Abstract
The TFAP2C transcription factor has been shown to downregulate transcription of the universal cell cycle inhibitor p21cip (CDKN1A). In examining the mechanism of TFAP2C-mediated repression, we have identified a ternary complex at the proximal promoter containing TFAP2C, the oncoprotein Myc, and the trimethylated lysine 4 of histone H3 (H3K4me3) demethylase, KDM5B. We demonstrated that while TFAP2C and Myc can downregulate the CDKN1A promoter independently, KDM5B acts as a corepressor dependent on the other two proteins. All three factors collaborate for optimal CDKN1A repression, which requires the AP-2 binding site at −111/−103 and KDM5B demethylase activity. Silencing of TFAP2C-KDM5B-Myc led to increased H3K4me3 at the endogenous promoter and full induction of CDKN1A expression. Coimmunoprecipitation assays showed that TFAP2C and Myc associate with distinct domains of KDM5B and the TFAP2C C-terminal 270 amino acids (aa) are required for Myc and KDM5B interaction. Overexpression of all three proteins resulted in forced S-phase entry and attenuation of checkpoint activation, even in the presence of chemotherapy drugs. Since each protein has been linked to poor prognosis in breast cancer, our findings suggest that the TFAP2C-Myc-KDM5B complex promotes cell cycle progression via direct CDKN1A repression, thereby contributing to tumorigenesis and therapy failure.
INTRODUCTION
The activation factor 2 (TFAP2) family consists of five homologous, developmentally regulated transcription factors, TFAP2A to -E, each encoded by a separate gene. Structurally, TFAP2 proteins contain a highly conserved C-terminal helix-span-helix motif required for dimerization, a basic DNA binding domain, and a third, less-conserved region toward the N terminus which contains a proline and glutamine-rich activation domain. These factors have been shown to bind to palindromic GC-rich DNA recognition sequences as either homo- or heterodimers and act as transcriptional activators or repressors in a promoter-specific manner (9).
Although studies in knockout mice (reviewed in reference 9) and of phenotypically related inherited human traits (25, 32) have shown that these factors have important functions during embryogenesis, they are minimally expressed in most adult tissues. However, expression of the TFAP2A and TFAP2C proteins has been demonstrated in a variety of solid tumors, including breast cancer and melanoma (reviewed in reference 24). TFAP2A expression in breast tumors has been associated with a favorable outcome and shows a positive correlation with expression of estrogen recepter α (ERα) and CDKN1A (14), while elevated expression of TFAP2C has generally been correlated with an adverse phenotype and resistance to hormone therapy (13, 16). Studies in vitro have shown that direct AP-2 transcriptional targets include many genes involved in tumor progression. Of particular interest is the regulation of the universal cell cycle inhibitor p21cip (CDKN1A), which can be activated through p53-dependent and -independent pathways to mediate cell cycle arrest (1). AP-2 factors bind a specific sequence in the proximal region of the promoter, and while TFAP2A positively regulates CDKN1A expression (27, 41), TFAP2C has been shown to repress its expression in breast cancer cells (36). These opposing activities at the CDKN1A locus may contribute to the contrasting phenotypes associated with tumors expressing these two AP-2 factors.
When mediating transcriptional activation, it has been shown that AP-2 factors associate with the CITED family of adapter proteins (Cited 2 and 4), which in turn recruit the histone acetyltransferases (HATs) CBP/p300 (2, 7). To date, no clear AP-2 corepressor proteins have been identified, although modification through sumoylation may be required for repressor activity (4, 10). TFAP2A has also been reported to interact with other nuclear factors, including Myc, pRB, and p53 (reviewed in reference 9), to regulate the transcription of target genes. The proto-oncogene MYC controls cellular growth, differentiation, and apoptosis, and its deregulation contributes to the development of a variety of cancers, including breast cancer. It encodes a basic region-helix-loop-helix-zipper (bHLHZ) transcription factor which forms a specific heterodimer with the small bHLHZ protein, Max. Myc-Max heterodimers bind with high affinity to the palindromic DNA sequence CACGTG (E box) but can also bind several other related sequences, leading to interaction at up to 15% of all promoters. While the genes activated by Myc include those required for cell growth and cell cycle progression, most of the genes it downregulates are involved in cell cycle arrest, and they notably include p21cip (CDKN1A) (11, 17). As with other activators of transcription, Myc is able to recruit HAT-containing coactivator complexes to its target genes (reviewed in reference 8), but the precise mechanism behind Myc-mediated gene repression is still disputed: Myc interaction occurs close to the transcriptional start site (TSS), but direct DNA binding, at least at the CDKN1A gene, is not required (12, 17).
The reported colocalization of TFAP2C and Myc at the proximal promoter of p21cip (CDKN1A) prompted us to investigate the possibility of a functional link between these two transcription factors in the repression of this gene in breast tumor cells. Moreover, recent studies have suggested that Myc forms a complex with the transcriptional corepressor KDM5B (lysine-specific demethylase 5B; also known as Jarid1B/PLU-1). Like TFAP2C, the KDM5B protein is overexpressed in breast cancer lines and tumors (22). Its proven ability to demethylate H3K4me3 (trimethylated lysine 4 of histone H3), an epigenetic marker particularly associated with the TSS of active promoters, is consistent with the transcriptional repressor role associated with this protein, and it has been proposed that KDM5B may promote tumorigenesis by downregulating expression of oncosuppressor genes, such as BRCA1 and HOXA5 (26, 39). In this work, we have investigated whether KDM5B regulates the expression of CDKN1A in cooperation with the TFAP2C and Myc proteins. Our data show that TFAP2C, Myc, and KDM5B form a functional protein complex in the vicinity of the TSS of the CDKN1A gene and corepress its expression in proliferating and drug-treated breast cancer cells, thus promoting cell cycle progression.
MATERIALS AND METHODS
Cell culture, transient transfection, and antibodies.
MCF-7, HepG2, and H1299 cells (ATCC) were grown in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum in 10% CO2 at 37°C. The shTFAP2C-MCF-7 line (36) was induced using 1 μg/ml doxycycline (Sigma). Transfection reagents were Genejuice (Merck) for plasmid DNA and Interferin (Polyplus) for small interfering RNA (siRNA), used according to the manufacturer's directions. Subconfluent MCF-7 or shTFAP2C-MCF-7 cells were transiently transfected with 20 nM nonsilencing control siRNA (no. 1022076; Qiagen) or with validated siRNA targeting KDM5B (J-009899-06; Darmacon) or Myc (6341; Cell Signaling). The following antibodies for Western blotting (Santa Cruz unless stated otherwise) were used: KDM5B (3273; Cell Signaling), Myc tag (2276; Cell Signaling), p53 (p53-DO-1, sc-126), TFAP2C (6E4, sc-53162), p21cip (2946; Cell Signaling), Myc (sc-788), β-actin (C-2, sc-8432), lamin A/C (4477; Cell Signaling), green fluorescent protein (GFP) (sc-8334), and Hsc70 (sc7298). Horseradish peroxidase-conjugated secondary antibodies were from DakoCytomation.
Plasmids and reporter assays.
