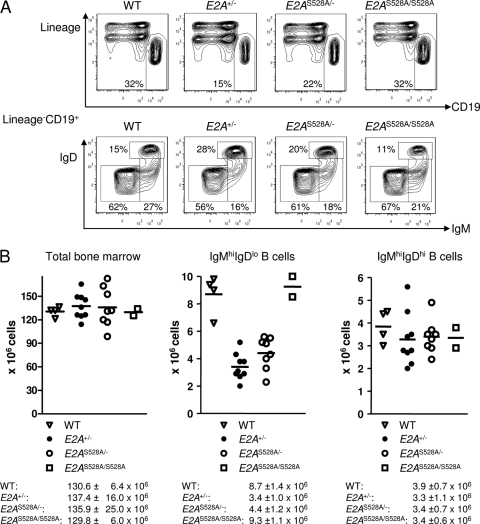
In order to determine, if PI3K-AKT-mediated phosphorylation of E2A is required to promote proper B cell development, E2AS528A mice were analyzed using flow cytometry. Specifically, bone marrow cells derived from WT, E2AS528A/S528A, E2A+/−, and E2AS528A/− mice were stained with antibodies directed against CD19, c-kit, CD25, IgM, and IgD. We found that both the pro-B (CD19+ IgM− IgD− c-kit+ CD25−) and pre-B (CD19+ IgM− IgD− c-kit− CD25+) cell compartments were not significantly altered in E2AS528A/S528A mice compared to WT mice (Fig. 4). Consistent with previous observations, E2A+/− mice showed a significant decrease in the pre-B cell compartment (34). However, replacement of S528 by an alanine residue in conjunction with a loss-of-function allele did not further decrease or rescue the proportion of pre-B cells compared to E2A+/− or WT mice (Fig. 4). The immature (CD19+ IgMhi IgDlo) and mature B (CD19+ IgMhi IgDhi) cell compartments in the bone marrow were not affected by replacement of serine residue 528 (Fig. 5). Cellularity was also not significantly perturbed in E2AS528A/S528A mice compared to WT mice (Fig. 5). Taken together, these data indicate that phosphorylation of E47 serine residue 528 is not essential for proper E47 function during B cell development.
Fig 4.
Analysis of early B cell development in E2AS528A mutant mice. (A) Representative FACS analysis of pro- and pre-B cells in the bone marrow of WT, E2A+/−, E2AS528A/−, and E2AS528A/S528A mice. Plots are gated on live CD19+ IgM− IgD− cells, and the numbers indicate the percentages of the depicted gates within the CD19+ IgM− IgD− fraction. (B) Total cell numbers of the indicated compartments are shown for individual mice. The horizontal line represents the mean. Means and standard deviations of total cell numbers are shown below the appropriate graphs.
Fig 5.
Analysis of B cell development in E2AS528A mutant mice. (A) Representative FACS analysis of immature and mature B cells in the bone marrow of WT, E2A+/−, E2AS528A/−, and E2AS528A/S528A mice. Plots are gated on live cells (top row) or lineage-negative CD19+ cells (bottom row). The numbers indicate the percentages of the gates within the depicted fraction. (B) Total cell numbers of the indicated compartments are shown for individual mice. The horizontal line represents the mean. Means and standard deviations of total cell numbers are shown below the appropriate graphs.
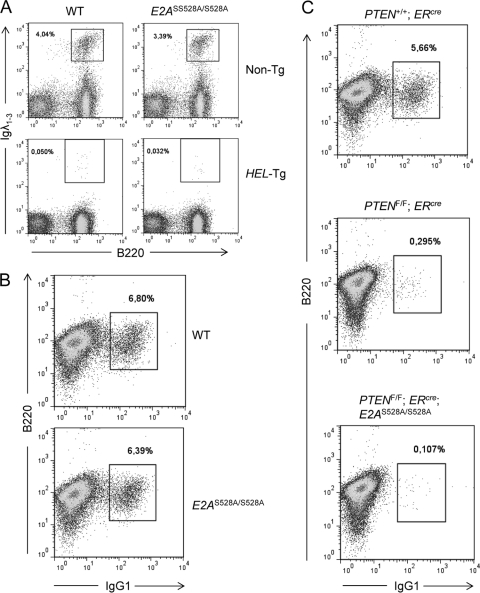
Previous studies have demonstrated that E47 plays a critical role in receptor editing, including rearrangement of the Ig light chain loci (35). To directly evaluate the role of E47-S528 in Igλ VJ rearrangements, we examined mature B cells of WT and E2AS528A/S528A mice for abnormalities in Igλ expression both in the presence and absence of the HEL transgene (HEL-Tg) (Fig. 6A). We identified normal numbers of Igλ+ B cells in the spleens of HEL-Tg; E2AS528A/S528A mice compared to HEL-Tg; E2A+/+ mice, indicating that E47-S528 does not play a critical role in modulating Igλ VJ rearrangement (Fig. 6A).
