Abstract
Autophagy is an evolutionarily conserved degradation pathway characterized by dynamic rearrangement of membranes that sequester cytoplasm, protein aggregates, organelles, and pathogens for delivery to the vacuole and lysosome, respectively. The ability of autophagosomal membranes to act selectively toward specific cargo is dependent on the small ubiquitin-like modifier ATG8/LC3 and the LC3-interacting region (LIR) present in autophagy receptors. Here, we describe a comprehensive protein-protein interaction analysis of TBC (Tre2, Bub2, and Cdc16) domain-containing Rab GTPase-activating proteins (GAPs) as potential autophagy adaptors. We identified 14 TBC domain-containing Rab GAPs that bind directly to ATG8 modifiers and that colocalize with LC3-positive autophagy membranes in cells. Intriguingly, one of our screening hits, TBC1D5, contains two LIR motifs. The N-terminal LIR was critical for interaction with the retromer complex and transport of cargo. Direct binding of the retromer component VPS29 to TBC1D5 could be titrated out by LC3, indicating a molecular switch between endosomes and autophagy. Moreover, TBC1D5 could bridge the endosome and autophagosome via its C-terminal LIR motif. During starvation-induced autophagy, TBC1D5 was relocalized from endosomal localization to the LC3-positive autophagosomes. We propose that LC3-interacting Rab GAPs are implicated in the reprogramming of the endocytic trafficking events under starvation-induced autophagy.
INTRODUCTION
Autophagy is a conserved cellular catabolic process that involves the formation of a membrane double layer, cargo sequestration, sealing, maturation, and eventual fusion with vacuoles and lysosomes. In Saccharomyces cerevisiae, the core machinery consists of 33 essential autophagy (Atg) genes, including two ubiquitin-like conjugation systems (47). The small ubiquitin-like modifier Atg8p (MAP1LC3/GABARAP in mammals) is processed and conjugated to phosphatidylethanolamine (PE) residing in a growing preautophagosomal structure (PAS) via sequential action of Atg4p, Atg7p, and Atg3p and cooperation with Atg10p, Atg5p—Atg12p (where — indicates covalent binding), and Atg16p. Lipidated Atg8p serves as a docking site for specific cargo receptors, such as the cytosol-to-vesicle targeting (cvt) protein Atg19p or the outer mitochondrial membrane protein Atg32p (52). In mammals, lipidated MAP1LC3 and GABARAP proteins recruit receptors for specific cargo, such as p62, NBR1, Nix, NDP52, and OPTN (22, 28, 31, 34, 48, 51), and adaptor proteins that regulate the movement of autophagosomes or their maturation (20, 33, 34). All known autophagy receptor and adaptor proteins bind to ATG8 modifiers via an LC3-interacting region (LIR) motif with a consensus sequence W/Y/F-X-X-I/L (40).
Though major signaling cascades leading to induction or suppression of autophagy are known, the membrane dynamics accompanying autophagy remain unclear. Several membrane compartments in the cell, including the endoplasmic reticulum (ER), Golgi apparatus, mitochondria, and plasma membrane, have been suggested as sources of incipient autophagic membranes (13, 16, 19). Functional endomembrane trafficking mediated by ESCRT (11), COPI (49), TRAPP complexes (25), and components of the secretory pathway, as well as the SNARE fusion machinery (24, 27, 29, 39), have been demonstrated to impact autophagy. However, what exactly links all those pathways is still unclear. Rab GTPase-activating proteins (Rab GAPs) contain the highly conserved TBC (Tre2, Bub2, and Cdc16) domain that regulates the activity of small Rab GTPases via a dual-finger mechanism (32). Hence, Rab GAPs are essentially implicated in the spatial and temporal dynamics of the cellular endomembrane system (4). In this study, we performed a comprehensive analysis of TBC domain-containing Rab GAPs in the context of autophagy. By screening a library of 36 human TBC proteins for binding to MAP1LC3 and GABARAP proteins, we identified 14 ATG8-interacting TBC proteins. Motif mapping revealed two LIR motifs in TBC1D5, which are both required for binding to and vesicular colocalization with human ATG8. Remarkably, the LIR1 motif in TBC1D5 was also required for retromer binding, cation-independent mannose-6-phosphate receptor (CI-M6PR) internalization, and trans-Golgi network (TGN) retrieval. LC3 and retromer interactions with TBCD5 were dependent on LC3 abundance, suggesting that TBC1D5 can switch from retromer-decorated endosomal to LC3-lipidated autophagosomal membranes upon autophagy induction. Moreover, CI-M6PR localization in the TGN appeared to be more pronounced under starvation conditions. Due to its implication in the regulation of autophagy and retrograde transport, TBC1D5 might be an important factor in preserving the balance between the endocytic pathway and remodeling of cellular membranes to generate functional autophagosomes.
MATERIALS AND METHODS
Plasmids and cell lines.
Open reading frame (ORF) clones were obtained from Francis Barr or from the human ORFeome collection (http://horfdb.dfci.harvard.edu/) or cloned from available full-length cDNAs (see Table S3 in the supplemental material). The VPS29 clone for bacterial expression was obtained from James Hurley (17). All human TBC proteins were employed as N-terminal green fluorescent protein (GFP) fusions in pEGFP-C2. ORFs for selected TBC proteins were subcloned into pMALC2, pGEX4T-1, and pGEX6P1 vectors (Clontech). Point mutations and LIR tetrapeptide motif deletion and domain-mapping fragments were generated by site-directed mutagenesis. Sequence-validated ORFs in pDONR223 were recombined into the Gateway destination vectors MSCV-Tet-FLAG-HA-IRES-PURO (which uses a tetracycline-inducible cytomegalovirus [CMV] promoter [iTAP]) for stable cell lines, pET60-DEST (Novagen) for recombinant expression, and pDEST-CMV-N-Myc and pHAGE-CMV-N-mCherry for transient transfection using λ recombinase. Virus was produced for the indicated ORFs by cotransfecting the respective iTAP, GAG/POL, and vesicular stomatitis virus Indiana G protein (VSVG) plasmids into 293T cells. After packaging, the viruses were used to infect the TREx-293 (Invitrogen) or U2OS cell line and subsequently selected for stable expression using 1 μg/ml puromycin.
Stable knockdown cell lines.
HeLa cells stably expressing two different short hairpin RNA (shRNA) oligonucleotides targeting all 3 isoforms of human TBC1D5 (GAGAACGAACAGATCACCATT and GCAAAGGTGATGGACACTCAT) were generated by lentiviral infection. Virus was produced for each shRNA oligonucleotide (Open Byosystems; pLKO) by cotransfecting the respective pLKO plasmid, together with Tat1b, Hgpm2, CMV-Rev, and VSVG plasmids, into 293T cells. After packaging, viral supernatants were used to infect HeLa cells and subsequently selected for stable expression using 1 μg/ml puromycin. An shRNA-resistant TBC1D5 variant (against GAGAACGAACAGATCACCATT) was generated by mutagenesis and transiently transfected in cells using pDEST-CMV-N-Myc. Control shRNA cells were generated using Mission nontarget shRNA control lentiviral particles obtained from Sigma-Aldrich.
Antibodies.
