Abstract
The recently identified Thanatos-associated protein (THAP) domain is an atypical zinc finger motif with sequence-specific DNA-binding activity. Emerging data suggest that THAP proteins may function in chromatin-dependent processes, including transcriptional regulation, but the roles of most THAP proteins in normal and aberrant cellular processes remain largely unknown. In this work, we identify THAP11 as a transcriptional regulator differentially expressed in human colon cancer. Immunohistochemical analysis of human colon cancers revealed increased THAP11 expression in both primary tumors and metastases. Knockdown of THAP11 in SW620 colon cancer cells resulted in a significant decrease in cell proliferation, and profiling of gene expression in these cells identified a novel gene set composed of 80 differentially expressed genes, 70% of which were derepressed by THAP11 knockdown. THAP11 was found to associate physically with the transcriptional coregulator HCF-1 (host cell factor 1) and recruit HCF-1 to target promoters. Importantly, THAP11-mediated gene regulation and its chromatin association require HCF-1, while HCF-1 recruitment at these genes requires THAP11. Collectively, these data provide the first characterization of THAP11-dependent gene expression in human colon cancer cells and suggest that the THAP11–HCF-1 complex may be an important transcriptional and cell growth regulator in human colon cancer.
INTRODUCTION
The Thanatos-associated protein (THAP) domain is an evolutionarily conserved C2-CH (C-X2-4-C-X35-50-C-X2-H) zinc finger motif with sequence-specific DNA-binding activity (5, 33–35). Twelve THAP proteins, each containing an N-terminally located THAP domain, have been identified in humans (THAP0 to THAP11), and a subset of these (THAP0, -1, -2, -4, -7, and -11) are also conserved in mice and rats (7).
THAP domains are approximately 80 to 90 amino acids in length and, in addition to zinc-coordinating residues, contain several conserved or invariant residues necessary for proper domain folding and DNA-binding activity (4, 5, 7, 35). The majority of conserved THAP proteins also contain a coiled-coil protein interaction domain adjacent to a host cell factor 1 (HCF-1)-binding motif (HBM) (26). The tetrapeptide HBM (E/DHXY, where X is any amino acid) facilitates the interaction of THAP proteins and other DNA-binding factors with the Kelch domain of HCF-1, a transcriptional coregulator and cell proliferation factor associated with a variety of enzymatic and histone-modifying activities, including SIN3/HDAC histone deacetylase, SET1/MLL histone methyltransferase, and MOF histone acetyltransferase (11, 22, 23, 26, 30, 39, 42).
Individual THAP proteins have been implicated in a diverse array of physiological processes, including cell proliferation, regulation of transcription, apoptosis, and maintenance of embryonic stem (ES) cell pluripotency (2, 3, 6, 9, 12, 24, 33, 45). The DNA- and HCF-1-binding properties of THAP proteins naturally suggest that these proteins may regulate normal or disease-specific physiological processes in a DNA- and chromatin-dependent manner. Indeed, mutations in the THAP1 gene which disrupt DNA binding have recently been identified as a genetic determinant of the neurological disorder DYT6 dystonia, suggesting that this disease may be a result of the perturbation of a THAP1-dependent gene expression program (12, 38). In addition, THAP1 has been shown to regulate the proliferation and cell cycle progression of vascular endothelial cells through HCF-1-dependent transcriptional regulation of RRM1 (ribonucleotide reductase 1), a gene known to be required for S-phase DNA synthesis (6, 26).
The murine homolog of human THAP11, termed RONIN, has recently been shown to be required for ES cell proliferation (9). Homozygous deletion of Ronin was found to be embryonically lethal to mice. The inner cell mass of Ronin null blastocysts failed to proliferate when the cells were cultured in vitro, while forced overexpression of RONIN in ES cells promoted teratocarcinoma formation in immunocompromised mice and also prevented spontaneous ES cell differentiation upon culture in the absence of leukemia inhibitory factor (9). The ability of RONIN/THAP11 to exert strong antidifferentiation effects in mouse ES cells was initially suggested to result from RONIN-dependent global transcriptional repression concomitant with the deposition of transcriptionally repressive histone modification (9). However, recent work in the same laboratory suggests that RONIN/THAP11 may activate, as well as repress, transcription (10). Contrasting with the role of THAP11 in ES cells, Zhu et al. have recently reported that THAP11 functions as a negative regulator of cell growth in human HepG2 hepatoma cells through transcriptional repression of the proto-oncogene MYC (45).