The pcDNA3.1-based TFAP2C and KDM5B expression plasmids, the pGL-3-CDKN1A luciferase reporter, and pGL-3-Mut4/Mut5 constructs have been previously described (26, 27). The fusion protein GFP-TFAP2C was generated by cloning the TFAP2C cDNA sequence in frame into the pEGFP-C1 vector (Clontech). The C-terminal deletion mutant (ΔC) was truncated at the internal SalI restriction site, retaining amino acids 1 to 180. The N-terminal deletion mutant (ΔN) fused sequences C-terminal of the SalI site (amino acids 181 to 450) to EGFP and retains the basic region and the helix-span-helix dimerization domains (5). To ensure efficient nuclear translocation of all fusion proteins, an oligonucleotide (5′-GA TCT CCA AAA AAG AAG AGA AAG GTT CA-3′), encoding a recognized nuclear localization signal (15), was ligated in frame between the GFP and wild-type (wt) or truncated TFAP2C sequences. The KDM5B deletion mutant constructs were as described previously (38). The pcDNA3-Myc expression plasmid was from Addgene. Reporter assays were controlled by cotransfection with the pSV-β-galactosidase control vector (Promega). Cells were harvested after 48 h and processed for Western blotting or assayed for luciferase and β-galactosidase activity (Promega).
RNA analysis.
Total RNA was extracted from subconfluent cells (RNeasy minikit; Qiagen), and 500 ng was reverse transcribed using the high-capacity reverse transcription kit (Applied Biosystems). The cDNA was analyzed by quantitative real-time PCR (q-RT-PCR) using TaqMan reaction mix and the StepOne real-time PCR system (Applied Biosystems) according to the manufacturer's instructions. TaqMan primer/probe sets were used to analyze the expression of TFAP2C (Hs00231476_m1), CDKN1A (Hs00355782_m1), KDM5B (Hs00366783_m1) and Myc (Hs0095030_m1), while a glyceraldehyde-3-phosphate dehydrogenase (GAPDH) TaqMan probe (no. 402869) was used as an internal control (Applied Biosystems). Analysis of q-RT-PCR data was carried out using the comparative ΔΔCT method. For each sample, the intensity of the amplimer was normalized against that of the GAPDH control, and the data were depicted as fold changes (2−ΔΔCT) with respect to the RNA level observed in the control experiments.
ChIP assay.
Subconfluent MCF-7 or shTFAP2C-MCF-7 cells were cross-linked by addition of formaldehyde directly to the culture medium to a final concentration of 1% for 10 min and assayed using the ChIP-IT Express chromatin immunoprecipitation (ChIP) kit (Active Motif). Chromatin was sheared to an average size of 500 bp using a Sonics Vibracell VCX500 sonicator and immunoprecipitated with ChIP-validated antibodies against KDM5B (3), TFAP2C (H77, sc-8977; Santa Cruz), Myc (sc-764), histone H3 (ab1791; Abcam), monomethylated lysine 4 of histone H3 (H3K4me1) (ab8895; Abcam), dimethylated lysine 4 of histone H3 (H3K4me2) (ab7766; Abcam) or H3K4me3 (ab8580; Abcam), and 2 μg purified rabbit IgG (sc2027). Quantification of immunoprecipitated DNA was carried out in triplicate using the StepOne real-time PCR system with SYBR green PCR master mix (Applied Biosystems). Primer sequences and calculation of values as fold enrichment compared with results for the IgG control were as detailed previously (36).
Coimmunoprecipitation assays.
Immunoprecipitation was carried out according to the manufacturer's protocol using the Universal Magnetic coimmunoprecipitation kit (Active Motif). Cells were lysed in nuclear extraction buffer supplemented with 5 μM trichostatin A, 1 μM phenylmethylsulfonyl fluoride (Sigma), and protease inhibitor cocktail (Roche). Nuclear extract (0.5 mg) was incubated with 50 μl of protein G-coated magnetic beads and 2 μg of either KDM5B (ab27689; Abcam), TFAP2C (6E4, H77 or AP25H01, sc-81181), Myc (sc-788), Myc tag (no. 2276; Cell Signaling), or control IgG (sc-2027) antibody overnight at 4°C. Immunoprecipitates were washed three times with the supplied wash buffer for 5 min at 4°C and resuspended in 45 μl of 2× SDS Western loading buffer. For GFP-tagged proteins, immunoprecipitation was carried out using the GFP-Trap kit (ChromoTek). Samples were resolved via SDS-PAGE and transferred onto polyvinylidene difluoride (PVDF) membranes for Western blot analysis. Additional Western blots established that the ΔC mutant is recognized only by the H77 antibody while the ΔN mutant interacts with all three TFAP2C antibodies.
Cell cycle analysis.
Asynchronous HepG2 cells were transiently transfected with expression vectors and treated with 1 mM hydroxyurea, 6 nM vinblastine, or vehicle alone (control). After incubation at 37°C for 16 h, cells were trypsinized, washed in phosphate-buffered saline, and fixed using ice-cold 70% ethanol. Fixed cells were incubated with 50 μl of 100 μg/ml RNase and stained with 300 μl of 50-μg/ml propidium iodide (Invitrogen) for 30 min at room temperature. Flow cytometry and cell cycle analysis were performed as previously described (36).
RESULTS
CDKN1A repression by TFAP2C, KDM5B, and Myc.
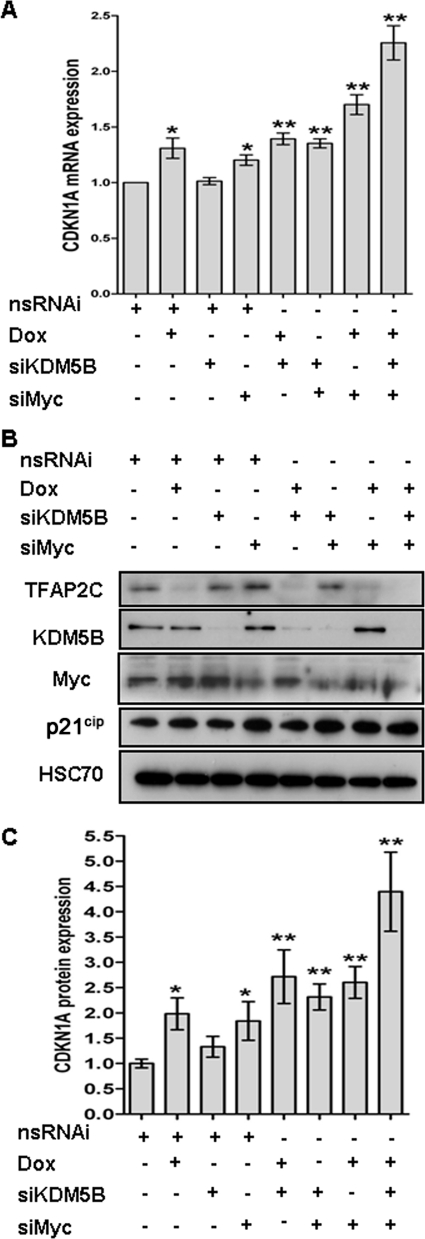
To study the relative activities of TFAP2C, KDM5B, and Myc in the repression of CDKN1A, we used the estrogen receptor-positive (ER+) MCF-7 breast cancer line, which expresses all three factors. We have previously described an MCF-7 derivative line with doxycycline-inducible short hairpin RNA (shRNA) targeting TFAP2C (shTFAP2C cells); induction of TFAP2C silencing leads to upregulation of CDKN1A and p53-independent G1/S arrest (36). Here, transient transfection of this line with validated specific siRNA against Myc and KDM5B allowed us to examine the expression level of the endogenous CDKN1A transcript and protein (Fig. 1A and B) after silencing each factor either individually or in combination. TFAP2C and Myc silencing, alone or together, induced expression of the CDKN1A transcript, while knockdown of KDM5B alone or with either TFAP2C or Myc had little effect. However, silencing all three proteins significantly increased CDKN1A mRNA induction (Fig. 1A, compare last 2 columns), indicating that KDM5B can collaborate with the two transcription factors to repress CDKN1A transcription in cycling cells. We observed a similar pattern of induction at the protein level (Fig. 1B and C).
