Abstract
The 5′ untranslated region (UTR) plays a central role in the regulation of mammalian translation initiation. Key components include RNA structure, upstream AUGs (uAUGs), upstream open reading frames (uORFs), and internal ribosome entry site elements that can interact to modulate the readout. We previously reported the characterization of two alternatively spliced 5′ UTR isoforms of the human elk-1 gene. Both contain two uAUGs and a stable RNA stem-loop, but the long form (5′ UTRL) was more repressive than the short form (5′ UTRS) for initiation at the ELK-1 AUG. We now demonstrate that ELK-1 expression arises by a combination of leaky scanning and reinitiation, with the latter mediated by the small uORF2 conserved in both spliced isoforms. In HEK293T cells, a considerable fraction of ribosomes scans beyond the ELK-1 AUG in a reinitiation mode. These are sequestered by a series of out-of-frame AUG codons that serve to prevent access to a second in-frame AUG start site used to express short ELK-1 (sELK-1), an N-terminally truncated form of ELK-1 that has been observed only in neuronal cells. We present evidence that all these events are fine-tuned by the nature of the 5′ UTR and the activity of the α subunit of eukaryotic initiation factor 2 and provide insights into the neuronal specificity of sELK-1 expression.
INTRODUCTION
The 5′ untranslated region (UTR) represents one of the key elements in the regulation of translation. This can occur at two levels, which we refer to as quantitative and qualitative control. In the former, changes in the 5′ UTR alter the amount of protein translated, whereas in the latter, changes in start site selection occur. Alterations in the 5′ UTR arise due to the use of alternative promoters and/or alternative splicing. Both serve to couple events in the nucleus to the proteomic readout in the cytoplasm. Estimates of the proportion of genes with alternative 5′ UTRs vary from 12 to 22% (29). Nonetheless, detailed genomic studies aimed at elucidating the functional role of transcript 5′ heterogeneity in mammalian cells are still very much in their infancy. This is further complicated by the fact that the precise delineation of the 5′ UTR is frequently problematic. Computational studies have demonstrated the conservation of elements that are thought to play a role in translational control, suggesting that at least some are functionally relevant (4). Elements within the 5′ UTR that modulate expression include length, RNA structure, upstream AUGs (uAUGs), upstream open reading frames (uORFs), and internal ribosomal entry site (IRES) elements (13).
ELK-1 belongs to the ternary complex factor (TCF) subfamily, whose members are part of the ETS transcription factors implicated in differentiation, development, transformation, and cellular proliferation. Their structure has been conserved during evolution (24). When activated they form a ternary complex with the serum response factor (SRF) on the serum response element (SRE) of the c-fos promoter. The human elk-1 gene contains seven exons (E1 to E7). The protein sequence is encoded within E3 to E7. E1, E2, and the first 34 nucleotides (nt) of E3 constitute the 5′ UTR (Fig. 1). The first exon is highly GC rich (>75%) and within the mature mRNA folds into a stable stem-loop (SL) with a ΔG° approaching −40 kcal/mol (2). In postmitotic neurons, ELK-1 is detected in both the nucleus and cytoplasm, including neuritic extensions, whereas in transiently transfected HeLa cells, it is exclusively nuclear. While studying the localization of ELK-1 in the rat brain, an isoform sELK-1 (short ELK-1) was identified (35). This 45-kDa protein was expressed uniquely in brain tissue and was exclusively nuclear. The 54-amino-acid (aa) N-terminal deletion removes most of the DNA binding domain, explaining why it is compromised in its ability to activate SRE genes (35). Curiously, in nerve growth factor (NGF)-treated PC12 cells, sELK-1 protein expression was turned on, and this correlated with both the redistribution of ELK-1 into the cytoplasm (giving a distribution similar to that observed in the mature brain) and cell differentiation (the morphology resembles that of synaptic neurons). Furthermore, overexpression of sELK-1 but not ELK-1 increased neurite extension in the PC12 model system (35). Preliminary studies on sELK-1 expression indicated that it arose due to a translation initiation event at the seventh AUG on the elk-1 mRNA (Fig. 1). Experiments eliminated both alternative splicing and cryptic promoters as the possible source of a second mRNA species (35). Furthermore, between the AUG of ELK-1 (AUGELK-1) and sELK-1 (AUGsELK-1), there are three AUG codons (two with good Kozak sequences), none of which are in frame with the ELK-1 open reading frame (ORF) (Fig. 1) (referred to here as internal AUG a/b/c [iAUGa/b/c]).
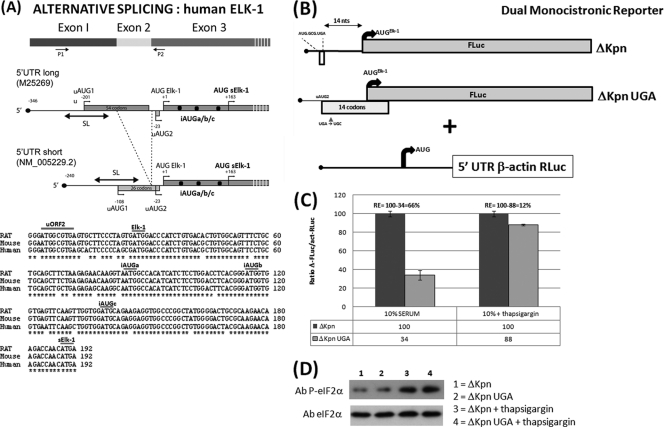
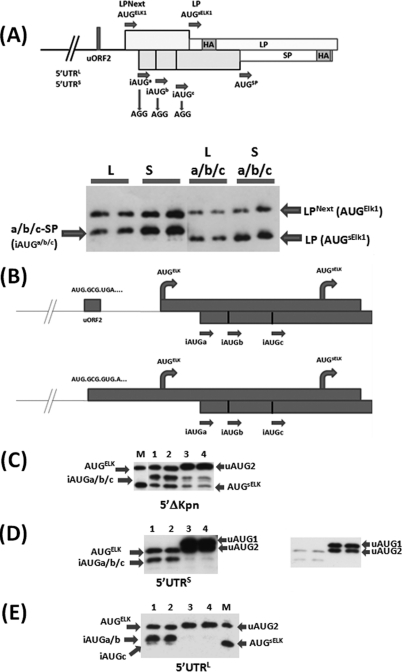
Fig 1.
Schematic representation of the elk-1 gene. (A) (Top) The first three exons, with P1 and P2 indicating the primers used to characterize the alternatively spliced exon 2. Also depicted is the ORF organization and positioning of the AUG codons upstream of the AUGsELK-1 in the alternatively spliced transcripts. Note that in the 5′ UTRS, uAUG1 and uAUG2 are in the same ORF. SL refers to a stable stem-loop structure in which the uAUG1 is embedded. (Bottom) Alignment of the human, rat, and mouse sequences between the uORF2 and AUGsELK-1. (B) ΔKpn-FLuc and β-actin RLuc monocistronic reporter constructs. The box below the line depicts the small uORF2 with the sequence indicated. The impact of the UGA/C mutation in uORF2 on the reading frame organization relative to the AUGELK-1 is also depicted. (C) HEK293T cells were cotransfected with each ΔKpn-FLuc (WT and UGA/C) plasmid construct and the β-actin–RLuc control (referred to as the normalization plasmid). At 4 h posttransfection, medium was removed and replaced with DMEM containing 10% fetal calf serum with and without thapsigargin (300 nM). Cells were harvested after 24 h, and reporter activities were measured. Normalization of reporter activities was performed as outlined in Materials and Methods. The normalized FLuc/RLuc ratios are depicted graphically, with the WT value set at 100 for each condition tested. The value (relative to the WT) obtained in the UGA/C background represents that fraction of AUGELK-1 initiation events arising due to leaky scanning and is indicated below the graph (the difference between the WT and UGA/C values reflects the loss of reinitiation-mediated events). Reinitiation (RE) frequency at the AUGELK-1 is depicted above the data. (D) Immunoblot with eIF2α- and phospho-eIF2α-specific antibodies (Ab) from extracts tested as described above. All transfections were performed in triplicate, and the bars indicate the SEM.