The antibodies used in this study were as follows: goat polyclonal anti-TBC1D5 (Santa Cruz), mouse monoclonal anti-LC3 (NanoTools), rabbit polyclonal anti-LC3 (MBL) and rabbit polyclonal anti-LC3B (a gift from Z. Elazar), rabbit polyclonal anti-p62 (Enzo Life Sciences) and mouse monoclonal anti-p62 (MBL), polyclonal rabbit anti VPS29 (Sigma-Aldrich; Atlas), goat polyclonal anti-VPS35 (LSBio), mouse monoclonal MEM-238 anti-mannose 6 phosphate receptor (cation independent) (Abcam), rabbit polyclonal anti-TGN46 (Sigma-Aldrich), rabbit polyclonal anti-Atg5 (Cell Signaling), rabbit polyclonal anti-GFP (Living Colors), mouse monoclonal anti-Flag M2 (Sigma-Aldrich), mouse monoclonal anti-myc (Santa Cruz), and mouse monoclonal antivinculin (Sigma-Aldrich). All corresponding secondary horseradish peroxidase (HRP)-conjugated antibodies were from Santa Cruz, and Cy5-, Cy3-, and fluorescein isothiocyanate (FITC)-conjugated secondary antibodies were from Jackson ImmunoResearch.
Antibody-feeding assay.
HeLa cells were grown on glass coverslips and transferred to Dulbecco's modified Eagle's medium (DMEM) with 2% fetal bovine serum (FBS) for 10 h prior to antibody feeding. After blocking with 5% bovine serum albumin (BSA) in phosphate-buffered saline (PBS) (containing Ca2+ and Mg2+) for 30 min, antibody against CI-M6PR was applied (10 μg/ml in 5% BSA in PBS) for 30 min. The cells were washed twice with PBS, and warm medium (DMEM-10% FBS) was added. After 10, 30, and 40 min of incubation at 37°C, the cells were fixed with 2% paraformaldehyde, permeabilized in 0.2% Triton X in PBS, and counterstained using secondary Cy5-conjugated donkey anti-mouse antibody. The plasma membrane fraction of the receptor (at time point 0) was stained on nonpermeabilized cells.
Protein purification and binding assay.
The indicated glutathione S-transferase (GST) fusion proteins were expressed in E. coli BL21 cells and purified using glutathione (GSH)-Sepharose. For lysate analysis, ORFs encoding TBC domain family proteins in pEGFP-C2 or N-Myc were transiently transfected in 293T cells using GeneJuice. After 20 h, whole-cell extracts were incubated with the indicated GST fusion proteins purified from bacteria. The washed beads were subjected to SDS-PAGE and immunoblotting using anti-GFP and anti-Flag antibodies, respectively. For in vitro analysis, recombinantly purified VPS29 and maltose-binding protein (MBP)-tagged MAP1LC3A were incubated with the indicated GST fusion proteins in binding buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 10% glycerol, 1% Triton-X, 25 mM NaF, 50 mM ZnCl2, 1 mM dithiothreitol [DTT]). After 2 h, the GSH resin was washed 5 times with binding buffer, and bound proteins were subjected to SDS-PAGE and immunoblotted using anti-MBP and anti-VPS29 antibodies.
Immunoprecipitation.
For Western blot analysis, TREx-293 cells stably expressing various TBC1D5 constructs (iTAP) and 293T cells transiently expressing human ATG8 family members (pEGFP-C2) were induced and transfected (GeneJuice; Novagen) for 24 h. Whole-cell lysates were incubated for 1 h with anti-Flag M2-coupled resin, anti-GFP-coupled beads, or anti-VPS35, followed by incubation with protein G-Sepharose. Samples were subjected to SDS-PAGE and immunoblotted with anti-Flag, anti-VPS34, anti-VPS29, anti-TBC1D5, and anti-GFP antibodies. For liquid chromatography coupled to tandem mass spectrometry (LC–MS-MS) analysis, four 15-cm tissue culture dishes of cells (∼80% confluence; ∼107 cells) stably expressing TBC1D5 and TBC1D25 were harvested and lysed with 3 ml lysis buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 0.5% Nonidet P40, and a Complete EDTA-free protease inhibitor tablet [Roche]). Centrifugation-cleared lysates (13,000 rpm) were filtered through 0.45-mm spin filters (Millipore Ultrafree-CL) and immunoprecipitated with 60 μl antihemagglutinin (anti-HA) (Sigma) resin. Resin containing immune complexes was washed five times with lysis buffer, followed by five PBS washes and elution with 150 μl of 250 mg/ml HA peptide in PBS.
Mass spectrometry.
The eluted immune complexes were precipitated with 20% trichloroacetic acid (TCA) (Sigma), and the pellets were washed once with 10% TCA and four times with cold acetone. The precipitated proteins were resuspended in 100 mM ammonium bicarbonate (pH 8.0) with 10% acetonitrile and incubated with sequencing-grade trypsin (Promega) at a concentration of 12.5 ng/ml at 37°C for 4 h. Trypsin reactions were quenched by addition of 5% formic acid, and peptides were desalted using the C18 StageTip method (37). For each LC–MS-MS run using an LTQ Velos linear ion trap mass spectrometer (Thermo Scientific), 4 μl was loaded onto an 18-cm by 125-μm (inside diameter [i.d.]) C18 column, and peptides were eluted using a 50-min 8% to 26% acetonitrile gradient. Spectra were acquired using a data-dependent Top-10 method. Each sample was shot twice in succession, followed by a wash with 70% acetonitrile and 30% isopropanol.
Confocal microscopy and live-cell imaging.
HeLa cells were transiently transfected with ORFs encoding TBC domain family or Rab proteins in pEGFP-C2, pDEST-CMV-N-Myc, or pHAGE-CMV-N-mCherry using GeneJuice (Novagen). After 20 h, cells were fixed on coverslips with 2% paraformaldehyde and counterstained for endogenous proteins with the indicated antibodies. Images were acquired with an LSM510 microscope (Zeiss) and processed using ImageJ software. Pearson's correlation coefficients of colocalizations after subtraction of background fluorescence were calculated with Volocity Demo software (Perkin Elmer), and graphs were created in Graph Pad Prism. The level of significance was calculated using a t test (Graph Pad Prism). Imaging of live U2OS cells, grown in 8-well Lab-Tek chambers, was performed on a Leica CTR7000 HS epifluorescence microscope. Movies were processed using Leica software, and separate images were exported as TIFF images following analysis with ImageJ (NIH).
RESULTS
TBC domain-containing Rab GAPs as novel ATG8-binding proteins.
To identify novel ATG8-interacting proteins implicated in autophagy, we performed a yeast two-hybrid screen using three different ATG8 proteins as bait: yeast Atg8p and two of its mammalian homologues, MAP1LC3B and GABARAPL2 (GATE-16). Among the ATG8-interacting proteins identified in this initial screening approach were NBR1 (22), Nix (31), and OPTN (51). Intriguingly, several TBC domain-containing proteins (TBC1D2B, TBC1D9, TBC1D9B, and TBC1D17) were highly abundant clones in all three screens (see Table S1A in the supplemental material). Moreover, several TBC domain-containing proteins, including TBC1D2B, TBC1D4, TBC1D5, TBC1D7, TBC1D9B, TBC1D15, TBC1D18, and TBC1D23, were also proteomically identified within autophagy interaction networks (5). Lastly, TBC1D25 (also known as OATL1) was recently identified as a novel ATG8-binding protein involved in regulation of Rab33B and autophagosome-lysosome fusion (20).