These findings suggest that THAP proteins likely function in DNA- and chromatin-dependent processes, including transcription. However, the transcriptional regulatory properties of most human THAP proteins and their role in physiological processes remain largely unknown. In this report, we identify a previously uncharacterized role for THAP11 as a transcription and cell growth regulator in human colon cancer cells. THAP11 was found to be differentially expressed in cell culture models of human colon cancer progression, and immunohistochemical analysis of tissue microarrays (TMAs) similarly revealed increased THAP11 expression in primary and metastatic tumors. Using microarray-based profiling of gene expression in SW620 THAP11 knockdown cells, we have determined that the majority of THAP11-regulated genes are derepressed upon THAP11 knockdown. We have performed extensive molecular characterization of THAP11-mediated transcriptional regulation and determined that THAP11 not only recruits but requires HCF-1 for stable chromatin association. Collectively, these data provide the first characterization of a directly regulated, THAP11-dependent gene expression program in human cancer cells and suggest that the THAP11–HCF-1 complex may be an important transcriptional and cell growth regulator in human colon cancer.
MATERIALS AND METHODS
Plasmids and cloning.
A Mammalian Gene Collection-verified full-length cDNA clone of human THAP11 was purchased from Open Biosystems, PCR amplified, and inserted into expression vectors pGEX-4T1 (GE Life Sciences), p3xFLAG-CMV14 (Sigma), and pCMX-Gal41-147 using standard molecular cloning procedures. The retroviral expression vector pBABE-puro has been described elsewhere (28). The vector pBABE-EGFP was generated from pBABE-puro by replacing the puromycin resistance cassette (HindIII/ClaI excised) with the enhanced green fluorescent protein (GFP) coding sequence, which was PCR amplified from pEGFP-C2 (Clontech).
The retroviral short hairpin RNA (shRNA) expression vector pSuper.Retro.Puro, here abbreviated pSRP, was purchased from Oligoengine, as was the nonsilencing control shRNA pSRP MAMM-X (designated shNS). THAP11 and HCF-1 shRNA targeting sequences were designed using Dharmacon siDESIGN Center (32). Synthetic oligonucleotides corresponding to shRNA targeting sequences were cloned into BglII/HindIII-linearized pSRP according to the manufacturer's instructions. An additional nonsilencing control shRNA (designated shNS2) targeting GFP was also cloned into pSRP. A summary of the shRNA targets and the corresponding synthesized oligonucleotide sequences used in this study is provided in the supplemental material.
An expression construct for nonsilenceable THAP11 (p3xFLAG-THAP11-Rescue) resistant to shRNAs was generated by two successive rounds of site-directed mutagenesis using p3xFLAG-THAP11 and the QuikChange site-directed mutagenesis kit (Stratagene). Each mutagenesis reaction introduced silent mutations into three consecutive codons within the shRNA targeting sequence using primers listed in the supplemental material. The p3xFLAG-THAP11-Rescue construct was further subcloned by PCR amplifying the nonsilenceable THAP11 coding sequence, including a 3xFLAG tag, and inserted into the EcoRI/SalI sites of pBABE-EGFP, generating pBABE-EGFP-THAP11-Rescue-3xFLAG. The sequence correctness of all constructs was verified by automated DNA sequencing.
Cell culture and treatment.
Cell lines 293T/17 (CRL-11268), HT-29 (HTB-38), HCT-116 (CCL-247), SW480 (CCL-228), SW620 (CCL-227), LoVo (CCL-229), DLD-1 (CCL-221), and Colo-320HSR (CCL-220.1) were purchased from the American Type Culture Collection. HT-29 and HCT-116 were maintained in McCoy's 5A medium with 10% fetal bovine serum. DLD-1 and Colo-320HSR cells were grown in RPMI 1640 medium containing 10% fetal bovine serum. SW480, SW620, and 293T/17 cells were maintained in Dulbecco's modified Eagle medium (high glucose) containing 10% fetal bovine serum. LoVo cells were grown in F12-K with 10% fetal bovine serum. All cells were grown without supplemental antibiotics in a humidified 37°C incubator containing 5% CO2.