Our lab has exploited the elk-1 mRNA as a model system to examine translational regulation mediated via the 5′ UTR. It is particularly intriguing because (i) we have characterized two alternatively spliced 5′ UTRs of the human gene (Fig. 1). The relative abundance of these isoforms showed tissue specificity (2). (ii) Both contain uORFs and RNA structure; however, we observed no IRES activity (2).
One potentially key regulatory element is the small uORF uORF2. It is found in both spliced isoforms and is conserved in position relative to the AUGELK-1 in all mammals (Fig. 1). Its small size, only two codons, suggests that it may promote downstream reinitiation events. Reinitiation efficiency varies depending on parameters such as uORF length (26, 28) and the distance between the stop codon and the next AUG (18). Reacquisition of the Met-accepting tRNA interference (Met-tRNAi) by the 40S ribosome posttermination is dependent on the levels of eukaryotic initiation factor 2 (eIF2)-GTP, with which it forms an eIF2-GTP-Met-tRNAi ternary complex (TC), and when low, the slow reacquisition can cause delayed reinitiation that bypasses the proximal downstream AUG. The best-characterized example of this phenomenon is the translational regulation of Saccharomyces cerevisiae GCN4. A crucial but vaguely understood feature of the GCN4 5′ UTR was the very divergent capacities of its four uORFs to permit efficient reinitiation. Sequences flanking the highly efficient uORF1 were reported to facilitate continued scanning posttermination (11). Recent studies have demonstrated that eIF3, positioned at the mRNA exit channel on the ribosome, is retained during translation of uORF1. An interaction between eIF3a and an upstream enhancer sequence on the mRNA served to stabilize the 40S ribosome-mRNA association after termination, thereby facilitating the resumption of scanning and downstream reinitiation (31). This opens the possibility that changes in the sequences flanking a uORF (in particular, regions 5′ to the stop codon) can modulate the readout. Indeed, experiments performed in the rabbit reticulocyte lysate in vitro system with a viral bicistronic construct carrying overlapping cistrons (AUGA) pointed to sequence elements upstream of the stop codon as regulators of the realignment/reinitiation event on the AUG start codon (27).
Studies have also demonstrated that delayed translational reinitiation plays a role in the regulation of a number of mammalian genes. ATF4 is a transcriptional activator whose expression is increased during endoplasmic reticulum stress. Such stress activates the kinase PERK, which in turn phosphorylates the α subunit of eIF2 (eIF2α). This leads to an accumulation of phospho-eIF2-GDP, which serves as a competitive inhibitor for the eIF2B exchange factor and as a consequence downregulates global cellular protein synthesis. Nonetheless, the ATF4 mRNA becomes associated with polysomes (12). This control is mediated by a series of uORFs in the 5′ UTR. The first contains only three codons and is followed by a longer second uORF of 59 codons that overlaps the ATF4 coding region. As in GCN4, the site of reinitiation downstream of the uORF1 is determined by the intracellular levels of eIF2-GTP (20, 36). When levels are high, reinitiation occurs mainly at the uORF2, but under the conditions induced by PERK activation, it is delayed and now occurs at the ATF4 ORF. A similar mechanism of control was also observed with another transcriptional regulator, ATF5 (41).
A particularly pertinent example of reinitiation (at least with regard to the ELK-1 model under investigation) is observed with the transcription factor CCAAT/enhancer binding protein β (C/EBPβ), in which the translational expression of a long form (liver activating protein [LAP]) and an N-terminally truncated short form (liver inhibitory protein [LIP]) is regulated via reinitiation events downstream of a uORF of 11 codons terminating 4 nt before the LAP AUG start codon (3). The N-terminal extension present on LAP contains trans-activating domains that induce differentiation and inhibit proliferation. The truncated LIP functions as a trans-dominant repressor for the C/EBPβ target genes. Changes in the LAP/LIP ratio have been associated with a number of human pathologies (38). In many respects, this model mimics the properties of the elk-1 gene, with LIP/LAP being replaced by ELK-1/sELK-1. However, superimposed upon this is an additional level of complexity arising by the alternative splicing of the exon 2 located within the elk-1 5′ UTR.
In the light of these observations, we hypothesized that the small elk-1 uORF2 could also modulate downstream initiation events in response to TC availability. In this article, we demonstrate that despite its proximity to the downstream ELK-1 ORF (separated by only 14 nt), greater than half of the initiation events at AUGELK-1 in a HEK293T cell background arise by uORF2-mediated reinitiation, with the remainder arising by leaky scanning. Expression from the AUGELK-1 increased when the uORF2-AUGELK-1 spacing was expanded, a result that arose due to increased reinitiation. Consistent with current models, expression from the AUGELK-1 also responded negatively to a reduction in TC availability. When examined within the context of the alternative 5′ UTRs (the long form [5′ UTRL] and the short form [5′ UTRS]), the effects mediated by both the spacer element and changes in TC availability were attenuated, with the effect being more marked in 5′ UTRL. Moreover, in both 5′ UTRs, initiation events downstream of the AUGELK-1, namely, at the iAUGa/b/c, arose exclusively by delayed reinitiation. In this manner, the iAUGa/b/c functioned as a translational repressor limiting ribosomal access to the AUGsELK-1. However, uORF2-mediated delayed reinitiation at the AUGsELK-1 could be observed when the amount of TC was reduced due to changes in the status of the S51 residue of eIF2α. Once again, small but significant differences in the behavior of the two 5′ UTRs were observed.
MATERIALS AND METHODS
Cell culture.
HEK293T cells (ATTC, CRL-11268) were cultured in Dulbecco's modified Eagle's medium (DMEM; Sigma) supplemented with 10% fetal calf serum (Brunschwig) and 1% penicillin-streptomycin (Gibco) in a humidified atmosphere containing 5% CO2.
DNA constructions.