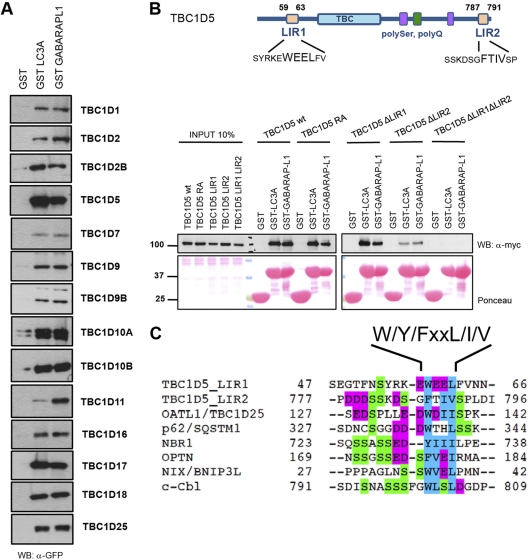
Thus, to generally probe TBC domain-containing proteins for their role in autophagy, we employed a transiently overexpressing library of human TBC proteins N-terminally tagged with GFP in 293T cells, together with GST pulldown assays to systematically screen for binding to human ATG8 proteins. Fourteen TBC proteins were specifically bound to GST-fused mammalian ATG8 proteins, but not to GST alone (Fig. 1A; see Fig. S1A to C in the supplemental material). Out of these 14 candidates, four TBC proteins colocalized with MAP1LC3B and p62 in HeLa cells (see Fig. S3A and Table S1B in the supplemental material). Since more than a dozen TBC proteins bind to ATG8 modifiers but differ in specificity for Rab GTPases, cellular localization, and domain architectures (see Fig. S3B in the supplemental material), we hypothesized that TBC Rab GAPs may have redundant functions in regulating different vesicular-trafficking pathways and bridge these pathways to autophagy in a stimulus-specific manner and in specific cellular compartments. To analyze whether the interactions of these four TBC proteins and mammalian ATG8 family members are direct, we purified full-length TBC proteins or fragments thereof from bacterial lysate and performed in vitro binding assays with purified MAP1LC3 and GABARAP family members. Direct binding between TBC1D2B, TBC1D5, TBC1D17, TBC1D25, and mammalian ATG8 proteins was readily detected (see Fig. S2A to D in the supplemental material). By performing a series of deletion mutageneses, we were able to map two LIR motifs in TBC1D5 (Fig. 1B).
Fig 1.
TBC proteins bind human ATG8 proteins via canonical LIR. (A) ATG8-interacting TBC proteins. HEK 293T cell lysates overexpressing GFP-tagged human TBC proteins were subjected to pulldown assays using GST, GST-MAP1LC3A, and GST-GABARAPL1. Coprecipitated TBC proteins were detected with anti-GFP antibody. Inputs are shown in Fig. S1A in the supplemental material. WB, Western blot. (B) (Top) Schematic representation of TBC1D5 with Pfam domains annotated and LIR motifs identified. (Bottom) The wild type and catalytic RA and LIR mutants of TBC1D5 were transiently expressed in 293T cells and subjected to pulldown assays with GST alone or GST-ATG8 proteins. GST fusion proteins were visualized by Ponceau S staining. (C) Alignments of canonical LIR motifs from annotated ATG8-interacting partners and LIR motifs mapped in TBC1D5. Magenta, acidic residues; blue, small and hydrophobic residues; green, serine residues.
TBC1D5 function is required for autophagy.
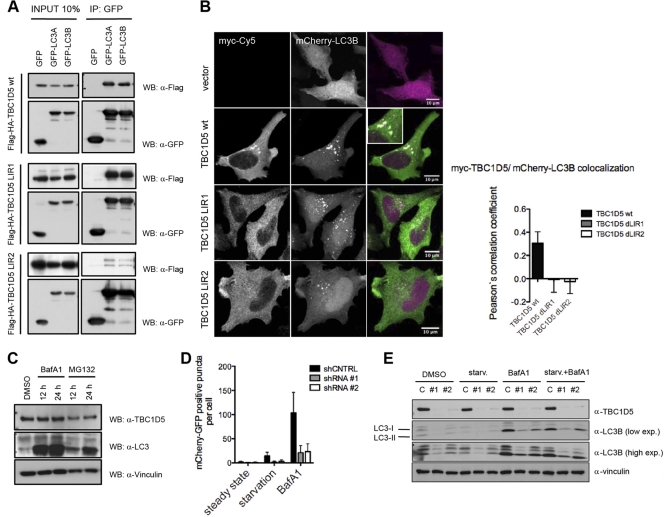
In contrast to the majority of established LIR-containing ATG8-interacting proteins (Fig. 1C), TBC1D5 contains two LIR motifs, which flank the TBC domain. Both TBC1D5 LIR motifs mediate binding to mammalian ATG8 in vitro (Fig. 1B). We performed a series of coimmunoprecipitations on lysates derived from cells transiently cotransfected with mammalian ATG8 proteins and a TBC1D5 ΔLIR1 or ΔLIR2 mutant to examine the contribution of each LIR motif to binding to MAP1LC3 in cells. Surprisingly, separate deletions of LIR1 and LIR2 led to substantially increased and decreased binding to MAP1LC3, respectively (Fig. 2A). Ectopically expressed TBC1D5 colocalized with mCherry-MAP1LC3B puncta (Fig. 2B), while TBC1D5 variants carrying LIR1 or LIR2 deletions did not colocalize with MAP1LC3-positive puncta (Fig. 2B). Thus, both LIR motifs of TBC1D5 seem to differentially contribute to the recruitment of TBC1D5 to autophagosomes.
Fig 2.
TBC1D5 is implicated in regulation of autophagy flux. (A) Coimmunoprecipitation (IP) of wild-type TBC1D5 and the ΔLIR1 and ΔLIR2 mutants with MAP1LC3A and MAP1LC3B proteins transiently overexpressed in 293T cells. (B) Immunofluorescence assay of myc-tagged TBC1D5 (wild type and ΔLIR1 and ΔLIR2 mutants) and mCherry-MAP1LC3B transiently overexpressed in HeLa cells. HeLa cells were cultured in regular medium, washed with PBS, fixed in 2% paraformaldehyde (PFA), and incubated with primary antibodies against myc tag. (C) Abundance of endogenous TBC1D5 and MAP1LC3 in HeLa cells 12 and 24 h after treatment with BafA1 (200 nM) and MG132 (10 μM), respectively. The indicated proteins were detected in whole-cell lysates by immunoblotting. DMSO, dimethyl sulfoxide. (D) shRNA control (shCNTRL) and stable knockdown cell lines expressing tandem mCherry-GFP-MAP1LC3B were subjected to starvation conditions or treated with BafA1 for 4 h. Autophagosomes were quantified in more than 100 cells in 3 independent experiments. A paired t test was used to indicate significance. The error bars represent standard deviations (SD) (GraphPad Prism). (E) shRNA control cells and two stable TBC1D5 knockdown HeLa cell lines were starved (starv.) in EBSS, treated with bafilomycin A1, or starved in the presence of BafA1 (4 h). Total cell lysates were analyzed by SDS-PAGE following immunoblotting of endogenous TBC1D5, MAP1LC3B, and vinculin (as a loading control). exp., expression.
To fully understand the role of TBC1D5 in autophagy, we monitored colocalization of endogenous TBC1D5 with MAP1LC3 and the autophagy receptor p62 in basal and stimulus-induced autophagy. TBC1D5 and MAP1LC3 readily colocalized moderately under basal autophagy. However, starvation substantially increased their colocalization (see Fig. S3A in the supplemental material). Next, we examined whether the observed recruitment of TBC1D5 to autophagosomes is concomitant with autophagosomal degradation of TBC1D5. Therefore, we monitored the abundance of TBC1D5 upon inhibition of lysosomal and proteasomal degradation. Bafilomycin A1 (BafA1) or MG132 treatment did not result in accumulation of TBC1D5 (Fig. 2C), suggesting that TBC1D5 is not an autophagy substrate. Moreover, stable shRNA-mediated depletion of TBC1D5 in cells led to an accumulation of p62-positive puncta (see Fig. S3B in the supplemental material), suggesting that TBC1D5 may play an important role in cargo engulfment, autophagosome biogenesis, and/or regulation of autophagosomal flux.