Fig 1.
TFAP2C, Myc, and KDM5B collaborate to repress CDKN1A expression. shTFAP2C-MCF-7 cells were transiently transfected with the indicated siRNA and/or treated with doxycycline (Dox) to induce TFAP2C silencing. All samples were additionally transfected with a nonsilencing control (nsRNAi); the amount was adjusted to keep the total level of siRNA constant across all samples. Cells were harvested 72 h after addition of siRNA/Dox. (A) Total RNA was analyzed by quantitative PCR (qPCR) and normalized to GAPDH. CDKN1A data are represented as fold change (2−ΔΔCT) from results for control cells (first column), averaged from 3 independent experiments ± standard error. *, P < 0.05; **, P < 0.01 (Student's t test). TFAP2C, Myc, and KDM5B silencing was confirmed by qPCR (not shown). Whole-cell lysates from each experiment were analyzed by Western blotting to monitor changes in protein levels; Hsc70 was used as a loading control. One experiment is shown in panel B; in panel C, the p21cip (CDKN1A) blots from all the experiments were scanned and normalized to the Hsc70 signal, and each condition is expressed as fold induction over control results (first column), as in panel A.
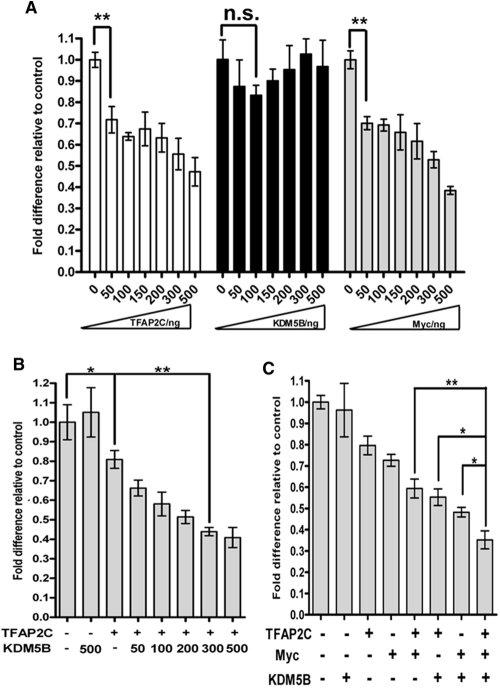
Reporter assays were used to confirm that CDKN1A repression by these factors occurs at the promoter level. Initially, HepG2 cells (which do not express AP-2 factors or KDM5B) were transiently transfected with a luciferase reporter construct containing the CDKN1A promoter (−2325/+8), and escalating amounts of an expression vector for each test factor individually. As shown previously, independent overexpression of either TFAP2C or Myc repressed CDKN1A promoter activity in a dose-dependent manner; however, addition of KDM5B alone did not significantly alter reporter activity (Fig. 2A). In contrast, when a constant, suboptimal dose of the TFAP2C construct was cotransfected with increasing amounts of the KDM5B expression construct, KDM5B corepressor activity was observed, since reporter activity declined as the dose of KDM5B plasmid increased (Fig. 2B). A similar result was also seen with a constant level of the Myc expression construct (data not shown). Moreover, by optimizing transfection input levels for all the constructs, we could demonstrate that coexpression of all three proteins significantly further repressed CDKN1A promoter activity (Fig. 2C), thus mirroring the induction of endogenous CDKN1A mRNA observed after cosilencing TFAP2C, KDM5B, and Myc in MCF-7 cells (Fig. 1A).
Fig 2.
TFAP2C, Myc, and KDM5B corepress CDKN1A promoter activity. (A) HepG2 cells were transfected with a CDKN1A-luc reporter construct (containing genomic sequences from −2325 to +8 relative to the TSS; 150 ng) plus increasing amounts of TFAP2C, KDM5B, or Myc expression vectors as indicated. (B) As in panel A, with TFAP2C fixed (50 ng) and/or KDM5B (escalating dose; ng) where indicated. (C) As in panel B, with fixed levels of TFAP2C (50 ng), KDM5B (300 ng), and/or Myc (50 ng) expression constructs as indicated. Luciferase values were corrected for transfection efficiency and are expressed as fold activity compared to the vector-alone (VA) control (first column), set at 1, using data from 3 independent experiments, each performed in triplicate; error bars indicate the standard errors. Student's t test was used to compare data from the indicated samples: *, P < 0.05; **, P < 0.001. n.s., not significant.
TFAP2C, Myc, and KDM5B form a nuclear complex in MCF-7 cells.
To address if negative regulation of CDKN1A by TFAP2C, Myc, and KDM5B occurs through complex formation between these factors, we first used confocal microscopy to show that all three proteins colocalize in the nuclear compartment of MCF-7 cells, and TFAP2C silencing did not alter the localization of KDM5B or Myc (see Fig. S1 in the supplemental material).
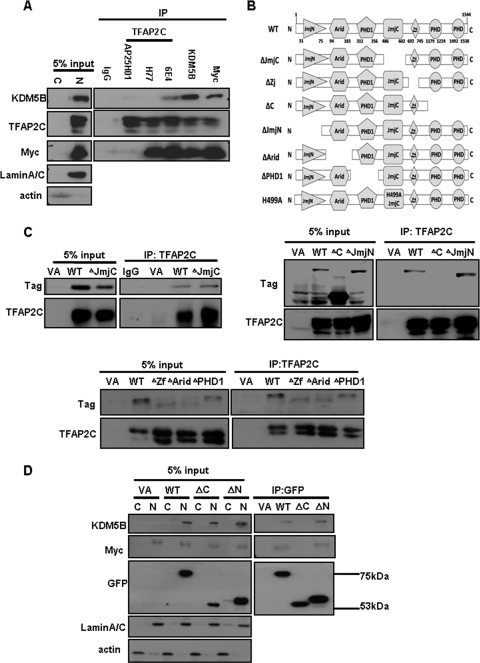
Next, we looked for physical interaction between these factors, examining endogenous proteins present in MCF-7 nuclear extracts by coimmunoprecipitation (coIP) assays. As seen in Fig. 3A, Myc and TFAP2C were both immunoprecipitated using an antibody specific for KDM5B (lane 5), and TFAP2C and KDM5B were also both detected in anti-Myc immunoprecipitates (lane 6). In order to detect endogenous KDM5B or Myc in TFAP2C immunoprecipitates, we tested antibodies specific for different regions along the 450-amino-acid (aa) protein. The H77 polyclonal antibody was raised against a peptide corresponding to amino acids 145 to 221; 6E4 recognizes an epitope within the C-terminal 20 aa (13), while AP25H01 was raised against a recombinant protein corresponding to amino acids 50 to 200 of TFAP2C. Interestingly, Myc was coprecipitated with both H77 and 6E4, while KDM5B was precipitated only with 6E4 (Fig. 3A, compare lanes 3 and 4). Although AP25H01 efficiently precipitated TFAP2C, it did not bring down either KDM5B or Myc (lane 2), suggesting that the epitope recognized by this antibody is not accessible when TFAP2C interacts with KDM5B and/or Myc. These assays confirm that endogenous TFAP2C, Myc, and KDM5B interact physically with each other to form a complex.
Fig 3.