Unless indicated, all clones were in a pcDNA3 backbone. The cloning of the 5′ UTRL-firefly luciferase (FLuc), 5′ UTRS-FLuc, and β-actin Renilla luciferase (RLuc) constructs has been previously described (2, 9). The ΔKpn 5′ UTR was generated by deleting all sequences 5′ to a unique KpnI site present in the 5′ UTRS at the exon 1-exon 3 boundary. To introduce the spacer sequences, a unique NheI site was first introduced downstream of the uORF2 by mutagenic PCR (AUGGCGUGAGCUAGC; the NheI site is underlined). The +13-, +26-, and +50-nt insertions were amplified from the 5′ UTR of β-actin (using the RLuc clone) with oligonucleotides carrying NheI extremities (with all inserts starting from the mRNA 5′ end). The insertions selected were in the positive orientation. The eIF2α ORF from the original eIF2α wild-type (WT), S51E, and S51A clones (16) was PCR amplified with oligonucleotides carrying BamHI/EcoRI ends (5′-ACGGATCCCCGGGTCTAAGTTGTAGATTTT-3′/5′-GCTCTAGATTAATCTTCAGCTTTGGCT-3′) and inserted between these sites in the plasmid pcDNA3 flag p53 (addgene plasmid 10838; replacing the p53), leaving an N-terminal Flag tag (10).
Firefly reporter assays.
Calcium phosphate-mediated DNA transfection was performed as described in reference 15. Transfections were done in triplicate in normal growth medium when the cells were no more than 50% confluent. Four hours later the medium was replaced with fresh growth medium, except in those experiments in which the serum concentration was altered. Extracts were prepared at 24 h posttransfection in passive lysis buffer according to the instructions of the supplier (Promega). At this stage the cells were not yet confluent. Protein concentrations were determined by Bradford assay (cytoskeleton). The activities of FLuc and RLuc were measured using a dual-luciferase reporter assay system (Promega) on a Gloma 20/20 luminometer (Promega). In the monocistronic dual-reporter assays, the individual FLuc and RLuc reporter values were normalized to RNA concentrations determined by quantitative reverse transcription-PCR (RT-qPCR) (see below) before estimating the FLuc/RLuc ratio.
Western blotting.
Thirty micrograms of protein was resolved on an SDS-polyacrylamide gel and electrotransferred to a polyvinylidene difluoride membrane. Antibodies used in this study were antihemagglutinin (anti-HA; clone 16B12; Covance), anti-Flag (M2 antibody; Sigma), antiactin (clone C4, Millipore), and goat anti-mouse or anti-rabbit horseradish peroxidase-conjugated secondary antibody (Bio-Rad). Blots were developed with SuperSignal substrate (Thermo Scientific) and quantified using the Quantity One software package (Bio-Rad).
RT-qPCR.
RNA was prepared directly from the passive lysis cell extracts using the TRIzol reagent (Invitrogen). RNA was treated with DNase (DNA free; Ambion), and quality was checked on an Agilent 2100 bioanalyzer. Reverse transcription was performed with PrimeScript reverse transcriptase (Takara). The real-time PCR was performed on an Applied Biosystems 7900HT apparatus using SYBR green, with technical triplicates used for each biological triplicate. Internal normalization controls were provided by eukaryote elongation factor 1A1 (EEF1A1) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (these procedures were performed at the Genomic Platform, University of Geneva). The primer sets used were Firefly (5′-ATTCAAAGTGCGCTGCTGGT-3′/5′-GCTTTTGGCGAAGAAGGAGAA-3′) and Renilla (5′-AAGGTGAAGTTCGTCGTCCAA-3′/5′-CTAACGGGATTTCACGAGGC-3′).
RESULTS
uORF2 directs downstream reinitiation at the AUGELK-1.
Although annotation of the 5′ UTR is frequently problematic, one feature that is clearly conserved in the mammalian elk-1 gene is the short uORF2 positioned 14 nt before the AUGELK-1 initiation codon (Fig. 1A). Our earlier studies indicated that within the context of the two human 5′ UTRs, the uAUG2 served as a translational repressor for ELK-1 expression (see Fig. S1 in the supplemental material) (2). However, its small size (2 codons) also suggested that it could mediate translational reinitiation events downstream (19). We therefore examined the reinitiation capacity of the isolated uORF2. This was rendered straightforward because the splicing of exon 2 generates a unique KpnI site at the splice junction. Deletion of all sequences 5′ to this site (referred to as ΔKpn) leaves uORF2 with only 12 upstream nucleotides from the elk-1 5′ UTR. Experiments exploited a monocistronic dual-reporter assay system in which the ΔKpn 5′ UTR was fused to FLuc and the β-actin 5′ UTR was fused to RLuc (initiation from this internal control is considered a paradigm of 5′ cap-dependent translation) (1) (Fig. 1B). To evaluate reinitiation frequency at the AUGELK-1, the uORF2 stop codon was changed to UGC (UGA/C), thereby extending this ORF (now 14 codons) such that it terminated 13 nt downstream of the AUGELK-1 (Fig. 1B). The overlapping ORF configuration precluded reinitiation; therefore, in the UGA/C background, FLuc activity arose only as a result of leaky scanning. Thus, by comparing the normalized FLuc/RLuc values in the WT versus UGA/C backgrounds, we could evaluate what fraction of initiation events was a product of uORF2-mediated reinitiation. This gave a value of 66% (it tended to vary between experiments, with an average of about 65%), with the remainder arising due to leaky scanning (Fig. 1C). We sought to confirm this by measuring the impact of reduced TC levels on the readout. This was achieved by repeating the assay in cells treated with the drug thapsigargin, a known ER stress agent that induces phosphorylation of eIF2α. In this context, the reinitiation frequency at the AUGELK-1 dropped dramatically (Fig. 1C).
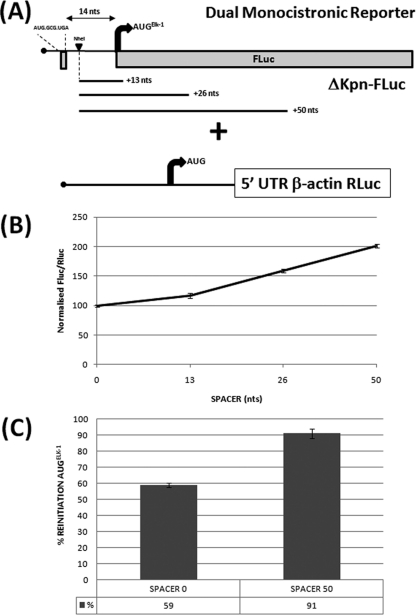
Only 14 nt separates the uORF2 stop codon from the AUGELK-1, a distance relatively short in terms of the current models for reinitiation (21). We therefore examined the impact of extending this distance by the insertion of spacer sequences immediately downstream of uORF2 in the dual FLuc/RLuc monocistronic reporter system (Fig. 2A). A gradual increase in normalized FLuc activity was observed upon the insertion of 13, 26, and 50 nt (Fig. 2B). We confirmed that this was due to increased reinitiation by measuring reinitiation frequencies in the SPACER 0 (S) and SPACER 50 (S50) backgrounds using the UGA/C mutation in uORF2 (Fig. 2C). This result was further validated using a bicistronic approach in which the ΔKpn 5′ UTR FLuc reporter was positioned as the first cistron (in either the WT or uORF2 UGA/C context carrying SPACER 0 or SPACER 50), while the second cistron was an RLuc reporter cloned downstream of the encephalomyocarditis virus IRES (see Fig. S2 in the supplemental material).
Fig 2.