Generation and purification of THAP11 antibody.
A custom rabbit polyclonal antibody was generated against the carboxyl terminus of human THAP11 (amino acids 132 to 313). Recombinant glutathione S-transferase (GST)–THAP11(132-313) was produced in Escherichia coli strain Rosetta-2 BL21(DE3) (Novagen) and purified using glutathione Sepharose as previously described (24). Protein was subjected to preparative-scale SDS-PAGE and Coomassie stained, and gel bands corresponding to GST–THAP11(132-313) were excised for use as an immunogen. Animal immunizations and serum collection were performed by a commercial facility (Covance). GST-specific antibodies were depleted by passing crude serum over a cross-linked GST-glutathione Sepharose column. Further affinity purification of anti-THAP11 antibodies was performed using the immunogen immobilized on nitrocellulose as described elsewhere (36). Briefly, GST–THAP11(132-313) was subjected to single-well SDS-PAGE, transferred to nitrocellulose, identified by Ponceau S staining, and excised. The nitrocellulose strip was blocked for 1 h with 3% bovine serum albumin (BSA) in phosphate-buffered saline (PBS) and then incubated overnight at 4°C with 1 ml of anti-THAP11 antiserum diluted 1:10 in PBS with 3% BSA. The antibody solution was discarded, and the nitrocellulose strip was washed with several changes of PBS at room temperature. The THAP11 antibody was eluted by incubating the nitrocellulose with 0.2 M glycine (pH 2.5) and immediately neutralized with 0.1 volume of 1 M Tris-HCl, pH 8.0. One-tenth volume of 10× PBS was added to the affinity-purified anti-THAP11 antibody, which was then concentrated by filtration using Microcon centrifugal filter devices according to the manufacturer's instructions (Millipore). The specificity of the affinity-purified THAP11 antibody was confirmed by immunoblotting.
Immunoblotting.
Whole-cell extracts were prepared from subconfluent cells using modified radioimmunoprecipitation assay (RIPA) buffer (20 mM Tris-HCl [pH 7.6], 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% IGEPAL CA-630, 1% sodium deoxycholate, 0.25% SDS). Extracts were clarified by centrifugation at 20,000 × g for 15 min at 4°C, and protein concentrations determined by bicinchoninic acid (BCA) assay (Pierce).
Thirty micrograms of whole-cell extract was separated on precast 8 to 16% polyacrylamide gels (Invitrogen), transferred to nitrocellulose membrane, and stained with Ponceau S to confirm equal protein loading. Membranes were blocked in PBS–0.05% Tween 20 (PBST) containing 5% nonfat dry milk and incubated overnight at 4°C with primary antibody diluted in PBST–5% nonfat dry milk. Membranes were subsequently washed with PBST, incubated with the appropriate horseradish peroxidase (HRP)-conjugated secondary antibody diluted in PBST–5% nonfat dry milk, and developed using enhanced chemiluminescence (ECL) plus detection reagent (GE Life Sciences). Immunoblotting for THAP11 was performed using our custom-generated THAP11 antibody. The commercially available primary antibodies used for immunoblotting included antibodies to β-actin (Sigma A5441), FLAG M2 (Sigma F1804), and HCF-1 (Bethyl Laboratories A301-399A). HRP-conjugated anti-mouse and anti-rabbit secondary antibodies were purchased from Sigma.
Coimmunoprecipitation and immunoblot analysis.