Fig 6.
Normal receptor editing, allelic exclusion, and class switch recombination in E2AS528A mice. (A) Analysis of Igλ light chain expression in E2AS528A mutant mice. Representative FACS analysis of Igλ-expressing B cells in the spleen of nontransgenic (Non-Tg) or BCR transgenic (HEL-Tg) mice expressing either the WT or the S528A mutant E47 protein. Plots are gated on live cells, and the numbers indicate the percentages of Igλ+ B cells among live splenocytes. (B) Analysis of class switch recombination of E2AS528A mutant peripheral B cells. Representative FACS analysis of IgG1-expressing B cells isolated from the spleen of WT or E2AS528A/S528A mice after in vitro stimulation with LPS and IL-4 for 3 days. Plots are gated on live cells, and the numbers indicate the percentages of IgG1+ B cells among the live cells. (C) B cells were isolated from the spleen of ERcre, PTENflox/flox; ERcre, and PTENflox/flox; ERcre; E2AS528A/S528A mice after 1 week of tamoxifen treatment. Shown are representative FACS plots of IgG1-expressing B cells after 3 days of in vitro stimulation with LPS and IL-4. Plots were gated on live cells. Numbers indicate the percentages of IgG1+ B cells among the live cells.
In previous studies we have demonstrated that E2A activates class switch recombination (CSR) (34, 40). This process is accompanied by an increase in E47 protein levels upon B cell stimulation; however, mRNA levels remain unchanged, indicative of a posttranslational regulation of the E47 protein (34). To determine whether B cells from E2AS528A/S528A mice have the ability to undergo CSR, splenic naïve B cells were isolated, stimulated in vitro for 3 days with lipopolysaccharide (LPS) and IL-4, and examined for the appearance of IgG1+-expressing B cells (Fig. 6B). We found that B cells derived from E2AS528A/S528A mice were not impaired in their ability to undergo CSR (Fig. 6B).
PTEN-deficient B cells, which exhibit increased AKT activity, have previously been shown to be impaired in their ability to undergo CSR (31). We therefore considered the possibility that PTEN and E2A are directly linked in activated B cells via the putative AKT phosphorylation site S528 to induce the expression of Aicda and CSR. Hence, we generated PTENflox/flox; ERcre as well as PTENflox/flox; ERcre; E2AS528A/S528A mice. After tamoxifen-induced PTEN deletion, naïve B cells were isolated and stimulated with LPS and IL-4 for 3 days in vitro and monitored for the generation of IgG1+ cells (Fig. 6C). Mice expressing the E47-S528A protein were not able to rescue the defect in CSR observed in PTEN-ablated B cells. In sum, these data indicate that replacement of E47 serine residue 528 does not significantly impair B cell development neither in the bone marrow nor in the periphery.
DISCUSSION
Numerous studies have documented distinct and essential roles for the E2A proteins throughout hematopoiesis (8, 25, 30). The E2A proteins are not merely indispensable for early B cell development but also continue to have critical functions in maturing B cells, including V(D)J rearrangement, receptor revision, follicular B cell development, and class switch recombination. These data bring into question how the E2A proteins are regulated and mediate such a diverse set of activities.
The best-characterized modulators of E2A activity are the Id proteins (4, 37). Four proteins have been identified, named Id1, Id2, Id3, and Id4. In vitro they all have the ability to interact with E proteins (4, 17, 20, 36, 44). Consistent with these observations are functional studies in which E- and Id-deficient mice exhibit opposite defects at multiple stages of lymphopoiesis. Furthermore, genetic studies using compound E protein and Id mutant mice have confirmed such interactions (24, 38, 47). However, it has remained to be determined whether E2A proteins interact indiscriminately with the entire spectrum of Id proteins. Here, using affinity chromatography and nuclear extracts derived from human embryonic kidney (HEK293T) as well as pro-B cells, we find that the Id proteins are among the most prominent factors that associate with E47. Intriguingly, we find that in HEK293T cells the entire ensemble of Id proteins associates with E47. These data do not imply that there are no subtle differences in affinity between the various Id members for E47. Nevertheless, they do indicate that E47 has the capacity to interact with all four members of the Id protein family in vivo. In pro-B cells only Id3 was found to associate with E47, consistent with it being the only Id member expressed in proliferating pro-B cells.