Impact of spacer sequences downstream of uORF2 on initiation events at the AUGELK-1. (A) Schematic representation of the ΔKpn-FLuc/β-actin–RLuc reporters. The NheI indicates a unique restriction site introduced just downstream of the uORF2. The spacer elements of 13, 26, and 50 nt were inserted at this site. The 14 nt is the normal distance between the uORF2 and AUGELK-1. (B) Constructs were cotransfected into HEK293T cells with the β-actin–RLuc control. Normalization of reporter activities was performed as outlined in Materials and Methods. For the 5′ UTR spacer series, the SPACER 0 (no insert) value was set at 100. (C) Reinitiation frequency was estimated for SPACER 0 and SPACER 50 by comparing the normalized FLuc/RLuc ratios for the WT and UGA/C constructs, as outlined in the legend to Fig. 1. The mean percent reinitiation frequency is indicated below each column. All transfections were performed in triplicate, and the bars indicate the SEM. In panel C, P = 0.001.
Impact of alternative 5′ UTRs.
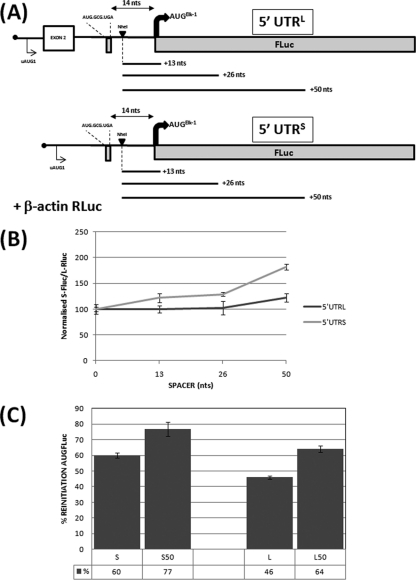
The experiments described above demonstrated that a significant fraction of the initiation events at the AUGELK-1 arose by uORF2-mediated reinitiation. This enhancement induced by the introduction of a spacer element indicates that the reinitiation frequency at this start site was limited by the capacity of the ribosome posttermination to recruit eIF2-GTP during scanning of the 14 nt that separates uORF2 and the AUGELK-1. We next asked what happens when the uORF2 is monitored in the context of the two alternative elk-1 5′ UTRs. As previously reported, both 5′ UTRs are highly repressive, with the effect being more marked with the 5′ UTRL (2). Nonetheless, significant uORF2-mediated reinitiation could be demonstrated in both (Fig. 3C). Although the measured reinitiation frequencies showed experimental variation, it was in the order ΔKpn ≥ 5′ UTRS > 5′ UTRL. Upon addition of the spacer sequences, the 5′ UTRS produced a pattern reminiscent of the one observed with ΔKpn: a gradual increase in the normalized FLuc/RLuc values attributable to higher reinitiation levels (Fig. 3B and C). However, the spacer effect on the normalized FLuc/RLuc ratios was attenuated in the 5′ UTRL (Fig. 3B). Curiously, despite this attenuation, we observed an increased reinitiation frequency in the SPACER 50, albeit at levels that remained lower than those observed with 5′ UTRS and ΔKpn (Fig. 3C). The results observed in the dual monocistronic assay were also validated using the bicistronic reporter (see Fig. S3 in the supplemental material), confirming that sequence elements upstream of uORF2 in the context of the 5′ UTRL are modulating the behavior of the ribosome downstream of uORF2.
Fig 3.
Analysis of the uORF2 in the context of the alternative elk-1 5′ UTRs. (A) Schematic representation of the 5′ UTRL-FLuc and 5′ UTRS-FLuc reporters cotransfected into HEK293T cells with the β-actin–RLuc normalization control. The NheI site and spacer insertions are also indicated. The alternatively spliced exon 2 is indicated by a box. (B) The effect of the spacer sequences was assayed as outlined in the legend to Fig. 2B. (C). Reinitiation frequency was estimated for SPACER 0 and SPACER 50 by comparing the normalized FLuc/RLuc ratios for the WT and UGA/C constructs as outlined in the legend to Fig. 1. The mean percent reinitiation frequency is indicated below each column. All transfections were performed in triplicate, and the bars indicate the SEM. The P values for the S/S50 and L/L50 assays depicted in panel C were 0.02 and 0.003, respectively.
The alternative 5′ UTRs modulate the response to changes in eIF2α activity.
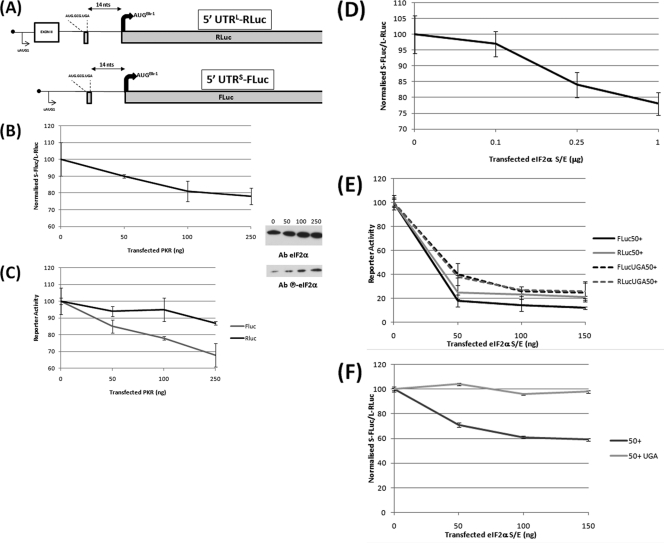
The GCN4 model for reinitiation posits that the effect of the spacer is associated with the rerecruitment of the active TC by the 40S ribosome posttermination of the uORF (21). As such, it mimics changes in cellular eIF2-GTP availability. Therefore, by extrapolation, the differential response of the 5′ UTRL and 5′ UTRS in the spacer assay suggests that they may respond differently to such changes. To assay this, each 5′ UTR was fused to a unique reporter (5′ UTRL-RLuc and 5′ UTRS-FLuc), and the dual activities were monitored in cells in which the amount of TC was modulated (Fig. 4A). We dosed plasmids expressing either the negative eIF2α S51E (eIF2αS/E) mutant or the kinase protein kinase R (PKR). Dosing of these plasmids was negative for both reporters in the SPACER 0 and SPACER 50 backgrounds (Fig. 4C and E). This global negative effect was an important control because, although we could monitor increased eIF2α phosphorylation upon dosing with plasmids expressing PKR (Fig. 4B and C), we were unable to detect expression of the eIF2αS/E mutant by immunoblotting, probably because it downregulates its own expression. However, the normalized reporter activity curve from the cotransfected 5′ UTRL-Rluc/5′ UTRS-FLuc demonstrated that the 5′ UTRL was less affected by changes in TC availability than the 5′ UTRS (Fig. 4B and D). We confirmed that this was associated with changes in uORF2-mediated reinitiation by repeating the eIF2αS/E assay with the two 5′ UTRs carrying the SPACER 50 with and without the uORF2 UGA/C mutation (Fig. 4F).
Fig 4.