To dissect potential roles of TBC1D5 in autophagy more rigorously, we examined autophagy flux in cells stably depleted of TBC1D5 by expressing the tandem mCherry-enhanced green fluorescent protein (eGFP)-LC3B “traffic light” reporter. In autophagosomes, mCherry-eGFP-LC3 produces yellow fluorescence, but upon transfer to the more acidic compartment of the (auto)lysosome, GFP fluorescence is specifically quenched due to pH-dependent unfolding, thereby rendering lysosomes mCherry positive. The ratio between yellow (autophagosome and [auto]lysosome) and red ([auto]lysosome) puncta provides a rapid means by which to determine autophagic flux. Upon starvation, autophagosomes and (auto)lysosomes accumulate, reflecting autophagic vesicle populations accompanying ongoing autophagy flux (Fig. 2D; see Fig. S3C in the supplemental material). However, depletion of TBC1D5 led to the almost complete disappearance of autophagic vesicles (Fig. 2D; see Fig. S3C in the supplemental material). Blockage of autophagosome clearance during basal or starvation-induced autophagy by BafA1 treatment yielded increased levels of lipidated LC3 (LC3-II) (Fig. 2E). Remarkably, TBC1D5 depletion substantially ablated BafA1-induced increase of LC3-II during both forms of autophagy (Fig. 2E). Thus, TBC1D5 seems to be critically required for efficient steady-state and starvation-induced formation of cargo-competent autophagosomes.
TBC1D5 binds the retromer in a LIR-dependent manner.
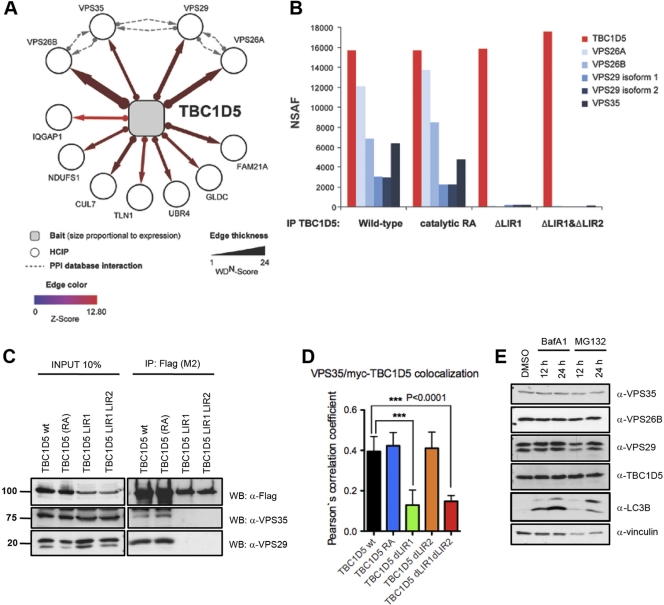
To probe the biological context in which TBC1D5 acts, we performed LC–MS-MS on trypsinized immune complexes derived from TREx-293 cells retrovirally expressing stable, doxycycline-inducible Flag-HA-tagged TBC1D5. All subunits of the retromer vesicle proteins sorting (VPS) subcomplex, VPS35, VPS26A, VPS26B, and VPS29, as well as the WASH complex component FAM21A, were detected as high-confidence candidate interaction proteins (HCIPs) in complex with TBC1D5 (Fig. 3A) (5, 45). Furthermore, known components of the retromer-sortin-nexin subcomplex, including SNX1, SNX5, and SNX6 were detected with subthreshold HCIP values (see Table S2A in the supplemental material).
Fig 3.
TBC1D5 requires LIR1 to bind to the retromer. (A) Proteomic analysis of TBC1D5. The association map shows identified HCIPs. The dashed lines indicate interactions annotated in the BIOGRID, MINT, and HPRD protein interaction databases. (B) Proteomic analysis of TBC1D5. Immune complexes from cells expressing wild-type and catalytic RA and ΔLIR mutant TBC1D5 proteins were subjected to IP–LC–MS-MS. Normalized spectral abundance factors (NSAF) were calculated based on total spectral counts (41). (C) Validation of TBC1D5-associated proteins. Expression of HA-Flag-tagged wild-type TBC1D5, TBC1D5 RA, TBC1D5 ΔLIR1, or TBC1D5 ΔLIR1 ΔLIR2 was induced in stable TREx293 cells for 24 h. Cell lysates were immunoprecipitated with Flag M2-coupled resin and immunoblotted with anti-Flag and anti-Vps29 antibody, respectively. Units for numbers (left) are kDa. (D) Immunofluorescence of endogenous Vps35 and transiently expressed myc-tagged TBC1D5 (wild type [wt], TBC1D5 RA, TBC1D5 ΔLIR1, TBC1D5 ΔLIR2, and TBC1D5 ΔLIR1 ΔLIR2) in HeLa cells. Colocalization of more than 100 cells was quantified in 3 independent experiments for each TBC1D5 construct using Volocity Demo software, and a paired t test was performed using GraphPad Prism. The error bars indicate SD. (E) Abundance of endogenous TBC1D5, MAP1LC3, and retromer subunits VPS35, VPS26B, and VPS29 in HeLa cells 12 and 24 h after treatment with BafA1 (100 nM) and MG132 (10 μM).
Next, we examined whether the association of TBC1D5 with the retromer complex depends on the LIR motifs in TBC1D5. Immune complexes derived from stable inducible TREx-293 cell lines expressing Flag-HA-tagged wild-type TBC1D5, a catalytic TBC1D5 mutant (RA), or TBC1D5 variants lacking either LIR1 (ΔLIR1) or both LIRs (ΔLIR1 ΔLIR2) were subjected to LC–MS-MS. Importantly, TBC1D5 bait peptides were detected at similar abundances across all four parallel proteomic experiments (see Table S2B in the supplemental material). Though association of TBC1D5 with retromer components was independent of TBC1D5 catalytic activity, as reported previously (14), binding of retromer components was abolished in single and double TBC1D5 LIR mutants (Fig. 3B).
To validate these results, we performed coimmunoprecipitation of Flag-HA TBC1D5 wild-type and mutant proteins from TREx-293 cells. Specific bands corresponding to endogenous VPS35 and VPS29 were present in the immunoprecipitated fraction of wild-type TBC1D5, as well as RA and ΔLIR2 TBC1D5 variants, but not in ΔLIR1 TBC1D5 or TBC1D5 W59A mutants (Fig. 3C; see Fig. S4A in the supplemental material). Consistently, reciprocal immunoprecipitation of endogenous VPS35 precipitated wild-type TBC1D5 and the RA mutant, but not TBC1D5 ΔLIR1 variants (see Fig. S4B in the supplemental material). Immunofluorescence microscopy revealed that myc-tagged wild-type TBC1D5 colocalized with the endogenous retromer subunit VPS35 in cells. While the catalytic RA and LIR2 mutants did not affect the subcellular distribution of TBC1D5, LIR1 deletions abolished localization with VPS35 and led to diffused cytoplasmic localization (Fig. 3D; see Fig. S4C in the supplemental material). Thus, the N-terminal TBC1D5 LIR1 motif is differentially employed in mediating MAP1LC3 and retromer binding, while LIR2 is required only for binding to mammalian ATG8 proteins. Finally, we also monitored the abundance of VPS retromer subunits upon inhibition of lysosomal and proteosomal degradation. BafA1 or MG132 treatment did not result in accumulation of VPS subunits, suggesting they are not autophagy substrates (Fig. 3E).
TBC1D5 partitions between LC3 and retromer-mediated functions.