Cells in 15-cm2 tissue culture dishes were rinsed three times with ice-cold PBS, scraped into PBS, and collected by centrifugation at 500 × g for 5 min at 4°C. Cells were then resuspended in 5 pellet cell volumes (PCV) of buffer A (10 mM HEPES-KOH [pH 7.6], 10 mM KCl, 1.5 mM MgCl2) containing Complete protease inhibitors (Roche) and allowed to swell for 10 min on ice. Cytoplasmic membranes were lysed by dropwise addition of IGEPAL CA-630 from a 10% stock solution to a final concentration of 0.5% while the cells were being gently mixed by vortexing at a half-maximum setting. Cells were incubated on ice for 5 min, and plasma membrane lysis was verified by trypan blue staining. Nuclei were isolated by centrifugation at 2,000 × g for 5 min at 4°C, washed once with buffer A, and resuspended in 1 PCV of buffer C (20 mM HEPES-KOH [pH 7.6], 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 25% glycerol) supplemented with Complete protease inhibitors (Roche). Nuclei were extracted for 45 min at 4°C with gentle inversion. Nuclear extracts were clarified by centrifugation (20,000 × g for 15 min at 4°C), diluted with 1 volume of buffer C without glycerol or NaCl, and adjusted to 0.5% IGEPAL CA-630. Nuclear extracts were reclarified by centrifugation to remove precipitates formed by dilution, and protein concentration was determined by BCA assay. Immunoprecipitations were performed with 0.5 to 1 mg of nuclear extract and 1 to 2 μg of either affinity-purified rabbit polyclonal anti-HCF-1 (Bethyl Laboratories A301-399A) or normal rabbit IgG (Sigma) overnight at 4°C with inversion. Protein G Dynabeads (20 μl) were added, and immunoprecipitation was continued for an additional 2 h. Beads were then washed four times with binding buffer (20 mM HEPES-KOH [pH 7.6], 150 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5% IGEPAL CA-630), and bound proteins were eluted by boiling in 2× Laemmli buffer. Immunoprecipitated proteins were resolved by SDS-PAGE and immunoblotted with HCF-1 and THAP11 antibodies as described above. Blots were developed by ECL using an anti-rabbit light-chain-specific, HRP-conjugated secondary antibody (Jackson ImmunoResearch 211-032-171) to minimize obscuring of the THAP11 signal by comigrating IgG heavy chains.
Immunofluorescence analysis.
For indirect immunofluorescence analysis, cells grown on glass coverslips were fixed with 4% paraformaldehyde in PBS for 10 min at room temperature. Fixed cells were rinsed three times with PBS and permeabilized with 0.2% Triton X-100 in PBS for 5 min at room temperature. Coverslips were washed twice with PBS, blocked with PBS–3% BSA for 30 min at 37°C, and incubated with affinity-purified anti-THAP11 antibody in PBS–3% BSA for 1 h at 37°C. Coverslips were then washed three times with PBS and incubated with Alexa Fluor 488-conjugated anti-rabbit IgG (1:1,000; Invitrogen) in PBS–3% BSA for 1 h at 37°C. Coverslips were again washed three times in PBS, counterstained with 4′,6-diamidino-2-phenylindole (DAPI), and mounted in Prolong antifade reagent (Invitrogen). Samples were analyzed using a Zeiss LSM 510 META laser scanning confocal microscope.
Immunohistochemistry and TMAs.
TMA slides prepared from formalin-fixed, paraffin-embedded samples were obtained from US Biomax. TMAs contained 33 normal colonic mucosa, 7 benign tubular adenoma, and 133 colon cancer samples. In addition, 3 liver samples and 37 samples from lymph nodes with metastatic diseases were also included. All cases contained tissue cores in triplicate. TMAs were deparaffinized in two changes of xylene and rehydrated in a graded alcohol series using standard procedures. Slides were subjected to heat-induced antigen retrieval by microwaving in citrate buffer (10 mM sodium citrate, 0.05% Tween 20, pH 6.0), followed by blocking of endogenous peroxidases with 3% hydrogen peroxide. Immunohistochemical staining for THAP11 was performed using affinity-purified anti-THAP11 antibody (1:400 dilution), Vectastain Elite ABC detection reagents, and 3,3′-diaminobenzidine tetrahydrochloride substrate (Vector Laboratories). Hematoxylin-counterstained slides were evaluated using a semiquantitative dual-scoring system as described elsewhere (41). Briefly, the intensity of THAP11 immunoreactivity in cell nuclei was scored numerically (0 = negative, 1 = weak, 2 = moderate, 3 = strong), as was the percentage of THAP11-immunopositive nuclei (0 = 0%, 1 = 1 to 10%, 2 = 11 to 50%, 3 = 51 to 100%). The intensity and percent immunopositivity scores were added, and samples were categorized into low/weak expression (combined score, ≤3) and high/strong expression (combined score, >3) groups. The statistical significance of differences between the combined scores of normal and primary cancer samples or normal and metastatic cancer samples was determined by the chi-square test.