The 5′ UTRL and 5′ UTRS respond differently to changes in eIF2α activity. (A) Schematic representation of the Renilla (RLuc) and firefly (FLuc) reporter constructs. (B) HEK293T cells were cotransfected with the 5′ UTRL-RLuc and 5′ UTRS-FLuc constructs in the presence of increasing amounts of a plasmid expressing the PKR kinase. Normalized FLuc/RLuc activities were plotted against the amount of PKR plasmid transfected, with 0 ng being set at 100. (C) Values for each reporter, with 0 ng being set at 100. The inset on the right demonstrates that the PKR dose produced a progressive increase in the levels of phospho-eIF2α, as determined by immunoblotting. (D) The dual-reporter assay was performed in the presence of increasing amounts of a plasmid clone expressing eIF2αS/E. The FLuc/RLuc-normalized value for 0 μg eIF2αS/E was set at 100. (E and F) The eIF2αS/E dosage experiment was performed with 5′ UTRL-RLuc and 5′ UTRS-FLuc reporters carrying the SPACER 50 insertion with or without the UGA/C mutation of the uORF2. (E) Values for each reporter; (F) normalized values. All transfections were performed in triplicate, and the bars indicate the SEM.
What is happening downstream of the AUGELK-1 in HEK293T cells?
The fact that reinitiation frequency can be increased by the addition of a spacer element (in the 5′ UTRS background, to as much as 80% in some experiments) indicates that in the WT background (SPACER 0) a significant number of reinitiating ribosomes bypass the AUGELK-1. This is not a mute point because, at least in rat neuronal cells, an N-terminal truncated form of ELK-1, sELK-1, arising from a de novo initiation event downstream of the AUGELK-1, has been reported (35). The AUGsELK-1 is conserved in all mammalian genes (Fig. 1A); however, we do not observe this protein in HEK293T cells even when HA-tagged versions of the human elk-1 gene carrying the alternative 5′ UTRs are overexpressed (data not shown). A possible source of this translational repression is the iAUGa/b/c positioned between the AUGELK-1 and AUGsELK-1 codons (Fig. 1A). To test this, we exploited a novel reporter developed specifically to examine leaky scanning/reinitiation within the C/EBPβ gene (39, 40). This system expresses two proteins (LP and SP) from overlapping ORFs each carrying an HA-epitope tag (Fig. 5A). The sequences upstream of the AUGsELK-1 were fused to these reporters such that AUGELK-1 and AUGsELK-1 were in the LP ORF (with protein products referred to as LPNext and LP, respectively) and the iAUGa/b/c were in the SP ORF (with protein products referred to as a-SP, b-SP, and c-SP). An N-terminal preprotrypsin (PPT) signal present in the original LP and SP reporters was mutated because we observed that when positioned internally in the LPNext and the a-SP, b-SP, and c-SP fusion proteins, this region induced efficient intracellular cleavage. This generated products that comigrated with the de novo-initiated LP/SP proteins, a phenomenon that seriously confused interpretation of the readout (see Fig. S4 in the supplemental material). Transient expression from both the 5′ UTRL and 5′ UTRS LPNext reporters produced two protein bands arising from initiation at the AUGELK-1 (LPNext) and iAUGa/b/c (these three proteins generally comigrated, although the c-SP product could occasionally be discerned). When the three iAUGa/b/c codons were mutated to AGG, the a-SP/b-SP/c-SP proteins were lost (confirming their origin) and a new faster-migrating protein corresponding to an initiation event at the AUGsELK-1 (LP) was observed (Fig. 5A). Therefore, the highly conserved iAUGa/b/c serve to limit ribosomal access to the AUGsELK-1 in HEK293T cells.
Fig 5.
The iAUGs serve to limit ribosomal access to the AUGsELK-1. (A) (Top) LP/SP reporter system that was adapted to monitor initiation events at the AUGELK-1, AUGsELK-1, and iAUG start codons. The position of the HA epitope in each ORF is indicated, as are the mutations introduced to remove the iAUG codons. The bars above and below the line indicate that LP and SP are in different ORFs. (Bottom) Immunoblot performed with the anti-HA antibody. HEK293T cells were transfected with the 5′ UTRL-LPNext (L), 5′ UTRS-LPNext (S), and the same 5′ UTRs carrying the iAUG → AGG changes (La/b/c and Sa/b/c). Transfections were performed in duplicate. (B) Schematic representation of the single nucleotide insertion within the uORF2 that fuses this ORF to the ORFs of AUGELK-1 and AUGsELK-1 (the LP ORF in the reporter). (C to E) Immunoblots performed with the anti-HA antibody for the three constructs tested. Lanes 1 and 2, WT constructs; lanes 3 and 4, iAUGa/b/c → AGG mutations. All clones were transfected in duplicate. The inset in panel D is a shorter exposure that demonstrates initiation products from both uAUG1 and uAUG2 (which are in the same ORF [Fig. 1A]). Lane M, a marker (the Sa/b/c construct depicted in panel A) for initiation products from the AUGELK-1 (LPNext) and AUGsELK-1 (LP).
Next we asked to what extent the a/b/c-SP protein products arise due to reinitiation as opposed to leaky scanning. The simple UGA/C mutation in uORF2 is less informative in this reporter context because, although the extended ORF ends after the AUGELK-1, it remains relatively short (12 codons) and terminates upstream of iAUGa/b/c (42 nt upstream of iAUGa [Fig. 6C]). Consequently, it could still potentially direct reinitiation events at these downstream sites (see below). To overcome this, we fused the uORF2 to the LP ORF by the insertion of a single nucleotide before the uORF2 stop codon (Fig. 5B). This suppresses all uORF2-mediated reinitiation events without impacting on the leakiness of the AUGs. A band above the LPNext was observed in all three 5′ UTR backgrounds arising from initiation at the uAUG2 codon (which has a good context). The intensity of this new band was also significantly increased (∼10-fold), consistent with the strong repression effect of the uORF2 (Fig. 5C to E; see Fig. S1 in the supplemental material). Furthermore, in the 5′ UTRS, an additional slow-migrating product could be observed due to initiation at the uAUG1 (which is in the same ORF as uAUG2 [Fig. 1A, top]). A similar pattern was reported earlier (2). Most significantly, the ORF fusion effectively ablated all iAUG expression in the 5′ UTRL and 5′ UTRS, demonstrating that they arise essentially by uORF2-mediated reinitiation (Fig. 5D and E; compare lanes 1 and 2 with lanes 3 and 4). In the ΔKpn background, the expression pattern was markedly different from the one observed with the intact 5′ UTRs. Protein products arising from the AUGELK-1, iAUGa/b/c, and AUGsELK-1 were all readily detectable in the WT background (Fig. 5C, lanes 1 and 2). Upon fusion of the uORF2, we could still detect initiation events at the iAUGs and the AUGsELK-1, albeit at levels >3-fold lower than in the WT, indicating that a fraction of these products arose by leaky scanning (Fig. 5C, lanes 3 and 4). This difference between the ΔKpn and the two intact 5′ UTRs serves to highlight the attenuating impact of the upstream 5′ sequences on the translational readout, most noticeably with regard to initiation at the AUGsELK-1 (compare lanes 1 and 2 in Fig. 5C with lanes 1 and 2 in Fig. 5D and E). It also opens the possibility that trans-acting cellular factors that alleviate this attenuation (e.g., cellular helicases acting on the secondary structural elements in the 5′ UTR [25]) may reproduce a ΔKpn phenotype that significantly facilitates 40S ribosome access to the AUGsELK-1. Furthermore, the fact that delayed reinitiation is the major source of expression from the iAUGa/b/c is also consistent with the effect of the spacer insertions with regard to reinitiation at the AUGELK-1.