Recent data also have clarified the mechanism by which E proteins mediate transcriptional activation and repression. Specifically, whereas E proteins have been shown to recruit p300 to activate transcription, they also have the ability to interact with members of the ETO family in order to repress downstream target gene expression (6, 33, 49). Here, we identify additional factors that copurify with E47 in HEK293T cells. Prominent among these are the histone methyltransferase, LSD1, the protein arginine N-methyltransferase 5 (PRMT5), and the corepressor CoREST. The recruitment of LSD1 by E47 is particularly interesting. Previous genome-wide studies have demonstrated that E47 occupancy is primarily associated with regions of H3K4 monomethylation (19). However, within close genomic proximity of E47 binding, the extent of methylated H3K4 is greatly reduced. The reduction in H3K4 methylation across E47 binding sites might be caused, in part, by nucleosome depletion, but it may also involve a reduction in the methylation of H3K4 by recruitment of LSD1. Further experiments will be required to determine whether and how E47 and LSD1 act coordinately to regulate specific programs of gene expression.
The phosphoproteomic analysis also revealed a number of E47 phosphorylated serine residues located across the N-terminal domains. Most prominent among these is a putative AKT substrate site (S528) located immediately N-terminal of the bHLH domain. Interestingly, residue S528 is present in E47 but absent in E12. Furthermore, two distinct acceptor splice sites flank the E47 exon, resulting in two distinct E47 isoforms, with one of them containing a putative AKT phosphorylation site and the other isoform lacking S528. These data indicate the presence of at least three E2A isoforms. However, mutation of S528 into an alanine residue in the murine germ line did not grossly affect B cell development. These data do not exclude the possibility that phosphorylation of S528 by AKT signaling is not essential for proper B cell development. It is plausible that mutation of the single serine residue (S528) enables AKT to phosphorylate adjacent serine or threonine residues to modulate E47 activity. Furthermore, it is conceivable that phosphorylation of S528 by AKT acts in concert with modulation of HEB or Id expression and/or activity to promote developmental progression, and further studies will be required to resolve the question as to how AKT signaling and E47 activity are linked in a common framework to modulate B cell development.
ACKNOWLEDGMENTS
We are grateful to Robert Rickert for the CD19Cre and PTENflox/flox mice. We thank members of the Murre laboratory for reagents and stimulating discussions.
R.T. and L.Y.T.W. were supported by a training grant from the NIH (Cellular and Molecular Genetics). K.B was supported by the NIH. This work was supported by grants from the NIH to C.M.
Footnotes
Published ahead of print 21 February 2012
REFERENCES
- 1. Adams MD, et al. 1991. Complementary DNA sequencing: expressed sequence tags and human genome project. Science 252:1651–1656 [DOI] [PubMed] [Google Scholar]
- 2. Bain G, et al. 1994. E2A proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements. Cell 79:885–892 [DOI] [PubMed] [Google Scholar]
- 3. Beck K, Peak MM, Ota T, Nemazee D, Murre C. 2009. Distinct roles for E12 and E47 in B cell specification and the sequential rearrangement of immunoglobulin light chain loci. J. Exp. Med. 206:2271–2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H. 1990. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell 61:49–59 [DOI] [PubMed] [Google Scholar]
- 5. Borghesi L, et al. 2005. E47 is required for V(D)J recombinase activity in common lymphoid progenitors. J. Exp. Med. 202:1669–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bradney C, et al. 2003. Regulation of E2A activities by histone acetyltransferases in B lymphocyte development. J. Biol. Chem. 278:2370–2376 [DOI] [PubMed] [Google Scholar]