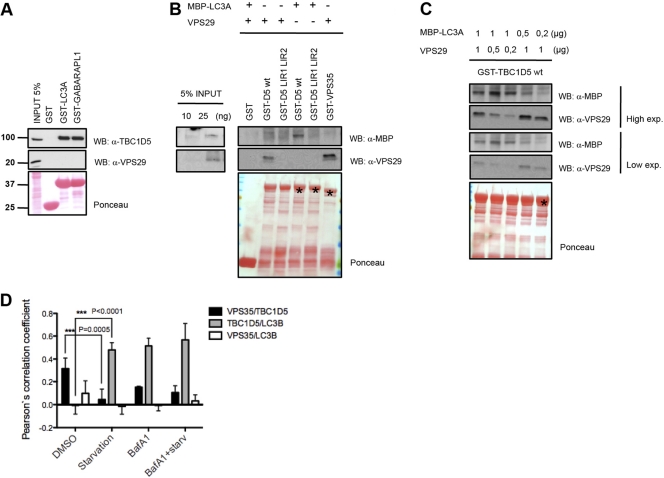
Since TBC1D5 interacts with MAP1LC3 and retromer components, we addressed whether these three proteins form a tripartite complex. Surprisingly, VPS29, which was proposed to mediate direct interaction with TBC1D5 (14), was not detected in MAP1LC3A-associated TBC1D5 in GST pulldown and coimmunoprecipitation experiments (Fig. 4A; see Fig. S4D in the supplemental material). Next, we rigorously examined binding of TBC1D5 to MAP1LC3 and retromer components in vitro using bacterial purified VPS29, MBP-MAP1LC3A, wild-type GST-TBC1D5, and the double LIR mutant. TBC1D5 bound directly to VPS29 and MAP1LC3A in a LIR-dependent manner (Fig. 4B). Incubating TBC1D5 with MAP1LC3A and various amounts of VPS29 or with VPS29 and various amounts of MAP1LC3A revealed that MAP1LC3A competed with VPS29 for binding to TBC1D5 (Fig. 4C). Based on our observations, we propose that TBC1D5 partitions between retromer-decorated endosomes and LC3-decorated autophagosomes, thereby potentially acting as a switch between these two vesicle-trafficking pathways. In that case, local increase in the LC3 concentration on the autophagosomal membrane upon autophagy induction might promote recruitment of TBC1D5 to autophagosomes from juxtaposed endosomes.
Fig 4.
TBC1D5 partitions between LC3 and retromer-mediated functions. (A) Lysates from nontransfected and HA-Flag-VPS35-, HA-Flag-VPS26A-, and HA-Flag-VPS26B-transfected 293T cells were incubated with GST, GST-MAP1LC3A, and GST-GABARAPL1. Copurifying proteins were detected by immunoblotting with anti-Flag, anti-VPS29, and anti-TBC1D5 antibodies. Units for numbers (left) are kDa. (B) In vitro binding assay with purified VPS29, MBP-coupled MAP1LC3A, and GST-TBC1D5 fusion protein. MAP1LC3A or VPS29 (1 μg) was used in a reaction volume of 400 μl (right). (C) In vitro competition assay. Concentrations of VPS29 and MAP1LC3A are indicated. GST-fused proteins TBC1D5, TBC1D5 ΔLIR1 ΔLIR2, and VPS35 are indicated by the asterisk on the Ponceau stain. (D) Immunofluorescence colocalization quantification of a double-stable U2OS cell line expressing mCherry-TBC1D5 and GFP-LC3B under normal conditions and starvation or bafilomycin A1 and starvation plus BafA1 treatment. Endogenous VPS35 was stained, and the colocalization of all three proteins in more than 200 cells was analyzed in Volocity Demo. The data were compared using a paired t test (GraphPad). The error bars indicate SD.
To test our hypothesis, we examined colocalization of all three components by employing cells stably expressing mCherry-TBC1D5 and GFP-MAP1LC3B, together with immunostaining of the endogenous retromer subunit VPS35. Since we did not observe colocalization of all three components, we quantified colocalization of VPS35 with TBC1D5, TBC1D5 with MAP1LC3B, and VPS35 with MAP1LC3B. VPS35 and TBC1D5 colocalized at basal autophagy, but their colocalization significantly decreased during starvation-induced autophagy. In contrast, colocalization of TBC1D5 and MAP1LC3B was not detectable at basal autophagy but significantly increased upon starvation (Fig. 4D; see Fig. S5A in the supplemental material). This shift in TBC1D5 colocalization from VPS35 to MAP1LC3B occurred to similar extents when cells undergoing basal autophagy or starvation-induced autophagy were treated with BafA1. Notably, VPS35 did not show any significant colocalization with MAP1LC3B (Fig. 4D; see Fig. S5A in the supplemental material). Finally, we monitored colocalization of TBC1D5 and MAP1LC3B during starvation-induced autophagy using live-cell imaging. Autophagosomes appeared overlying and moving on microtubules with TBC1D5-positive vesicles shortly after autophagy induction with Earle's balanced salt solution (EBSS) (see Movie S1 in the supplemental material). Two hours after starvation induction, larger vesicles containing double staining on the membrane could be observed, indicating possible transfer of TBC1D5 from endosomes to autophagosomes (see Fig. S6B and Movie S2 in the supplemental material).
TBC1D5 function is required for retrograde transport.
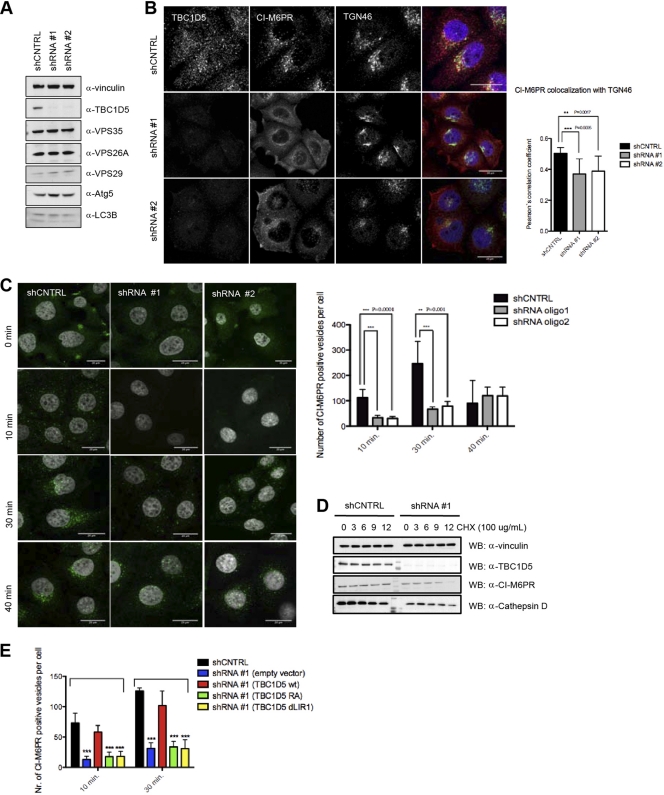
The retromer complex plays an essential role in retrograde transport from endosomes to the TGN (reviewed in references 3 and 6). TBC1D5 binds components of the retromer complex and has been implicated in regulating retromer detachment from endosomes (43). However, TBC1D5 depletion did not alter retromer localization (14) or the stability of VPS retromer components (Fig. 5A). To start dissecting the specific role of TBC1D5 in retrograde transport, we monitored trafficking of the endosomal cargo receptor CI-M6PR upon TBC1D5 depletion. While CI-M6PR was enriched in the TGN, marked by trans-Golgi network protein 46 (TGN46) in shRNA control cells, CI-M6PR appeared partially dispersed in the cytosol, as well as membrane localized in TBC1D5-depleted cells (Fig. 5B). Next, we performed a complementary anti-CI-M6PR antibody-feeding assay to examine CI-M6PR retrieval to the TGN. Consistently, TBC1D5 depletion yielded significantly reduced internalization of anti-CI-M6PR (Fig. 5C). Furthermore, during a 12-h cycloheximide chase, levels of CI-M6PR and mature cathepsin D decreased in TBC1D5-depleted cells (Fig. 5D), suggesting that protein sorting is compromised upon TBC1D5 depletion, as reflected by lysosomal targeting of CI-M6PR instead of TGN retrieval and excretion of cathepsin D instead of lysosomal targeting (1, 2, 42).