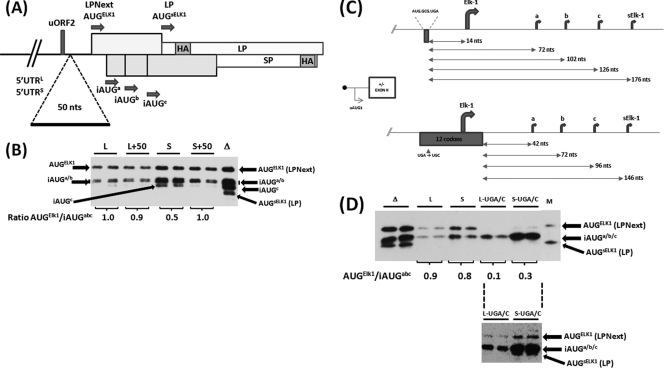
Fig 6.
The LP/SP reporter system demonstrates differences in the behavior of the 5′ UTRs. (A) Schematic representation of the SPACER 50 insertion in the context of 5′ UTRL-LPNext (L) and 5′ UTRS-LPNext (S). (B) The SPACER 0 (WT, represented by L and S) and SPACER 50 (L+50 and S+50) constructs were expressed in HEK293T cells. Proteins were visualized by immunoblotting with an anti-HA antibody. Lane Δ, a cell extract prepared from ΔKpn-LPNext-transfected cells that provides a marker for protein products arising from all the AUG codons depicted in panel A. Transfections were performed in duplicate. The protein bands in each individual transfection arising from the AUGELK-1 and iAUGa/b/c were quantified. The value of their ratio in each transfection is indicated below the gel (averaged from the duplicate samples). (C) Schematic representation of the impact of the uORF2 UGA/C mutation in the 5′ UTRL-LPNext and 5′ UTRS-LPNext reporter system. The sequence of the uORF2 is indicated, as is the distance between the stop codon of this ORF and the downstream AUG codons (arrows). (D) HEK293T cells were transfected with ΔKpn-LPNext (lanes Δ), 5′ UTRL-LPNext (lanes L), 5′ UTRS-LPNext (lanes S), and the long and short 5′ UTRs carrying the UGA/C mutation (lanes L-UGA/C and S-UGA/C). Protein products were detected by immunoblotting with the anti-HA antibody. Lane M, the same marker for LPNext and LP as outlined in the legend to Fig. 5. The AUGELK-1/iAUGa/b/c levels within each duplicate transfection were determined, and the averaged value is indicated below the gel. (Lower inset) An overexposure of the L-UGA/C and S-UGA/C lanes demonstrating the presence of a protein product arising from the AUGELK-1.
Further insights into the role of uORF2-mediated reinitiation could be gleaned by analyzing the impact of the SPACER 50 and the uORF2 UGA/C mutation on the relative ratios of the AUGELK-1 and iAUGa/b/c proteins expressed (Fig. 6A). By measuring the relative ratios of products generated from the same mRNA within a single transfection, we effectively eliminated the necessity to quantify cellular transcript levels, a process that would have been essential if we wished to compare across transfections. In the context of the 5′ UTRL, the introduction of the spacer had only a marginal impact on the relative products observed (Fig. 6B; compare lanes L and L+50), whereas in the 5′ UTRS, the levels of LPNext increased twofold relative to the a/b/c-SP products upon addition of the SPACER 50 (Fig. 6B; compare lanes S and S+50). This is consistent with increased initiation at the AUGELK-1 codon, with fewer 40S ribosomes reaching the iAUGa/b/c. The more attenuated response to the presence of the spacer observed in the 5′ UTRL background correlates with our observations made using the dual luciferase reporter assay (i.e., downstream initiation events are largely insensitive) (Fig. 3B). The differences in the absolute values observed in the two assays (e.g., in Fig. S3 in the supplemental material, the S/S50 difference is 1.45, whereas in Fig. 6B it is 2-fold) probably reflect differences in the nature of the two assays employed. In the LPNext reporter, increased initiation at the AUGELK-1 impacts negatively on the downstream initiation events at the iAUGa/b/c, the products against which its expression is normalized. This is clearly not the case in the dual FLuc/RLuc assays, in which the readout from the RLuc reporter used for the normalization is independent of events occurring on FLuc. Likewise, the uORF2 UGA/C mutation (Fig. 6C and D) severely reduced initiation from the AUGELK-1 codon in both 5′ UTRL and 5′ UTRS backgrounds but significantly increased the relative abundance of products arising from iAUGa/b/c (Fig. 6D). This suggests that the new extended 12-codon ORF has not only prevented reinitiation at the AUGELK-1 but also permitted reinitiation events at the iAUGa/b/c.
What is astounding is that a large number of 40S ribosomes are scanning downstream of the AUGELK-1 in all the 5′ UTR backgrounds tested. What does this mean in terms of elk-1 gene expression?
What about the protein sELK-1?
A question that remains unresolved is, how can ribosomes access the AUGsELK-1? Our results indicate that, at least in HEK293T cells, the ribosomes scanning downstream of the AUGELK-1 are mainly, if not exclusively, in a reinitiation mode, suggesting that they could reach this start site by delayed reinitiation if the amount of TC was reduced. To test this model, we cotransfected the ΔKpn, 5′ UTRS, and 5′ UTRL LPNext constructs with increasing amounts of the eIF2αS/E plasmid. In the 5′ UTRL and 5′ UTRS backgrounds, we observed a gradual reduction in the expression of LPNext (initiation at the AUGELK-1) with increasing eIF2αS/E plasmid dose. Concomitant with this loss of expression from AUGELK-1, there was an initial increase in initiation at the iAUGc codon followed by expression from the AUGsELK-1 codon (Fig. 7B and C). This is consistent with a progressive shift in delayed reinitiation toward the mRNA 3′ as TC levels were reduced. Although the patterns from both 5′ UTRs looked similar, some differences were discernible. (i) Products from the 5′ UTRL background tended to be less affected by the increasing dose of eIF2αS/E relative to products from the 5′ UTRS background, a result consistent with the findings from the reporter assays depicted in Fig. 4. (ii) In the 5′ UTRL, the AUGsELK-1 product arose later but continued to accumulate even at the highest eIF2αS/E level tested. The response profile observed in the ΔKpn background was somewhat different, in part, because initiation from the AUGsELK-1 is already observed without altering TC availability but also because initiation events arise by both reinitiation and leaky scanning. Nonetheless, there was a clear increase in AUGsELK-1-mediated initiation events at the lower eIF2αS/E doses, and this codon became the major initiation site at the highest dose tested (bypassing AUGELK-1 and iAUGa/b/c) (Fig. 7A).
Fig 7.

Expression from the AUGsELK-1 can be observed in HEK293T cells upon dosing of eIF2αS/E. The ΔKpn-LPNext (A), 5′ UTRL-LPNext (B), and 5′ UTRS-LPNext (C) constructs were transfected in the presence of increasing amounts (indicated in ng above each blot) of a plasmid expressing the negative eIF2αS/E. Each transfection was performed in duplicate. Proteins were visualized by immunoblotting with an anti-HA antibody. Lane C, a mock-transfected control; lane M, a marker for LPNext (AUGELK-1) and LP (AUGsELK-1). (D) Negative effect of the eIF2αS/E dosage on an LP-expressing plasmid with a short 5′ UTR. The LP protein was detected by immunoblotting with an anti-HA antibody. The steady-state levels of eIF2α were followed with an anti-eIF2α antibody.