KDM5B forms a complex with TFAP2C and Myc. (A) Cytoplasmic (C) and nuclear (N) extracts were prepared from MCF-7 cells (see Materials and Methods). The nuclear extracts were subjected to immunoprecipitation (IP) with antibodies against Myc, KDM5B, or TFAP2C (6E4, H77, or AP25H01, as marked) prior to Western blotting (WB) for the indicated proteins (lanes 1 to 6). C and N proteins, equivalent to 5% of the amount used for IP, were also analyzed by WB to control for protein size and extract integrity. (B) Schematic representation of the domain structure of KDM5B and the series of peptide-tagged deletion mutants. (C) H1299 cells (lacking TFAP2C and KDM5B expression) were cotransfected with TFAP2C and tagged wt or mutant KDM5B proteins or vector alone (VA) as a control. Nuclear extracts were subjected to IP for TFAP2C (6E4) followed by WB for TFAP2C or the peptide Tag to reveal mutant KDM5B proteins, as indicated. (D) GFP-tagged TFAP2C wild-type (WT) or a ΔN or ΔC deletion mutant was cotransfected with KDM5B, and nuclear extracts were immunoprecipitated using the GFP-trap system prior to WB to detect each protein plus endogenously expressed Myc. Additional blotting for actin and lamin A/C was used to confirm the integrity of the cytoplasmic and nuclear extracts, respectively; the 6E4 antibody was used for all TFAP2C blots.
Protein domains required for interaction and KDM5B corepressor activity.
KDM5B is a 1,544-aa protein which comprises several conserved domains, including an ARID (A/T-rich interacting domain) domain that, in the case of KDM5 family members, binds GC-rich DNA sequences (26, 34), a C5HC2 zinc finger (Zf), three plant homeobox (PHD) domains, a Jumonji N (JmjN) domain, and the Jumonji C (JmjC) demethylase catalytic domain (Fig. 3B) (38). Previous studies of the KDM5B Drosophila melanogaster ortholog, Lid, and mammalian KDM5A/B established that Myc can interact independently with both the JmjC and Zf domains (20, 28). To determine which KDM5B domains mediate interaction with TFAP2C, we expressed wild-type TFAP2C together with a series of tagged KDM5B deletion mutants (Fig. 3B) (38) in H1299 cells, which do not express either factor. After 48 h, nuclear lysates were prepared for coIP with the TFAP2C antibody, 6E4. Proteins lacking the JmjN, Arid, JmjC, Zf, or PHD1 domain still bound TFAP2C, while removal of the two C-terminal PHD domains abolished TFAP2C interaction (Fig. 3C, mutant ΔC). Thus, distinct KDM5B domains are required for its interaction with TFAP2C and Myc.
For the reciprocal experiment, GFP fusion proteins with full-length or truncated TFAP2C sequences were generated (see Materials and Methods): deletion of the C-terminal region (ΔC) retained the proline-rich activation domain, while ΔN retained just 40 amino acids of N-terminal sequence plus the C-terminal basic DNA binding and helix-span-helix dimerization domains. Each fusion protein was expressed in H1299 cells, together with full-length, tagged KDM5B, and nuclear extracts were assayed for interacting proteins using the GFP-trap system. As shown in Fig. 3D, only the full-length and ΔN fusion proteins bound KDM5B and endogenously expressed Myc. Assessing this finding together with the coIP data with TFAP2C antibodies (Fig. 3A) suggests that the basic-helix-span-helix domains and the 40 amino acids immediately N-terminal of this region are necessary for TFAP2C interaction with both Myc and KDM5B.
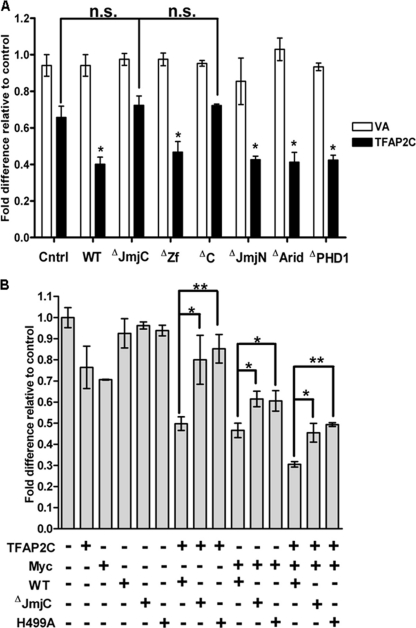
To identify the KDM5B domains required for corepressor function, we transiently transfected HepG2 cells with the (−2325/+8) CDKN1A luciferase reporter construct and the series of KDM5B deletion mutants (Fig. 3B) either with or without a TFAP2C expression vector. As before, expression of KDM5B alone had no effect on reporter activity but was able to cooperate with TFAP2C to repress the CDKN1A promoter (Fig. 4A, compare first and second black columns). In this assay, the majority of the KDM5B mutants also showed significant, wt levels of corepression activity (denoted by asterisks; Fig. 4A). The exceptions were constructs lacking either the C-terminal PHD domains (ΔC) or the JmjC domain (ΔJmjC) (Fig. 4A; nonsignificant [n.s.] difference from the control). Since it was unclear whether the demethylase activity itself or the physical requirement of the JmjC domain for protein-protein interactions was essential for corepressor activity, we additionally tested a point mutation, H499A, known to abrogate demethylase activity (38). Compared to wt KDM5B, the H499A mutant and the JmjC mutant both showed significantly reduced corepressor activity when expressed with either TFAP2C or Myc or both factors (Fig. 4B). Together these data suggest that KDM5B-mediated repression at the CDKN1A promoter requires its functional interaction with TFAP2C/Myc proteins plus an intact, functional JmjC domain.
Fig 4.
Identification of KDM5B domains required for corepressor activity. (A) HepG2 cells were cotransfected with 150 ng of CDKN1A-luc and 50 ng TFAP2C expression plasmid or vector-alone (VA) control, as indicated, plus 300 ng of wt or mutant KDM5B, as labeled. (B) As in panel A, with TFAP2C and/or Myc (50 ng) in combination with wt or KDM5B JmjC domain mutants (see Fig. 3B), as indicated. Efficient expression of all KDM5B constructs was assessed by blotting (see Fig. S2A in the supplemental material). Luciferase values, corrected for transfection efficiency, are expressed as fold activity compared to that for the vector-alone control, set at 1, using data from 3 independent experiments, each performed in triplicate; error bars indicate the standard errors. Student's t test was used to compare data from the indicated samples: n.s., not significant; *, P < 0.05; **, P < 0.001.
AP-2 binding at the CDKN1A promoter is critical for repression by the TFAP2C/Myc/KDM5B ternary complex.
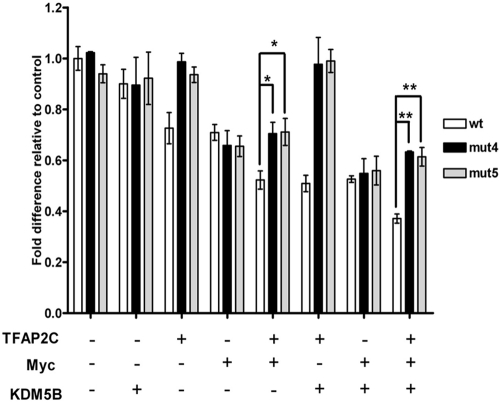
We have previously identified an AP-2 DNA binding site (GCCCCGGGG) at −111/−103 in the CDKN1A promoter (27). In contrast, there is no consensus Myc binding site within the proximal promoter, and it is thought that Myc is recruited to the locus by “piggy-backing” on other factors, such as Miz-1 or Sp1, that bind to sites within 100 bp of the CDKN1A TSS (17, 29). To investigate the importance of the AP-2 binding site for repression by the TFAP2C/Myc/KDM5B complex, we performed further reporter assays using additional CDKN1A luciferase constructs mutated to abolish AP-2 binding (mut4 and mut5) (27). Mutation of the AP-2 binding site clearly abrogated repression by TFAP2C alone and with KDM5B (Fig. 5, column groups 3 and 6). Myc-mediated repression was less affected by mutation of the AP-2 binding site (Fig. 5, column groups 4 and 7), but intriguingly, the degree of repression was significantly attenuated on mutant promoters when Myc was coexpressed with TFAP2C, either alone or with KDM5B (Fig. 5, column groups 5 and 8). This suggests, therefore, that while Myc does not require an intact AP-2 site to interact with and repress the CDKN1A locus, when TFAP2C is present, its binding at −111/−103 allows optimal recruitment of Myc and KDM5B to the proximal promoter and more-efficient repression.