Fig 5.
TBC1D5 regulates retrograde transport and the autophagy pathway. (A) Total cell lysates from shRNA control and TBC1D5 knockdown cells were analyzed on SDS-PAGE and blotted for TBC1D5 and retromer subunits, respectively. (B) CI-M6PR localization is changed upon TBC1D5 knockdown. shRNA control cells and TBC1D5 knockdown cells were fixed and costained with primary antibodies against CI-M6PR and TGN46. Colocalization of both was analyzed in more than 200 cells in 3 independent experiments using Volocity Demo. The error bars indicate SD. (C) TBC1D5 knockdown affects CI-M6PR internalization. An antibody-feeding assay was performed as described in Materials and Methods, and CI-M6PR-positive vesicles were quantified in more than 300 cells using an ImageJ particle analysis plug-in. (D) TBC1D5 is important for stability of CI-M6PR and cathepsin D sorting. A cycloheximide (CHX) chase was performed on shRNA control and TBC1D5 knockdown cells, using 100 μg/ml of CHX and chase for 12 h. Total cell lysates were blotted against endogenous CI-M6PR, cathepsin D, and TBC1D5. (E) Rescue experiment for CI-M6PR retrieval to the TGN. shRNA control cells were subjected to an antibody-feeding assay as described for panel C; at the 10- and 30-min time points, cells were fixed, permeabilized, and incubated with primary antibody for TGN46 and then stained with secondary antibodies for both primary antibodies, CI-M6PR and TGN46. Colocalization of the internalized fraction of CI-M6PR with TGN46 was analyzed using Volocity Demo. Knockdown cells were transiently transfected with myc-tagged shRNA-resistant clones of TBC1D5, the wild type as well as RA and ΔLIR1 mutants. An antibody-feeding assay was performed 16 h posttransfection, and cells were fixed and stained using the same procedure described above. The graph presents quantification of CI-M6PR-positive vesicles quantified for more than 200 transfected cells. An unpaired t test was performed using GraphPad Prism. ***, P < 0.0005.
To address whether the observed protein-sorting defect indeed specifically depends on TBC1D5 and whether LIR1-mediated retromer binding is involved in this process, we generated myc-tagged shRNA-resistant variants of wild-type TBC1D5 and the RA and ΔLIR1 mutants. After transient expression of shRNA-resistant clones, we performed a CI-M6PR antibody-feeding assay and determined the numbers of receptor-positive vesicles, as well as colocalization of CI-M6PR with the TGN. Intriguingly, only wild-type TBC1D5, but not the RA and LIR1 mutant forms of TBC1D5, rescued the retrieval of CI-M6PR to the TGN (Fig. 5E; see Fig. S6A in the supplemental material), suggesting that Rab GAP activity and LIR1-mediated retromer binding are required for the proper function of TBC1D5 in the context of retrograde transport.
Interdependence between autophagy and retrograde transport.
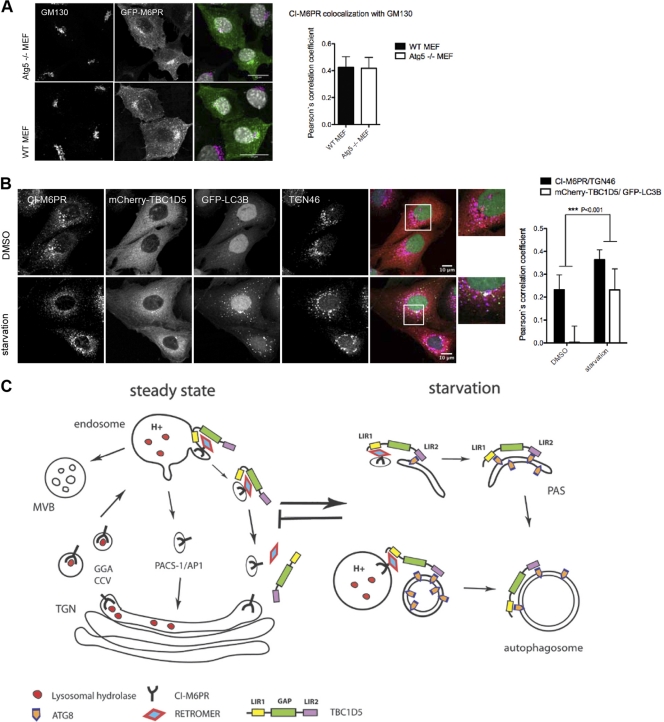
TBC1D5 appears to be involved in two pathways: autophagy and retrograde transport. Interestingly, ATG9 has been reported to traffic between the TGN and endosomes in a ULK1- and starvation-dependent manner, suggesting retrograde transport is critical for autophagosome formation (53). To explore whether these two pathways are interconnected, we monitored localization of CI-M6PR in autophagy-deficient and -induced cells. First, colocalization of GFP-tagged CI-M6PR to Golgi networks and of myc-tagged TBC1D5 to the retromer subunit VPS35 was unaffected in ATG5−/− mouse embryonic fibroblasts (Fig. 6A; see Fig. S6B in the supplemental material), indicating that basal autophagy does not contribute to retrograde transport. Next, colocalization of endogenous CI-M6PR to the TGN increased significantly upon starvation in cells stably expressing mCherry-TBC1D5 and GFP-MAP1LC3B (Fig. 6B). This suggested that exit from the TGN or retrieval of receptor to the TGN may be enhanced upon starvation. Though we could show that TBC1D5 is critical for retrograde transport, whether clathrin-, PACS-1/GGA3-, and TIP47/Rab9-dependent pathways of M6PR transport are likewise affected upon TBC1D5 depletion remains to be addressed. Taking the data together, we conclude that TBC1D5 can act as a molecular switch between endosomal and autophagosomal transport and is ultimately involved in reprogramming vesicle trafficking upon autophagy induction. Overlapping binding of retromer and mammalian ATG8 proteins to TBC1D5's LIR1 might be the basis of TBC1D5 translocation from endosomes to autophagosomes upon starvation-induced autophagy. Further investigations are needed to address whether TBC1D5 translocation is part of a negative regulation of retrograde transport or actively participates in autophagosome formation (Fig. 6C).
Fig 6.
Autophagy and retrograde transport are interdependent. (A) Localization of GFP-CI-M6PR and Golgi marker GM130 in WT and ATG5 knockout (KO) mouse embryonic fibroblastes (MEFs) and quantification of GFP-CI-M6PR colocalization with Golgi marker GM130 in WT and ATG5 KO MEFs. Colocalization with Golgi marker GM130 was analyzed in more than 60 transfected MEFs using Volocity Demo. The error bars indicate SD. (B) Immunofluorescence analysis of localization of CI-M6PR and TGN46 in double-stable U2OS cell lines expressing mCherry-TBC1D5 and GFP-MAP1LC3B. Cells were starved for 4 h in EBSS, fixed, and stained with corresponding endogenous antibodies. Colocalization of CI-M6PR/TGN46 and TBC1D5/MAP1LC3B was analyzed on more than 200 cells using Volocity Demo, and a paired t test was performed in GraphPad Prism. (C) Working model for TBC1D5 function in retrograde transport of CI-M6PR and autophagy. In steady state, TBC1D5 is localized on endosomes via binding to the retromer complex, regulating the dynamics of retrograde transport of cargo–CI-M6PR. Upon starvation-induced autophagy, TBC1D5 binds to autophagosomes via LIR2 and is transferred to the autophagosomal membrane via LIR1 and LIR2.