These results demonstrate that ribosomes can access the AUGsELK-1, despite the presence of iAUGa/b/c, when the availability of the TC is reduced. In the context of the two 5′ UTRs, this is achieved exclusively by modulation of delayed reinitiation events downstream of uORF2 in a manner that appears to be characteristic of each 5′ UTR.
DISCUSSION
Genomic analysis has estimated that approximately 50% of human 5′ UTRs contain one or more uORFs (8, 17, 30). Moreover, studies have reported that they are conserved between species during evolution (4, 14). uORFs inhibit translation by restricting the access of ribosomes to the start codon of the main coding ORF (22). As a consequence, reinitiation in combination with leaky scanning is employed to significantly increase the complexity of the mammalian proteome (37). In this study, we have demonstrated that the tightly conserved uORF2, retained in both alternatively spliced elk-1 5′ UTRs, traps scanning ribosomes. In so doing, it serves both as a translational repressor for downstream initiation events and as a generator of reinitiating ribosomes. The latter feature serves to couple subsequent initiation events to intracellular levels of eIF2-GTP. Analyzed independently of the rest of the elk-1 5′ UTR (in the ΔKpn background), the uORF2 can direct initiation events at the AUGELK-1, iAUGa/b/c, and even the AUGsELK-1 using a combination of leaky scanning and delayed reinitiation (Fig. 5C). However, this behavior is modified within the context of the two 5′ UTRs.
With regard to initiation at the AUGELK-1, the 5′ UTRL was already demonstrated to be more repressive than the 5′ UTRS, in large part because of increased RNA structure (with the uAUGs being conserved in both; Fig. 1B) (2). We now demonstrate an additional layer of control which modulates initiation events downstream of uORF2. The introduction of spacer sequences and the modulation of TC levels have indicated subtle differences in the behavior of the two 5′ UTRs, with uORF2-mediated reinitiation frequency being more attenuated in the 5′ UTRL background relative to the 5′ UTRS background (Fig. 3). The attenuation of reinitiation would also explain the observation that the 5′ UTRL was less affected by changes in eIF2-GTP availability than the 5′ UTRS (Fig. 4), suggesting that the latter may be more fine-tuned into intracellular pathways that target TC levels.
In both 5′ UTRs, few ribosomes pass beyond the AUGELK-1 in the leaky scanning mode, despite the fact that leaky scanning-mediated initiation has been observed at the 4th or 5th AUG codon in certain transcripts (37). What is striking is the number of ribosomes scanning downstream of the AUGELK-1 in a uORF-2-mediated delayed reinitiation mode, indicating that the short distance before the AUGELK-1 (14 nt) is not compatible with optimal reinitiation. This explains the enhanced reinitiation frequency at the AUGELK-1 observed with the spacer insertions and the concomitant reduction in delayed reinitiation events at the downstream iAUGa/b/c (Fig. 6). In this context, it should also be noted that in the HEK293T cell background, reinitiation events at the AUGELK-1 did not respond to the overexpression of the constitutively active eIF2α S51A (eIF2αS/A) mutant, indicating that in these cells ELK-1 expression was not limited by endogenous TC levels (see Fig. S5 in the supplemental material).
Overall, the current results and those from our earlier study (2) indicate that the elk-1 gene employs alternative splicing in the 5′ UTR to control the number of ribosomes that reach uORF2 (with 5′ UTRL being more repressive than 5′ UTRS). Furthermore, the same splicing event serves to modulate ribosome behavior downstream of uORF2. The uAUG2 has an optimal context and as a consequence serves to further repress ELK-1 expression, and at the same time, the uORF2 permits a fraction of the posttermination 40S ribosomes to remain associated with the RNA and to resume scanning. Those 40S ribosomes that initiate at the AUGELK-1 reacquire the TC before reaching it. The remainder continue to scan downstream. Such a scenario serves to couple translational readout to physiological events that modulate delayed reinitiation. The other reported protein product of the elk-1 gene is sELK-1. However, this has been observed only in neuronal cells and NGF-differentiated PC12 cells, wherein its expression has been reported to potentiate the neurite outgrowth possible by antagonizing the proproliferative effect of ELK-1 (34, 35). Clearly, sELK-1 expression in HEK293T cells is inhibited under normal growth conditions by the iAUGa/b/c, which serve to mop up reinitiating ribosomes, preventing them from reaching the AUGsELK-1. Indeed, their distance from the uORF2 would fall within a window considered optimal for reinitiation (18), and their scattering between the AUGELK-1 and AUGsELK-1 suggests that they function as a reinitiation rheostat to ensure tight sELK-1 expression, despite changing TC levels. Nonetheless, the experiments performed with the eIF2αS/E mutant not only demonstrate that changes in TC availability can permit initiation at AUGsELK-1 (even in HEK293T cells) but once again highlight differences in the behavior of the two 5′ UTRs (Fig. 7). Modulation of initiation events downstream of a uORF by upstream sequence elements is novel but not unique. The high reinitiation frequency observed with the uORF1 of GCN4 is associated with an enhancer element positioned −21 to −181 nt relative to the uORF1 AUG codon. This region interacts with eIF3a on the posttermination ribosome to promote resumption of scanning (23, 31). However, in the case of the elk-1 5′ UTRs, these upstream sequence elements seem to be functioning as attenuators rather than enhancers.
These observations begin to provide clues into how sELK-1 expression may take place in neuronal cells. A number of studies indicate that a change in eIF2-GTP levels plays a central role in the translational regulation of specific mRNA subsets in neuronal cells (5, 7). Mild physiological changes in the amounts of TC are sufficient to alter expression of ATF4, a transcript whose translational readout is also coupled to reinitiation (6, 12). With regard to the PC12 model system, NGF-induced neurite outgrowth is associated with translational changes in the preexisting mRNA pool (i.e., the response is not transcriptional) (32, 33). Our results point to delayed reinitiation coupled to changes in TC levels as the mechanism behind sELK-1 expression, a model that is under investigation in a neuronal cell context.
Supplementary Material
ACKNOWLEDGMENTS
Plasmids expressing eIF2α WT, S51E, and S51A were kindly provided by Randal Kaufman and Janet Mitchell (HHMI). The PKR clone was kindly provided by Nahum Sonenberg and Olivier Donzé (McGill University, Canada). We thank Bernadino Conrad for his constructive comments about the work.
The work was supported by grants from the Swiss National Science Foundation and the Schmidheiny Foundation.