Fig 5.
The AP-2 binding site at −111/−103 is required for optimal CDKN1A repression by the ternary complex. HepG2 cells were transfected, as described for Fig. 5, with the wt or versions of the CDKN1A-luc reporter construct mutated within the AP-2 binding site (mut4 and mut5) (27), plus expression constructs for TFAP2C, Myc, and KDM5B, as indicated. Luciferase values, corrected for transfection efficiency, are expressed as fold activity compared to that for the vector-alone control, set at 1, using data from 3 independent experiments, each performed in triplicate; error bars indicate the standard errors. Student's t test was used to compare data from the indicated samples; *, P < 0.05; **, P < 0.001.
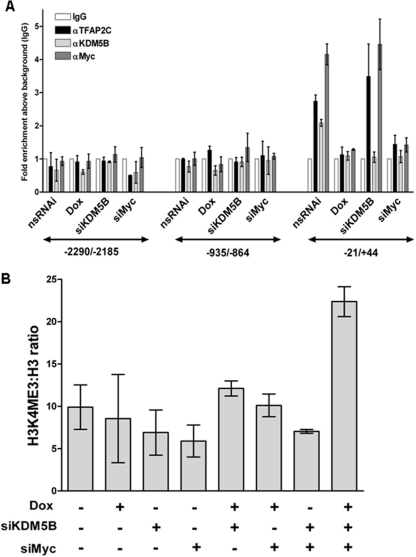
We next used ChIP assays to verify the colocalization of TFAP2C, Myc, and KDM5B at the endogenous CDKN1A locus. In agreement with results of the reporter assays described above, the proximal region of the CDKN1A promoter (primers at −21/+44), but not regions further upstream (−2290/−2185 or −935/−864), was immunoprecipitated specifically using antibodies to TFAP2C, KDM5B, or Myc (Fig. 6A). Functional interrelationships between these factors at the CDKN1A locus were tested by comparing ChIP assays with control cells and those silenced for each factor in turn. Treatment with doxycycline for 72 h reduced TFAP2C binding, as expected; however, the localization of Myc and KDM5B at the promoter was also reduced to background levels in TFAP2C-silenced cells (Fig. 6A). Similar results were obtained when cells were treated for 24 or 120 h (data not shown). These data cannot be explained by changes in the Myc or KDM5B protein level, since these remained constant in doxycycline-treated cells (Fig. 1B). siRNA-mediated silencing of Myc also reduced binding of both TFAP2C and KDM5B to the CDKN1A proximal promoter, while knockdown of KDM5B did not influence interaction by the other two factors, although KDM5B localization to the promoter was reduced as expected (Fig. 6A). Therefore, not only does TFAP2C assist the binding of Myc to this locus, as suggested by the reporter assays with mutated AP-2 binding sites (Fig. 5), but Myc also facilitates TFAP2C binding.
Fig 6.
Colocalization and functional interrelation between endogenous TFAP2C, Myc, and KDM5B at the CDKN1A locus. (A) shTFAP2C-MCF-7 cells were transfected with nonsilencing control siRNA (nsRNAi) or siRNA against Myc (siMyc) or KDM5B (siKDM5B) or treated with doxycycline (Dox) to induce shTFAP2C and harvested 72 h later for use in ChIP with antibodies against IgG (control), TFAP2C, (KDM5B, or Myc. Precipitates were analyzed by qPCR using primer pairs specific to 3 distinct regions (−2290/−2185, −935/−864, and −21/+44) across the CDKN1A 5′ sequence, as indicated by the double-headed arrows. (B) ChIP was performed as described for panel A using antibodies specific for histone 3 and H3K4-me3. qPCRs were performed using the −21/+44 CDKN1A proximal promoter-specific primers. Results are shown as fold enrichment above the IgG (background) control (A) or as an H3K4me3/H3 ratio (B). Data were averaged from three independent experiments, ± standard errors.
Since KDM5B is a specific di- and trimethyl-H3K4 demethylase (39), we tested whether silencing of TFAP2C or MYC, with the resulting reduction in KDM5B localization at the CDKN1A locus, would lead to changes in H3K4 methylation status within the proximal promoter. Silencing individual factors or pairs of factors did not significantly alter the H3K4 methylation level; in particular, loss of KDM5B alone did not promote H3K4 trimethylation, since this would require the recruitment of a COMPASS-like methyltransferase complex, which can occur only during transcriptional activation (31). However, when all three factors were cosilenced (a situation that does lead to CDKN1A activation [Fig. 1]), we observed a robust increase in the H3K4me3 signal (Fig. 6B). In contrast, levels of H3K4me1 did not alter significantly, while there was a general decline in the H3K4me2 signal (data not shown). Taken together, these results demonstrate that TFAP2C and Myc, but not KDM5B, are required for the localization and stabilization of the complex at the CDKN1A proximal promoter. Furthermore, while TFAP2C and Myc can downregulate CDKN1A, the demethylase activity of KDM5B is required for maximal repression of both transiently expressed reporter constructs and the endogenous gene, and coordinate loss of all three factors led to increased H3K4me3 levels, in keeping with the activation of CDKN1A expression (Fig. 1A).
Effect of overexpression of the TFAP2C-Myc-KDM5B complex on cell cycle progression and checkpoint activation.
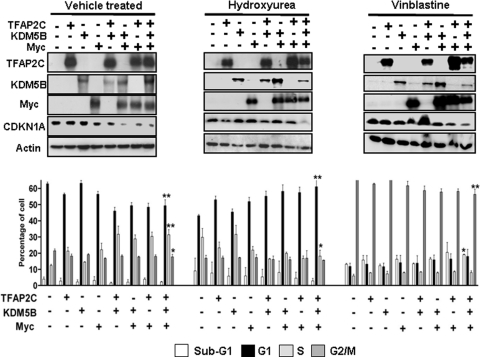
Since p21cip (CDKN1A) functions mainly as a regulator of cell cycle progression by inhibiting CDK activity and is required for activation of G1/S and G2/M checkpoints when cells are exposed to DNA damage or replication or mitotic stress, we questioned whether manipulating levels of TFAP2C, Myc, and/or KDM5B could affect cell cycle distribution and activation of specific checkpoints. HepG2 cells, which express wild-type CDKN1A, were transiently transfected with the TFAP2C, Myc, and/or KDM5B expression vectors and 48 h later were subjected to cell cycle analysis. Western blotting was used to confirm expression of exogenous proteins. Overexpression of KDM5B alone had little effect on CDKN1A expression but enhanced repression when cells were cotransfected with TFAP2C and/or Myc (Fig. 7, top left panel, vehicle treated), consistent with the reporter assays described above (Fig. 2). Cell cycle distribution analysis showed that transient overexpression of TFAP2C or Myc but particularly both proteins together forced a higher percentage of cells into S phase. Overexpression of KDM5B alone had a minimal effect on cell cycle distribution; however, cotransfection with TFAP2C and/or Myc led to a significant shift from G1 to S phase (Fig. 7, lower left panel).
Fig 7.