Retrovirus production.
Vesicular stomatitis virus G glycoprotein (VSVG)-pseudotyped retrovirus was produced in 293T/17 cells (∼70% confluent in 10-cm2 dishes) by cotransfection with pCMV-VSVG (4 μg), pMLV-GagPol (8 μg), and a retroviral construct (12 μg) using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Following overnight transfection, cells were given fresh medium (Dulbecco's modified Eagle's medium [DMEM]–10% fetal bovine serum [FBS]) and allowed to equilibrate for several hours in a 37°C cell culture incubator prior to being shifted to a 32°C 5% CO2 humidified cell culture incubator for an additional 24 to 30 h. Retroviral supernatants were harvested ∼48 h from the start of transfection, cleared of residual 293T/17 cells by centrifugation (1,000 × g, 5 min), and either used immediately or aliquoted and stored at −80°C for future use.
Retroviral transduction and stable cell production.
To generate pools of SW620 and Colo-320HSR cells stably expressing either control or THAP11 shRNA, cells in six-well plates (∼20% confluent) were transduced with a 1:1 mixture of viral supernatant and fresh medium, adjusted to 8 μg/ml Polybrene, and spin infected at 500 × g for 2 h at 32°C. Following spin infection, cells were returned to the 37°C incubator for 2 h and then given fresh growth medium. Two days postransduction, cells from individual wells of the six-well plate were split into 10-cm2 dishes containing 2 μg/ml puromycin. Cells were grown under selection for an additional 2 days, after which mock-transduced cells exhibited complete cell death. For rescue experiments using nonsilenceable THAP11, SW620 cells were first transduced with pSRP shRNA virus and transduced 24 h later with the indicated pBABE-EGFP virus. Doubly expressing cells were then selected with puromycin and sorted for GFP expression using fluorescence-activated cell sorting (FACS).
RNA isolation and quantitative reverse transcription (RT)-PCR.
Total RNA was isolated by using the Qiagen RNeasy minikit according to the manufacturer's instructions. Total RNA (1 μg) was reverse transcribed using qScript cDNA synthesis mix (Quanta Biosciences) containing both random hexamer and oligo(dT) primers. Quantitative PCR (qPCR) was performed with diluted cDNA using an ABI PRISM 7900HT 384-well real-time PCR machine (Applied Biosystems) in a final volume of 20 μl using SYBR green PCR master mix (Applied Biosystems) and gene-specific primers. Fold changes in mRNA levels were determined using the ΔΔCT method normalized to β-actin.
Microarray gene expression analysis.
Analysis of gene expression in SW620 cells stably expressing either control (pSRP-shNS, pSRP-shNS2) or THAP11 (pSRP-T11A, pSRP-T11C) shRNAs was done using Nimblegen Homo sapiens 385K oligonucleotide microarrays. Total RNA from two independent pools of SW620 cells per shRNA were isolated using Qiagen RNeasy minikits as described above. RNA quality was verified using an Agilent 2100 Bioanalyzer and provided to Nimblegen for subsequent cDNA synthesis, labeling, and microarray hybridization.
Nuclear run-on assay.