DISCUSSION
Collectively, our work indicates that TBC domain-containing Rab GAPs contribute to autophagy dynamics by direct binding to small ubiquitin-like modifiers of the LC3 and GABARAP family. TBC Rab GAPs are conserved from yeast to human. Together with guanine nucleotide exchange factors (GEF), GAPs tightly regulate the nucleotide status of small Rab GTPases, which are spatially organized in distinct membranes and act as small switches, alternating between GDP-bound (inactive) and GTP-bound (active) conformations. Rab GTPases are involved in the organization of almost all cellular membrane trafficking processes (reviewed in reference 46). For example, the activity of Rab GTPases residing in endocytic and secretory compartments has been shown to be critical for organizing specific regions for recruitment of tethering factors preceding docking and eventual fusion of donor and acceptor membrane compartments or progression from early to late endosomal compartments (10, 35, 50).
Our screen revealed a set of ATG8-interacting proteins from diverse branches within the phylogenetic tree of TBC domain-containing Rab GAPs (12) covering endocytosis (TBC1D2B and TBC1D5) (7), mTOR regulation (TBC1D7) (30), Rab33 (TBC1D25) (20), Rab35 and exocytosis (TBC1D10A and TBC1D10B) (18), and the Rab6 and Mad2 spindle checkpoint (TBC1D11/GAPCenA) (8, 26), as well as the Akt pathway (TBC1D1) (54), suggesting potential regulatory links between autophagy and these diverse cellular pathways. Notably, since ATG8 proteins can be hijacked for unconventional vesicle-trafficking events under certain conditions, as recently reported for coronaviruses (9, 38), ATG8-interacting Rab GAPs might also function in nonautophagic processes.
As the autophagic compartment is not permanently present to the same extent in the cell but rather is induced upon specific stimuli, we envision that its appearance needs to be followed by remodeling of distinct permanent membrane compartments in the cell. Therefore, the formation of preautophagosomal structures (PAS) is driven, while other trafficking processes may be redirected or downregulated so that the balance in the cellular lipid content is preserved. Under autophagy-stimulated conditions, different membrane compartments can contribute to form PAS (13, 16, 19). Though autophagy stimulation can be driven by many different stress factors, such as lack of nutrients, pathogen entry, or a defect in protein folding or the stability of organelles, many different signaling pathways may be involved in regulating this highly dynamic and complex set of processes leading to PAS formation, expansion, and cargo sequestration.
We focused on understanding how TBC1D5 functions in autophagy and retrograde transport. The retromer complex is important for efficient sorting of CI-M6PR to the trans-Golgi network (2), as well as Wntless-mediated secretion of Wnt (3, 15). Interestingly, processing and localization of amyloid precursor protein (APP) has been also shown to be regulated by the retromer, implicating the retromer as a critical player in the pathogenesis of Alzheimer's disease (44), and multiple VPS35 mutations have been reported in Parkinson's disease (55). We observed that the requirement for TBC1D5 in the CI-M6PR TGN retrieval pathway is dependent on TBC1D5 catalytic activity and its ability to bind to the retromer complex. Moreover, we started to dissect how this pathway may be affected under autophagy-induced conditions. TBC1D5 employs two different LIR motifs to switch from being localized on endosomes via binding to the VPS retromer subcomplex to localize on autophagosomes via ATG8 binding upon starvation-induced autophagy. We observed that loading of TBC1D5 on LC3-positive vesicles depends on both LIR motifs, suggesting that TBC1D5 first must be localized in the endosomal compartment by binding to the retromer in order to be efficiently transferred to lipidated MAP1LC3 on autophagosomes upon starvation. A functional consequence of this switch seems to be pronounced colocalization of CI-M6PR with the TGN upon autophagy induction. Intriguingly, WIPI-1 involvement in both starvation-induced autophagy (36) and retrograde transport of CI-M6PR (21) has been demonstrated. Whether CI-M6PR retrieval to the TGN is regulated by PACS-1 clathrin-mediated trafficking and TIP47/Rab9-dependent trafficking or retromer-dependent Wntless-mediated Wnt secretion is regulated interdependently on starvation-induced autophagy remains to be addressed. Furthermore, the exact role of TBC1D5 on autophagosomal membranes awaits elucidation in future studies. However, defects in efficient cargo-competent autophagosome formation indicate a critical role of TBC1D5 in regulating starvation-induced autophagy.
Taken together, this study provides a systematic analysis of the TBC domain-containing family of proteins in the context of their interaction with LC3/GABARAP autophagy modifiers. Importantly, we observed binding of mammalian ATG8 proteins to TBC domain-containing GAPs that regulate a variety of different trafficking pathways, revealing another level of complexity by which autophagy dynamics might be regulated. Alternatively, these ATG8-binding TBC GAPs might be employed in positioning the autophagic compartment within the dynamic network of endomembrane remodeling mediated by small Rab GTPases and their direct regulators. Tissue-specific expression, regulation by posttranslational modifications, the requirement for each TBC GAP, and their possible roles in development or cargo-specific autophagy are questions we wish to systematically address in our future studies in the context of cellular localization, protein-interacting networks, and specific Rab GTPase-GAP pairs.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to F. Barr for providing a cDNA library of 36 human TBC proteins, J. Hurley for providing human VPS29 in a vector for bacterial expression, and Z. Elazar for LC3B antibody. ATG5 knockout mouse embryonic fibroblasts were a kind gift from Naboru Mizushima (23).
This work was supported by NIH grant GM095567 (J.W.H.), the Cluster of Excellence “Macromolecular Complexes” of the Goethe University Frankfurt (EXC115), Loewe Center for Cell and Gene Therapy Frankfurt, a European Research Council Advanced Grant (I.D.), a DFG/Emmy Noether Grant, and a European Research Council Starting Grant (C.B.).