TFAP2C/KDM5B/MYC overexpression attenuates drug-activated checkpoints and forces cells to enter S phase. HepG2 cells were transfected with TFAP2C, KDM5B, or Myc expression vectors in combination, as shown. After 24 h, transfected cells were treated with normal medium (vehicle treated, LH panels), hydroxyurea (middle panels), or vinblastine (RH panels) for a further 16 h. Cells were then either lysed and assayed by Western blotting for the proteins indicated (upper panels) or ethanol fixed and stained with propidium iodide for cell cycle analysis (lower panels). Graphs show percentages of cells from each condition in sub-G1, G1, S, or G2/M phase. Representative experiments from 3 repeats (each done in triplicate) are shown; *, P < 0.05; **, P < 0.01 (Student's t test; significant differences between control [1st group of columns] and 3-way-transfected cells [last group of columns] for cycling [vehicle treated] or drug-treated cells).
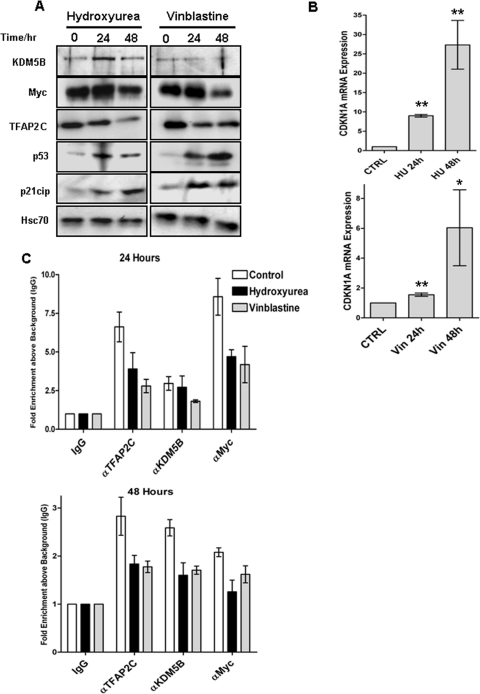
Since overexpression of the TFAP2C-Myc-KDM5B complex induced G1/S transition in cycling cells, we next investigated whether the complex could also affect activation of CDKN1A at the S-phase or G2/M checkpoints induced by treatment with hydroxyurea (HU) or vinblastine, respectively. We first assayed the effects on endogenous protein expression after drug treatment for 24 and 48 h in MCF-7 cells. Induction of CDKN1A mRNA and protein was observed with both drugs in concert with upregulation of p53 levels (Fig. 8A and B). Expression of TFAP2C and Myc was generally downregulated following treatment with vinblastine or HU, both at the mRNA and protein levels (Fig. 8A and data not shown), although levels of KDM5B remained more constant. The binding of each protein to the proximal region of the CDKN1A promoter was assessed by ChIP assays with drug-treated cells and showed that treatment for 24 or 48 h reduced binding of all three factors to the CDKN1A locus (Fig. 8C). This demonstrates that drug activation of the endogenous CDKN1A gene was accompanied by reduced expression of Myc and TFAP2C and coordinate release of these transcriptional repressors from the proximal promoter.
Fig 8.
HU and vinblastine induce CDKN1A and reduce TFAP2C/KDM5B/Myc occupancy at the proximal promoter. MCF-7 cells were treated with hydroxyurea (HU) or vinblastine (Vin) or vehicle control (CTRL) for 24 and 48 h. (A) Whole-cell extracts from treated or control cells, blotted for the indicated proteins. (B) q-RT-PCR analysis of CDKN1A expression in control and treated cells. Data represent the means and standard errors for 3 independent experiments. Significant differences (*, P < 0.05; **, P,0.01) between induced and uninduced cells are given (Student's t test). (C) ChIP assay for TFAP2C, KDM5B, and Myc factors was performed as for Fig. 6 using primers to the −21/+44 region of CDKN1A. Values are averages for three independent ChIP experiments; error bars represent standard errors between the repeats.
Since drug induction of CDKN1A was accompanied by loss of the TFAP2C-Myc-KDM5B complex, we investigated whether forced overexpression of these proteins could repress CDKN1A expression and thereby attenuate the cytostatic effect induced by either HU or vinblastine treatment. Arrest in S or G2/M phase was clearly seen after cell cycle analysis of control cells treated with HU or vinblastine, respectively (Fig. 7, bottom panels, compare the first group of columns for each condition). However, overexpression of TFAP2C and/or Myc decreased the extent of the S or G2/M block, allowing a proportion of cells to reenter the cycle. Overexpression of KDM5B alone did not affect HU- or vinblastine-induced checkpoints, but KDM5B was able to cooperate with either TFAP2C or Myc to relieve the cell cycle block, with the most significant effects being observed when all three proteins were expressed together, particularly for HU-treated cells, where the cell cycle profiles resembled those of untreated cells (Fig. 7, compare the last series of columns for the HU-treated condition with the first series of columns for the control condition). For each treatment, repression of p21cip expression was maximal when all three proteins were coexpressed (Fig. 7, upper panels, last lane).
In summary, since its overexpression is sufficient to induce entry into S phase of the cell cycle and activate proliferation of normal cycling cells, TFAP2C resembles Myc in these assays and acts as an oncogene. KDM5B has little effect alone but can amplify responses to the other two factors. Activation of S or G2/M checkpoints is attenuated when TFAP2C or Myc is overexpressed, with and without KDM5B overexpression, suggesting that elevated levels of these proteins in cancer cells may collaborate to confer resistance to genotoxic or mitotic stress via direct downregulation of the universal cell cycle inhibitor CDKN1A.
DISCUSSION
As with TFAP2C (13, 16) and Myc (37), KDM5B overexpression has been associated with a poorer outcome in breast cancer patients (22), and all three proteins have been shown to promote cell proliferation by facilitating G1/S transition (36, 39), a checkpoint particularly sensitive to cellular levels of p21cip. Repression of CDKN1A, mediated by Myc or TFAP2C, has been documented previously (see the introduction), and the related KDM5A demethylase was shown to regulate H3K4 methylation status at this locus in gastric tumor cells, although its site and mode of binding to the promoter were not defined (40). Our data suggest that in breast tumor cells, where it is expressed predominantly (19), KDM5B is the key H3K4me3 demethylase which acts in a ternary complex with Myc and TFAP2C to regulate CDKN1A expression, thus uniting many separate observations and providing a mechanistic rationale for how deregulated expression of these factors may permit breast cancer cells to sustain their proliferation and survival.
Since Myc repression of CDKN1A does not involve heterodimerization with Max or direct DNA binding (12), a number of studies have looked at the requirement of Myc to access the promoter through interaction with other DNA binding proteins, particularly Sp1 and Miz-1. The precise mechanism remains unclear, possibly because more than one pathway is active, with cell type-specific factors undoubtedly playing a role (reviewed in reference 17). Here we have explored the interdependence of Myc and TFAP2C. Investigating the binding of these proteins at the endogenous CDKN1A gene in silenced MCF-7 cells by ChIP (Fig. 6) revealed that each factor appeared to stabilize and/or facilitate the binding of the other to the proximal promoter. This was partly recreated using reporter assays where maximal repression by all three factors could be achieved only on promoter constructs with an intact AP-2 binding site (Fig. 5). Taking the ChIP and reporter assay data together, therefore, this suggests that TFAP2C and Myc interact with the CDKN1A locus at separate sites, with only the AP-2 site at −111/−103 for TFAP2C defined, and together they recruit the histone demethylase KDM5B to form a repressive ternary complex required for optimal downregulation of CDKN1A (Fig. 9, diagram). Interestingly, a similar mutual interdependence has recently been suggested to allow the stable binding of both TFAP2C and FoxA1 to specific chromatin sites, leading to the recruitment of ERα and estrogen-responsive gene expression in breast cell lines (33).