Footnotes
Published ahead of print 21 February 2012
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Andreev DE, et al. 2009. Differential contribution of the m7G-cap to the 5′ end-dependent translation initiation of mammalian mRNAs. Nucleic Acids Res. 37:6135–6147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Araud T, Genolet R, Jaquier-Gubler P, Curran J. 2007. Alternatively spliced isoforms of the human elk-1 mRNA within the 5′ UTR: implications for ELK-1 expression. Nucleic Acids Res. 35:4649–4663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Calkhoven CF, Muller C, Leutz A. 2000. Translational control of C/EBPalpha and C/EBPbeta isoform expression. Genes Dev. 14:1920–1932 [PMC free article] [PubMed] [Google Scholar]
- 4. Churbanov A, Rogozin IB, Babenko VN, Ali H, Koonin EV. 2005. Evolutionary conservation suggests a regulatory function of AUG triplets in 5′-UTRs of eukaryotic genes. Nucleic Acids Res. 33:5512–5520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Costa-Mattioli M, et al. 2007. eIF2alpha phosphorylation bidirectionally regulates the switch from short- to long-term synaptic plasticity and memory. Cell 129:195–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Costa-Mattioli M, Sossin WS, Klann E, Sonenberg N. 2009. Translational control of long-lasting synaptic plasticity and memory. Neuron 61:10–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Costa-Mattioli M, et al. 2005. Translational control of hippocampal synaptic plasticity and memory by the eIF2[alpha] kinase GCN2. Nature 436:1166–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Davuluri RV, Suzuki Y, Sugano S, Zhang MQ. 2000. CART classification of human 5′ UTR sequences. Genome Res. 10:1807–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Genolet R, Rahim G, Gubler-Jaquier P, Curran J. 2011. The translational response of the human mdm2 gene in HEK293T cells exposed to rapamycin: a role for the 5′-UTRs. Nucleic Acids Res. 39:989–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gjoerup O, Zaveri D, Roberts TM. 2001. Induction of p53-independent apoptosis by simian virus 40 small t antigen. J. Virol. 75:9142–9155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Grant CM, Miller PF, Hinnebusch AG. 1995. Sequences 5′ of the first upstream open reading frame in GCN4 mRNA are required for efficient translational reinitiation. Nucleic Acids Res. 23:3980–3988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Harding HP, et al. 2000. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 6:1099–1108 [DOI] [PubMed] [Google Scholar]
- 13. Hinnebusch AG. 2011. Molecular mechanism of scanning and start codon selection in eukaryotes. Microbiol. Mol. Biol. Rev. 75:434–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Iacono M, Mignone F, Pesole G. 2005. uAUG and uORFs in human and rodent 5′untranslated mRNAs. Gene 349:97–105 [DOI] [PubMed] [Google Scholar]
- 15. Jordan M, Schallhorn A, Wurm FM. 1996. Transfecting mammalian cells: optimization of critical parameters affecting calcium-phosphate precipitate formation. Nucleic Acids Res. 24:596–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kaufman RJ, Davies MV, Pathak VK, Hershey JW. 1989. The phosphorylation state of eucaryotic initiation factor 2 alters translational efficiency of specific mRNAs. Mol. Cell. Biol. 9:946–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kochetov AV, et al. 2008. uORFs, reinitiation and alternative translation start sites in human mRNAs. FEBS Lett. 582:1293–1297 [DOI] [PubMed] [Google Scholar]
- 18. Kozak M. 1987. Effects of intercistronic length on the efficiency of reinitiation by eucaryotic ribosomes. Mol. Cell. Biol. 7:3438–3445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kozak M. 2001. Constraints on reinitiation of translation in mammals. Nucleic Acids Res. 29:5226–5232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lu PD, Harding HP, Ron D. 2004. Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J. Cell Biol. 167:27–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Merrick WC. 2010. Eukaryotic protein synthesis: still a mystery. J. Biol. Chem. 285:21197–21201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Morris DR, Geballe AP. 2000. Upstream open reading frames as regulators of mRNA translation. Mol. Cell. Biol. 20:8635–8642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Munzarova V, et al. 2011. Translation reinitiation relies on the interaction between eIF3a/TIF32 and progressively folded cis-acting mRNA elements preceding short uORFs. PLoS Genet. 7:e1002137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Oikawa T, Yamada T. 2003. Molecular biology of the Ets family of transcription factors. Gene 303:11–34 [DOI] [PubMed] [Google Scholar]
- 25. Parsyan A, et al. 2011. mRNA helicases: the tacticians of translational control. Nat. Rev. Mol. Cell Biol. 12:235–245 [DOI] [PubMed] [Google Scholar]
- 26. Peabody DS, Berg P. 1986. Termination-reinitiation occurs in the translation of mammalian cell mRNAs. Mol. Cell. Biol. 6:2695–2703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Poyry TAA, Kaminski A, Connell EJ, Fraser CS, Jackson RJ. 2007. The mechanism of an exceptional case of reinitiation after translation of a long ORF reveals why such events do not generally occur in mammalian mRNA translation. Genes Dev. 21:3149–3162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rajkowitsch L, Vilela C, Berthelot K, Ramirez CV, McCarthy JE. 2004. Reinitiation and recycling are distinct processes occurring downstream of translation termination in yeast. J. Mol. Biol. 335:71–85 [DOI] [PubMed] [Google Scholar]
- 29. Resch A, Ogurtsov A, Rogozin I, Shabalina S, Koonin E. 2009. Evolution of alternative and constitutive regions of mammalian 5′ UTRs. BMC Genomics 10:162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Suzuki Y, et al. 2000. Statistical analysis of the 5′ untranslated region of human mRNA using “oligo-capped” cDNA libraries. Genomics 64:286–297 [DOI] [PubMed] [Google Scholar]
- 31. Szamecz B, et al. 2008. eIF3a cooperates with sequences 5′ of uORF1 to promote resumption of scanning by post-termination ribosomes for reinitiation on GCN4 mRNA. Genes Dev. 22:2414–2425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Twiss JL, Shooter EM. 1995. Nerve growth factor promotes neurite regeneration in PC12 cells by translational control. J. Neurochem. 64:550–557 [DOI] [PubMed] [Google Scholar]
- 33. Twiss JL, Smith DS, Chang B, Shooter EM. 2000. Translational control of ribosomal protein L4 mRNA is required for rapid neurite regeneration. Neurobiol. Dis. 7:416–428 [DOI] [PubMed] [Google Scholar]
- 34. Vanhoutte P, Caboche J. 2002. Elk-1: an important regulator of immediate early gene expression in the brain, p 287–308 Kaczmarck L, Robertson HJ. (ed), Handbook of chemical neuroanatomy, vol. 19 Elsevier, New York, NY [Google Scholar]
- 35. Vanhoutte P, et al. 2001. Opposing roles of Elk-1 and its brain-specific isoform, short Elk-1, in nerve growth factor-induced PC12 differentiation. J. Biol. Chem. 276:5189–5196 [DOI] [PubMed] [Google Scholar]
- 36. Vattem KM, Wek RC. 2004. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 101:11269–11274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang XQ, Rothnagel JA. 2004. 5′-Untranslated regions with multiple upstream AUG codons can support low-level translation via leaky scanning and reinitiation. Nucleic Acids Res. 32:1382–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wethmar K, et al. 2010. C/EBPbetaDeltauORF mice—a genetic model for uORF-mediated translational control in mammals. Genes Dev. 24:15–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wiesenthal V, Leutz A, Calkhoven CF. 2006. A translation control reporter system (TCRS) for the analysis of translationally controlled processes in the vertebrate cell. Nucleic Acids Res. 34:e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wiesenthal V, Leutz A, Calkhoven CF. 2006. Analysis of translation initiation using a translation control reporter system. Nat. Protoc. 1:1531–1537 [DOI] [PubMed] [Google Scholar]
- 41. Zhou D, et al. 2008. Phosphorylation of eIF2 directs ATF5 translational control in response to diverse stress conditions. J. Biol. Chem. 283:7064–7073 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.