- 7. Chen M, Manley JL. 2009. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat. Rev. Mol. Cell Biol. 10:741–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. de Pooter RF, Kee BL. 2010. E proteins and the regulation of early lymphocyte development. Immunol. Rev. 238:93–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dias S, Mansson R, Gurbuxani S, Sigvardsson M, Kee BL. 2008. E2A proteins promote development of lymphoid-primed multipotent progenitors. Immunity 29:217–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Greenbaum S, Zhuang Y. 2002. Identification of E2A target genes in B lymphocyte development by using a gene tagging-based chromatin immunoprecipitation system. Proc. Natl. Acad. Sci. U. S. A. 99:15030–15035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Inlay MA, et al. 2009. Ly6d marks the earliest stage of B-cell specification and identifies the branchpoint between B-cell and T-cell development. Genes Dev. 23:2376–2381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Inlay MA, Tian H, Lin T, Xu Y. 2004. Important roles for E protein binding sites within the immunoglobulin κ chain intronic enhancer in activating Vκ Jκ rearrangement. J. Exp. Med. 200:1205–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Johnson SE, Wang X, Hardy S, Taparowsky EJ, Konieczny SF. 1996. Casein kinase II increases the transcriptional activities of MRF4 and MyoD independently of their direct phosphorylation. Mol. Cell. Biol. 16:1604–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. King AM, Van der Put E, Blomberg BB, Riley RL. 2007. Accelerated Notch-dependent degradation of E47 proteins in aged B cell precursors is associated with increased ERK MAPK activation. J. Immunol. 178:3521–3529 [DOI] [PubMed] [Google Scholar]
- 15. Kwon K, et al. 2008. Instructive role of the transcription factor E2A in early B lymphopoiesis and germinal center B cell development. Immunity 28:751–762 [DOI] [PubMed] [Google Scholar]
- 16. Lakso M, et al. 1996. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc. Natl. Acad. Sci. U. S. A. 93:5860–5865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Langlands K, Yin X, Anand G, Prochownik EV. 1997. Differential interactions of Id proteins with basic-helix-loop-helix transcription factors. J. Biol. Chem. 272:19785–19793 [DOI] [PubMed] [Google Scholar]
- 18. Lazorchak A, Jones ME, Zhuang Y. 2005. New insights into E-protein function in lymphocyte development. Trends Immunol. 26:334–338 [DOI] [PubMed] [Google Scholar]
- 19. Lin YC, et al. 2010. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat. Immunol. 11:635–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Loveys DA, Streiff MB, Kato GJ. 1996. E2A basic-helix-loop-helix transcription factors are negatively regulated by serum growth factors and by the Id3 protein. Nucleic Acids Res. 24:2813–2820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mandel EM, Grosschedl R. 2010. Transcription control of early B cell differentiation. Curr. Opin. Immunol. 22:161–167 [DOI] [PubMed] [Google Scholar]
- 22. Massari ME, Murre C. 2000. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol. Cell. Biol. 20:429–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mercer EM, Lin YC, Murre C. 2011. Factors and networks that underpin early hematopoiesis. Semin. Immunol. 23:317–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Miyazaki M, et al. 2011. The opposing roles of the transcription factor E2A and its antagonist Id3 that orchestrate and enforce the naive fate of T cells. Nat. Immunol. 12:992–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Murre C. 2005. Helix-loop-helix proteins and lymphocyte development. Nat. Immunol. 6:1079–1086 [DOI] [PubMed] [Google Scholar]
- 26. Murre C, McCaw PS, Baltimore D. 1989. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell 56:777–783 [DOI] [PubMed] [Google Scholar]
- 27. Muslin AJ, Tanner JW, Allen PM, Shaw AS. 1996. Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell 84:889–897 [DOI] [PubMed] [Google Scholar]
- 28. Nie L, Xu M, Vladimirova A, Sun XH. 2003. Notch-induced E2A ubiquitination and degradation are controlled by MAP kinase activities. EMBO J. 22:5780–5792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Novak R, Jacob E, Haimovich J, Avni O, Melamed D. 2010. The MAPK/ERK and PI3K pathways additively coordinate the transcription of recombination-activating genes in B lineage cells. J. Immunol. 185:3239–3247 [DOI] [PubMed] [Google Scholar]
- 30. Nutt SL, Kee BL. 2007. The transcriptional regulation of B cell lineage commitment. Immunity 26:715–725 [DOI] [PubMed] [Google Scholar]
- 31. Omori SA, et al. 2006. Regulation of class-switch recombination and plasma cell differentiation by phosphatidylinositol 3-kinase signaling. Immunity 25:545–557 [DOI] [PubMed] [Google Scholar]