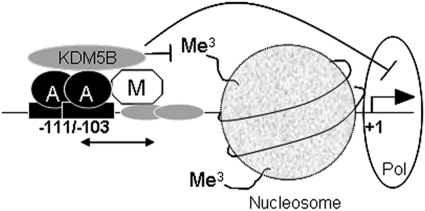
Fig 9.
Schematic diagram of CDKN1A repression by TFAP2C/Myc/KDM5B. The TSS (+1) and proximal promoter region of CDKN1A are represented with the AP-2 site at −111/−103, bound by a TFAP2C dimer (A). Myc (M) is shown binding adjacent regions through interaction with another undefined DNA binding factor (shown as gray ovals). Myc and TFAP2C stabilize each other's binding to the promoter (double-headed arrow) and together recruit KDM5B, which has a negative effect on chromatin structure via demethylation of H3K4me3, leading to suppression of RNA polymerase II (Pol)-mediated transcription of the gene.
The majority of studies examining the activity of the KDM5 family of histone demethylases have relied on the analysis of global H3K4 methylation status, which suggested that these enzymes can act on a broad range (tri, di, mono) of methylation states at this site. However, a small number of specifically regulated genes have been identified which, as shown here for CDKN1A, all exhibited KDM5B binding close to their TSS. Moreover, modulation of KDM5B expression led to consistent alterations in H3K4me3 levels at these loci, but dimethylation (me2) and monomethylation (me1) levels remained relatively constant (39). This again mirrors our ChIP results at the CDKN1A locus, where cosilencing of the ternary complex proteins led to induction of gene transcription (Fig. 1) and a concomitant increase in H3K4me3 but little alteration in me2 or me1 levels (Fig. 6B and data not shown). This suggests that the observations made on global H3K4 methylation status may not always reflect the activities of KDM5 demethylases bound at the promoters of their directly regulated genes.
The C-terminal region of KDM5B contains two PHD fingers, one of which has been shown to bind H3K4me3 and contribute to binding to active genes (reviewed in reference 18). This C-terminal domain was also found here to be required for interaction with TFAP2C (Fig. 3). Studies primarily of the Drosophila KDM5 homologue Lid have shown that the JmjC and Zf motifs were independently required for Myc interaction (20, 28), and thus these two factors interact with distinct domains of KDM5B. In addition, analysis of TFAP2C deletion mutants plus data from coIP with a range of TFAP2C antibodies (Fig. 3) defines amino acids 181 to 430 as the region of TFAP2C able to bind KDM5B and Myc. This includes the basic-helix-span-helix domain, suggesting that TFAP2C dimerization may be required for factor interaction, although actual binding may be mediated by a 40-aa region immediately N-terminal of the basic domain. The sequences required for Myc/KDM5B interaction therefore overlap those previously shown to mediate TFAP2C binding to CITED2/p300 (6) during gene activation, suggesting that competition by distinct complexes for an overlapping interaction domain may determine whether TFAP2C will activate or repress transcription.
Reporter assays confirmed the requirement for the C-terminal KDM5B PHD fingers for corepressor activity with TFAP2C. An intact JmjC domain was also essential, with even a single amino acid substitution (H499A), known to prevent demethylase activity, neutralizing corepressor function (Fig. 4B). Superficially this suggests that demethylase activity is required for corepression, which would agree with our ChIP data on histone methylation status at the CDKN1A locus (Fig. 6). However, previously reported domain structure analyses for KDM5B, again using the global H3K4 methylation level as the readout, concluded that disruption of any of the domains within the N-terminal half of the protein will ablate demethylase activity. This is thought to be due to spatial constraints on the folding of the JmjN and JmjC domains to form a functional enzyme (38, 39). Since other domains were not found to be required for corepressor activity with TFAP2C, it suggests either that there are differences between demethylase activity monitored on total histone versus observations at gene-specific sites or the intact JmjC domain alone is required for optimal localization of KDM5B at the CDKN1A promoter and even the H499A mutation is sufficient to disrupt this.
In Drosophila, the interaction between dMyc and the JmjC domain of Lid was shown to negatively regulate demethylase activity and thereby activate gene expression (20, 28). The increased level of H3K4me3 we observe at the CDKN1A locus upon silencing of TFAP2C/Myc/KDM5B suggests, however, that here TFAP2C and Myc recruit active demethylase that contributes to repression by removal of the H3K4me3 epigenetic mark. This agrees with many other studies confirming a role in dynamic gene silencing for the KDM5 family; notably, the recruitment of KDM5A by the core transcription factor RBP-J to silence target genes during Notch signaling has several parallels with our findings here (21). Nevertheless, repression by the ternary complex is likely to involve additional mechanisms, since we have previously reported that loss of histone deacetylases (HDACs) and increased histone acetylation also occur during induction of CDKN1A upon TFAP2C silencing in MCF-7 cells (36). However, KDM5B has also been shown to interact with HDACs through its PHD fingers (3), and the homologue KDM5A has been purified in HDAC corepressor complexes (35).
We have extended our examination of the interaction of TFAP2C/Myc/KDM5B with each other and the CDKN1A locus by using functional experiments to demonstrate that overexpression of these factors can perturb normal cell cycle control, both in cycling cells and in those incubated with drugs to activate p53-dependent checkpoints. In each case, the most efficient reversal of drug-induced CDKN1A activation occurred when all three ternary complex proteins were coexpressed, and this was sufficient to force arrested cells back into cycle (Fig. 7). Moreover, both TFAP2C and MYC are estrogen-responsive genes, and loss of this regulation in breast cancer has been associated with the lack of CDKN1A induction in response to antioestrogen therapy, thus contributing to resistance (13, 23, 36). Significantly, the ubiquitously expressed demethylase homologue KDM5A was found to be upregulated in drug-tolerant subpopulations of cells from several tumor origins treated with a range of anticancer agents (30). Thus, this family of histone demethylases may be particularly implicated in the epigenetic heterogeneity within cancer cell populations that can contribute to the development of treatment resistance and tumor progression. This suggests, therefore, that the association of TFAP2C, KDM5B, and Myc allows these proteins to cooperate when overexpressed in breast tumors, leads to a poor patient prognosis, and provides a rationale for the further investigation of histone demethylase activity as a therapeutic target in this disease.
Supplementary Material
ACKNOWLEDGMENTS
We thank Joyce Taylor-Papadimitriou and Charlie Degui Chen for kindly providing KDM5B antibodies and constructs and Richard Grose for his comments on the manuscript.
This work was supported by Cancer Research UK program grant no. C6775/A6250 (to H.C.H.) and Breast Cancer Campaign grant no. 2009MayPR60 (to A.G.S.). K.V.C. was supported by a Cancer Research UK studentship. P.-P.W. was funded by MRC93277.