Footnotes
Published ahead of print 21 February 2012
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Adachi A, et al. 2010. Golgi-associated GSK3beta regulates the sorting process of post-Golgi membrane trafficking. J. Cell Sci. 123:3215–3225 [DOI] [PubMed] [Google Scholar]
- 2. Arighi CN, Hartnell LM, Aguilar RC, Haft CR, Bonifacino JS. 2004. Role of the mammalian retromer in sorting of the cation-independent mannose 6-phosphate receptor. J. Cell Biol. 165:123–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Attar N, Cullen PJ. 2010. The retromer complex. Adv. Enzyme Regul. 50:216–236 [DOI] [PubMed] [Google Scholar]
- 4. Barr F, Lambright DG. 2010. Rab GEFs and GAPs. Curr. Opin. Cell Biol. 22:461–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Behrends C, Sowa ME, Gygi SP, Harper JW. 2010. Network organization of the human autophagy system. Nature 466:68–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bonifacino JS, Hurley JH. 2008. Retromer. Curr. Opin. Cell Biol. 20:427–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chotard L, et al. 2010. TBC-2 regulates RAB-5/RAB-7-mediated endosomal trafficking in C. elegans. Mol. Biol. Cell. 21:2285–2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cuif MH, et al. 1999. Characterization of GAPCenA, a GTPase activating protein for Rab6, part of which associates with the centrosome. EMBO J. 18:1772–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. de Haan CA, Molinari M, Reggiori F. 2010. Autophagy-independent LC3 function in vesicular traffic. Autophagy 6:994–996 [DOI] [PubMed] [Google Scholar]
- 10. Del Conte-Zerial P, et al. 2008. Membrane identity and GTPase cascades regulated by toggle and cut-out switches. Mol. Syst. Biol. 4:206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Filimonenko M, et al. 2007. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J. Cell Biol. 179:485–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gao X, Jin C, Xue Y, Yao X. 2008. Computational analyses of TBC protein family in eukaryotes. Protein Pept. Lett. 15:505–509 [DOI] [PubMed] [Google Scholar]
- 13. Hailey DW, et al. 2010. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell 141:656–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Harbour ME, et al. 2010. The cargo-selective retromer complex is a recruiting hub for protein complexes that regulate endosomal tubule dynamics. J. Cell Sci. 123:3703–3717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Harterink M, et al. 2011. A SNX3-dependent retromer pathway mediates retrograde transport of the Wnt sorting receptor Wntless and is required for Wnt secretion. Nat. Cell Biol. 13:914–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hayashi-Nishino M, et al. 2009. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat. Cell Biol. 11:1433–1437 [DOI] [PubMed] [Google Scholar]
- 17. Hierro A, et al. 2007. Functional architecture of the retromer cargo-recognition complex. Nature 449:1063–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hsu C, et al. 2010. Regulation of exosome secretion by Rab35 and its GTPase-activating proteins TBC1D10A-C. J. Cell Biol. 189:223–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ishihara N, et al. 2001. Autophagosome requires specific early Sec proteins for its formation and NSF/SNARE for vacuolar fusion. Mol. Biol. Cell 12:3690–3702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Itoh T, Kanno E, Uemura T, Waguri S, Fukuda M. 2011. OATL1, a novel autophagosome-resident Rab33B-GAP, regulates autophagosomal maturation. J. Cell Biol. 192:839–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jeffries TR, Dove SK, Michell RH, Parker PJ. 2004. PtdIns-specific MPR pathway association of a novel WD40 repeat protein, WIPI49. Mol. Biol. Cell 15:2652–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kirkin V, et al. 2009. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell 33:505–516 [DOI] [PubMed] [Google Scholar]
- 23. Kuma A, et al. 2004. The role of autophagy during the early neonatal starvation period. Nature 432:1032–1036 [DOI] [PubMed] [Google Scholar]
- 24. Longatti A, Tooze SA. 2009. Vesicular trafficking and autophagosome formation. Cell Death Differ. 16:956–965 [DOI] [PubMed] [Google Scholar]
- 25. Lynch-Day MA, et al. 2010. Trs85 directs a Ypt1 GEF, TRAPPIII, to the phagophore to promote autophagy. Proc. Natl. Acad. Sci. U. S. A. 107:7811–7816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Miserey-Lenkei S, et al. 2006. A role for the Rab6A′ GTPase in the inactivation of the Mad2-spindle checkpoint. EMBO J. 25:278–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Moreau K, Ravikumar B, Renna M, Puri C, Rubinsztein DC. 2011. Autophagosome precursor maturation requires homotypic fusion. Cell 146:303–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mostowy S, et al. 2011. p62 and NDP52 proteins target intracytosolic Shigella and Listeria to different autophagy pathways. J. Biol. Chem. 286:26987–26995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nair U, et al. 2011. SNARE proteins are required for macroautophagy. Cell 146:290–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nakashima A, et al. 2007. Identification of TBC7 having TBC domain as a novel binding protein to TSC1-TSC2 complex. Biochem. Biophys. Res. Commun. 361:218–223 [DOI] [PubMed] [Google Scholar]
- 31. Novak I, et al. 2010. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 11:45–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pan X, Eathiraj S, Munson M, Lambright DG. 2006. TBC-domain GAPs for Rab GTPases accelerate GTP hydrolysis by a dual-finger mechanism. Nature 442:303–306 [DOI] [PubMed] [Google Scholar]
- 33. Pankiv S, et al. 2010. FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport. J. Cell Biol. 188:253–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pankiv S, et al. 2007. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282:24131–24145 [DOI] [PubMed] [Google Scholar]
- 35. Poteryaev D, Datta S, Ackema K, Zerial M, Spang A. 2010. Identification of the switch in early-to-late endosome transition. Cell 141:497–508 [DOI] [PubMed] [Google Scholar]
- 36. Proikas-Cezanne T, et al. 2004. WIPI-1alpha (WIPI49), a member of the novel 7-bladed WIPI protein family, is aberrantly expressed in human cancer and is linked to starvation-induced autophagy. Oncogene 23:9314–9325 [DOI] [PubMed] [Google Scholar]
- 37. Rappsilber J, Mann M, Ishihama Y. 2007. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2:1896–1906 [DOI] [PubMed] [Google Scholar]
- 38. Reggiori F, et al. 2010. Coronaviruses hijack the LC3-I-positive EDEMosomes, ER-derived vesicles exporting short-lived ERAD regulators, for replication. Cell Host Microbe 7:500–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Renna M, et al. 2011. Autophagic substrate clearance requires activity of the syntaxin-5 SNARE complex. J. Cell Sci. 124:469–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rozenknop A, et al. 2011. Characterization of the interaction of GABARAPL-1 with the LIR motif of NBR1. J. Mol. Biol. 410:477–487 [DOI] [PubMed] [Google Scholar]
- 41. Sardiu ME, et al. 2008. Probabilistic assembly of human protein interaction networks from label-free quantitative proteomics. Proc. Natl. Acad. Sci. U. S. A. 105:1454–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Seaman MN. 2007. Identification of a novel conserved sorting motif required for retromer-mediated endosome-to-TGN retrieval. J. Cell Sci. 120:2378–2389 [DOI] [PubMed] [Google Scholar]
- 43. Seaman MN, Harbour ME, Tattersall D, Read E, Bright N. 2009. Membrane recruitment of the cargo-selective retromer subcomplex is catalysed by the small GTPase Rab7 and inhibited by the Rab-GAP TBC1D5. J. Cell Sci. 122:2371–2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Small SA. 2008. Retromer sorting: a pathogenic pathway in late-onset Alzheimer disease. Arch. Neurol. 65:323–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sowa ME, Bennett EJ, Gygi SP, Harper JW. 2009. Defining the human deubiquitinating enzyme interaction landscape. Cell 138:389–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Stenmark H. 2009. Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 10:513–525 [DOI] [PubMed] [Google Scholar]
- 47. Suzuki K, Ohsumi Y. 2007. Molecular machinery of autophagosome formation in yeast, Saccharomyces cerevisiae. FEBS Lett. 581:2156–2161 [DOI] [PubMed] [Google Scholar]
- 48. Thurston TL, Ryzhakov G, Bloor S, von Muhlinen N, Randow F. 2009. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat. Immunol. 10:1215–1221 [DOI] [PubMed] [Google Scholar]
- 49. Tooze SA, Razi M. 2009. The essential role of early endosomes in autophagy is revealed by loss of COPI function. Autophagy 5:874–875 [DOI] [PubMed] [Google Scholar]
- 50. Wegner CS, et al. 2010. Ultrastructural characterization of giant endosomes induced by GTPase-deficient Rab5. Histochem. Cell Biol. 133:41–55 [DOI] [PubMed] [Google Scholar]
- 51. Wild P, et al. 2011. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science 333:228–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yang Z, Klionsky DJ. 2009. An overview of the molecular mechanism of autophagy. Curr. Top. Microbiol. Immunol. 335:1–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Young AR, et al. 2006. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J. Cell Sci. 119:3888–3900 [DOI] [PubMed] [Google Scholar]
- 54. Zhou QL, et al. 2008. Akt substrate TBC1D1 regulates GLUT1 expression through the mTOR pathway in 3T3-L1 adipocytes. Biochem. J. 411:647–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zimprich A, et al. 2011. A mutation in VPS35, encoding a subunit of the retromer complex, causes late-onset Parkinson disease. Am. J. Hum. Genet. 89:168–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.