- 32. Page JL, Wang X, Sordillo LM, Johnson SE. 2004. MEKK1 signaling through p38 leads to transcriptional inactivation of E47 and repression of skeletal myogenesis. J. Biol. Chem. 279:30966–30972 [DOI] [PubMed] [Google Scholar]
- 33. Qiu Y, Sharma A, Stein R. 1998. p300 mediates transcriptional stimulation by the basic helix-loop-helix activators of the insulin gene. Mol. Cell. Biol. 18:2957–2964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Quong MW, Harris DP, Swain SL, Murre C. 1999. E2A activity is induced during B-cell activation to promote immunoglobulin class switch recombination. EMBO J. 18:6307–6318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Quong MW, et al. 2004. Receptor editing and marginal zone B cell development are regulated by the helix-loop-helix protein, E2A. J. Exp. Med. 199:1101–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Riechmann V, van Cruchten I, Sablitzky F. 1994. The expression pattern of Id4, a novel dominant negative helix-loop-helix protein, is distinct from Id1, Id2 and Id3. Nucleic Acids Res. 22:749–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rivera R, Murre C. 2001. The regulation and function of the Id proteins in lymphocyte development. Oncogene 20:8308–8316 [DOI] [PubMed] [Google Scholar]
- 38. Rivera RR, Johns CP, Quan J, Johnson RS, Murre C. 2000. Thymocyte selection is regulated by the helix-loop-helix inhibitor protein, Id3. Immunity 12:17–26 [DOI] [PubMed] [Google Scholar]
- 39. Ruzinova MB, Benezra R. 2003. Id proteins in development, cell cycle and cancer. Trends Cell Biol. 13:410–418 [DOI] [PubMed] [Google Scholar]
- 40. Sayegh CE, Quong MW, Agata Y, Murre C. 2003. E-proteins directly regulate expression of activation-induced deaminase in mature B cells. Nat. Immunol. 4:586–593 [DOI] [PubMed] [Google Scholar]
- 41. Schwartz R, Engel I, Fallahi-Sichani M, Petrie HT, Murre C. 2006. Gene expression patterns define novel roles for E47 in cell cycle progression, cytokine-mediated signaling, and T lineage development. Proc. Natl. Acad. Sci. U. S. A. 103:9976–9981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Semerad CL, Mercer EM, Inlay MA, Weissman IL, Murre C. 2009. E2A proteins maintain the hematopoietic stem cell pool and promote the maturation of myelolymphoid and myeloerythroid progenitors. Proc. Natl. Acad. Sci. U. S. A. 106:1930–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sloan SR, Shen CP, McCarrick-Walmsley R, Kadesch T. 1996. Phosphorylation of E47 as a potential determinant of B-cell-specific activity. Mol. Cell. Biol. 16:6900–6908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sun XH, Copeland NG, Jenkins NA, Baltimore D. 1991. Id proteins Id1 and Id2 selectively inhibit DNA binding by one class of helix-loop-helix proteins. Mol. Cell. Biol. 11:5603–5611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tzivion G, Avruch J. 2002. 14-3-3 Proteins: active cofactors in cellular regulation by serine/threonine phosphorylation. J. Biol. Chem. 277:3061–3064 [DOI] [PubMed] [Google Scholar]
- 46. Welinder E, et al. 2011. The transcription factors E2A and HEB act in concert to induce the expression of FOXO1 in the common lymphoid progenitor. Proc. Natl. Acad. Sci. U. S. A. 108:17402–17407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yan W, et al. 1997. High incidence of T-cell tumors in E2A-null mice and E2A/Id1 double-knockout mice. Mol. Cell. Biol. 17:7317–7327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yang Q, et al. 2008. E47 controls the developmental integrity and cell cycle quiescence of multipotential hematopoietic progenitors. J. Immunol. 181:5885–5894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang J, Kalkum M, Yamamura S, Chait BT, Roeder RG. 2004. E protein silencing by the leukemogenic AML1-ETO fusion protein. Science 305:1286–1289 [DOI] [PubMed] [Google Scholar]
- 50. Zhang SH, Kobayashi R, Graves PR, Piwnica-Worms H, Tonks NK. 1997. Serine phosphorylation-dependent association of the band 4.1-related protein-tyrosine phosphatase PTPH1 with 14-3-3β protein. J. Biol. Chem. 272:27281–27287 [DOI] [PubMed] [Google Scholar]
- 51. Zhuang Y, Jackson A, Pan L, Shen K, Dai M. 2004. Regulation of E2A gene expression in B-lymphocyte development. Mol. Immunol. 40:1165–1177 [DOI] [PubMed] [Google Scholar]
- 52. Zhuang Y, Soriano P, Weintraub H. 1994. The helix-loop-helix gene E2A is required for B cell formation. Cell 79:875–884 [DOI] [PubMed] [Google Scholar]