Footnotes
Published ahead of print 27 February 2012
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Abbas T, Dutta A. 2009. p21 in cancer: intricate networks and multiple activities. Nat. Rev. Cancer 9:400–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bamforth SD, et al. 2001. Cardiac malformations, adrenal agenesis, neural crest defects and exencephaly in mice lacking cited2, a new tfap2 co-activator. Nat. Genet. 29(4):469–474 [DOI] [PubMed] [Google Scholar]
- 3. Barrett A, et al. 2007. Breast cancer associated transcriptional repressor PLU-1/JARID1B interacts directly with histone deacetylases. Int. J. Cancer 121:265–275 [DOI] [PubMed] [Google Scholar]
- 4. Berlato C, et al. 2011. Alternative TFAP2A isoforms have distinct activities in breast cancer. Breast Cancer Res. 13:R23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bosher JM, Totty NF, Hsuan JJ, Williams T, Hurst HC. 1996. A family of AP-2 proteins regulates c-erbB-2 expression in mammary carcinoma. Oncogene 13:1701–1707 [PubMed] [Google Scholar]
- 6. Braganca J, et al. 2003. Physical and functional interactions among AP-2 transcription factors, p300/CREB-binding protein, and CITED2. J. Biol. Chem. 278:16021–16029 [DOI] [PubMed] [Google Scholar]
- 7. Braganca J, et al. 2002. Human CREB-binding protein/p300-interacting transactivator with ED-rich tail (CITED) 4, a new member of the CITED family, functions as a co-activator for transcription factor AP-2. J. Biol. Chem. 277:8559–8565 [DOI] [PubMed] [Google Scholar]
- 8. Cowling VH, Cole MD. 2006. Mechanism of transcriptional activation by the Myc oncoproteins. Semin. Cancer Biol. 16:242–252 [DOI] [PubMed] [Google Scholar]
- 9. Eckert D, Buhl S, Weber S, Jager R, Schorle H. 2005. The AP-2 family of transcription factors. Genome Biol. 6:246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Eloranta JJ, Hurst HC. 2002. Transcription factor ap-2 interacts with the sumo-conjugating enzyme ubc9 and is sumolated in vivo. J. Biol. Chem. 277(34):30798–30804 [DOI] [PubMed] [Google Scholar]
- 11. Gartel AL, Radhakrishnan SK. 2005. Lost in transcription: p21 repression, mechanisms, and consequences. Cancer Res. 65:3980–3985 [DOI] [PubMed] [Google Scholar]
- 12. Gartel AL, et al. 2001. Myc represses the p21(WAF1/CIP1) promoter and interacts with Sp1/Sp3. Proc. Natl. Acad. Sci. U. S. A. 98:4510–4515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gee JM, et al. 2009. Overexpression of TFAP2C in invasive breast cancer correlates with a poorer response to anti-hormone therapy and reduced patient survival. J. Pathol. 217:32–41 [DOI] [PubMed] [Google Scholar]
- 14. Gee JM, Robertson JF, Ellis IO, Nicholson RI, Hurst HC. 1999. Immunohistochemical analysis reveals a tumour suppressor-like role for the transcription factor AP-2 in invasive breast cancer. J. Pathol. 189:514–520 [DOI] [PubMed] [Google Scholar]
- 15. Goldfarb DS, Gariepy J, Schoolnik G, Kornberg RD. 1986. Synthetic peptides as nuclear localization signals. Nature 322:641–644 [DOI] [PubMed] [Google Scholar]
- 16. Guler G, et al. 2007. Wwox and Ap2γ expression levels predict tamoxifen response. Clin. Cancer Res. 13:6115–6121 [DOI] [PubMed] [Google Scholar]
- 17. Herkert B, Eilers M. 2010. Transcriptional repression: the dark side of Myc. Genes Cancer 1:580–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Islam AB, Richter WF, Lopez-Bigas N, Benevolenskaya EV. 2011. Selective targeting of histone methylation. Cell Cycle 10:413–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Krishnakumar R, Kraus WL. 2010. PARP-1 regulates chromatin structure and transcription through a KDM5B-dependent pathway. Mol. Cell 39:736–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li L, Greer C, Eisenman RN, Secombe J. 2010. Essential functions of the histone demethylase lid. PLoS Genet. 6:e1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liefke R, et al. 2010. Histone demethylase KDM5A is an integral part of the core Notch-RBP-J. repressor complex. Genes Dev. 24:590–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lu PJ, et al. 1999. A novel gene (PLU-1) containing highly conserved putative DNA/chromatin binding motifs is specifically up-regulated in breast cancer. J. Biol. Chem. 274:15633–15645 [DOI] [PubMed] [Google Scholar]
- 23. Mukherjee S, Conrad SE. 2005. c-Myc suppresses p21WAF1/CIP1 expression during estrogen signaling and antiestrogen resistance in human breast cancer cells. J. Biol. Chem. 280:17617–17625 [DOI] [PubMed] [Google Scholar]
- 24. Pellikainen JM, Kosma VM. 2007. Activator protein-2 in carcinogenesis with a special reference to breast cancer—a mini review. Int. J. Cancer 120:2061–2067 [DOI] [PubMed] [Google Scholar]
- 25. Satoda M, et al. 2000. Mutations in TFAP2B cause Char syndrome, a familial form of patent ductus arteriosus. Nat. Genet. 25:42–46 [DOI] [PubMed] [Google Scholar]
- 26. Scibetta AG, et al. 2007. Functional analysis of the transcription repressor PLU-1/JARID1B. Mol. Cell. Biol. 27:7220–7235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Scibetta AG, Wong PP, Chan KV, Canosa M, Hurst HC. 2010. Dual association by TFAP2A during activation of the p21cip/CDKN1A promoter. Cell Cycle 9:4525–4532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Secombe J, Li L, Carlos L, Eisenman RN. 2007. The Trithorax group protein Lid is a trimethyl histone H3K4 demethylase required for dMyc-induced cell growth. Genes Dev. 21:537–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Seoane J, Le HV, Massague J. 2002. Myc suppression of the p21(Cip1) Cdk inhibitor influences the outcome of the p53 response to DNA damage. Nature 419:729–734 [DOI] [PubMed] [Google Scholar]
- 30. Sharma SV, et al. 2010. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 141:69–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shilatifard A. 2008. Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr. Opin. Cell Biol. 20:341–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stoetzel C, et al. 2009. Confirmation of TFAP2A gene involvement in branchio-oculo-facial syndrome (BOFS) and report of temporal bone anomalies. Am. J. Med. Genet. A 149A:2141–2146 [DOI] [PubMed] [Google Scholar]
- 33. Tan SK, et al. 2011. AP-2gamma regulates oestrogen receptor-mediated long-range chromatin interaction and gene transcription. EMBO J. 30:2569–2581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tu S, et al. 2008. The ARID domain of the H3K4 demethylase RBP2 binds to a DNA CCGCCC motif. Nat. Struct. Mol. Biol. 15:419–421 [DOI] [PubMed] [Google Scholar]
- 35. Vermeulen M, et al. 2010. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell 142:967–980 [DOI] [PubMed] [Google Scholar]
- 36. Williams CM, et al. 2009. AP-2gamma promotes proliferation in breast tumour cells by direct repression of the CDKN1A gene. EMBO J. 28:3591–3601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wolfer A, et al. 2010. MYC regulation of a “poor-prognosis” metastatic cancer cell state. Proc. Natl. Acad. Sci. U. S. A. 107:3698–3703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Xiang Y, et al. 2007. JARID1B is a histone H3 lysine 4 demethylase up-regulated in prostate cancer. Proc. Natl. Acad. Sci. U. S. A. 104:19226–19231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yamane K, et al. 2007. PLU-1 is an H3K4 demethylase involved in transcriptional repression and breast cancer cell proliferation. Mol. Cell 25:801–812 [DOI] [PubMed] [Google Scholar]
- 40. Zeng J, et al. 2010. The histone demethylase RBP2 is overexpressed in gastric cancer and its inhibition triggers senescence of cancer cells. Gastroenterology 138:981–992 [DOI] [PubMed] [Google Scholar]
- 41. Zeng YX, Somasundaram K, el-Deiry WS. 1997. AP2 inhibits cancer cell growth and activates p21WAF1/CIP1 expression. Nat. Genet. 15:78–82 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.