Modified nuclear run-on assays using 5-bromouridine (BrU)-labeled nascent RNA were performed as previously described, with minor modifications (8). SW620 cells (∼1 × 108) expressing either pSRP-shNS, pSRP-shT11A, or pSRP-shT11C were rapidly cooled by rinsing in ice-cold PBS, scraped into ice-cold PBS, and collected by centrifugation (500 × g, 5 min, 4°C). Cell pellets were resuspended in hypotonic lysis buffer (10 mM Tris-HCl [pH 7.6], 10 mM NaCl, 3 mM MgCl2) and immediately centrifuged as before. Cell pellets were loosened by gentle vortexing (setting 6), resuspended in hypotonic lysis buffer containing 0.5% IGEPAL CA-630, and incubated on ice for 5 min. Cell lysis was routinely >90%, as determined by trypan blue staining and hemacytometer counting. Nuclei were recovered by centrifugation (1,000 × g, 5 min, 4°C), washed once in hypotonic lysis buffer containing 0.5% IGEPAL CA-630, and centrifuged as before. Recovered nuclei were resuspended in 1 ml of freezing buffer (50 mM Tris-HCl [pH 8.0], 40% glycerol, 5 mM MgCl2, 0.1 mM EDTA, 40 U RNasinPlus [Promega]) and aliquoted into 1.5-ml tubes at 200 μl (∼107 nuclei) per tube, snap-frozen in liquid nitrogen, and stored at −80°C.
For run-on transcription, thawed nuclei were mixed with an equal volume (200 μl) of 2× run-on buffer (10 mM Tris-HCl [pH 8.0]; 5 mM MgCl2; 300 mM KCl; 1% Sarkosyl; 160 U RNasinPlus; 5 mM dithiothreitol [DTT]; 1 mM each ATP, CTP, GTP, and BrUTP) and incubated at 30°C for 30 min. Following run-on transcription, samples were digested with DNase I and proteinase K. Total RNA was extracted using acid phenol-chloroform-isoamyl alcohol (25:24:1), ethanol precipitated, and resuspended in nuclease-free water. Total RNA was then further purified using Qiagen RNeasy minispin columns according to the RNA cleanup procedure described by the manufacturer. Anti-BrdU monoclonal antibody (Sigma; 5 μl per immunoprecipitation, 15 μl total) was preincubated with 90 μl of protein G Dynabeads in 1 ml binding buffer (10 mM Tris-HCl [pH 7.6], 100 mM NaCl, 1 mM EDTA, 0.05% Tween 20, 0.1% IGEPAL CA-630, 15 μg yeast tRNA) for 1 h at 4°C. Dynabeads were washed three times in binding buffer without supplemental tRNA and resuspended in 90 μl of binding buffer. To immunoprecipitate BrU-labeled nascent RNA, 25 μg of nuclear run-on RNA, 2.5 μg of yeast tRNA, and 40 U of RNasinPlus were added to 500 μl of binding buffer on ice. Anti-BrdU antibody-bound Dynabeads (30 μl per immunoprecipitation) were added, and immunoprecipitations was continued for 1 h at 4°C with end-over-end rotation. Beads were subsequently washed three times in binding buffer and eluted by the addition of 300 μl of buffer RLT (Qiagen RNeasy kit). Immunoprecipitated RNAs were purified using the Qiagen RNeasy minikit, reverse transcribed, and analyzed by quantitative RT-PCR as described above.
ChIP.
Chromatin immunoprecipitation (ChIP) was performed essentially as previously described, with minor modifications (21). SW620 cells (∼2 × 107) or those stably expressing the indicated retroviral constructs were cross-linked by the addition of 1/10 volume of freshly prepared formaldehyde cross-linking buffer (11% formaldehyde, 10 mM HEPES-KOH [pH 7.6], 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA) to tissue culture dishes and incubated for 10 min at room temperature. Cross-linking was terminated by the addition of 1/20 volume of 2.5 M glycine. Cross-linked cells were washed three times with ice-cold PBS, scraped into PBS, and recovered by centrifugation (1,500 × g, 10 min, 4°C). Cell pellets were resuspended in 1 ml of lysis buffer 1 (50 mM HEPES-KOH [pH 7.6], 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% IGEPAL-CA630, 0.25% Triton X-100, 1× protease inhibitors) and incubated for 10 min at 4°C with gentle rocking. Nuclei were recovered by centrifugation, resuspended in 1 ml of lysis buffer 2 (10 mM Tris-HCl [pH 8.0], 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 1× protease inhibitors), and extracted for 10 min at room temperature with gentle inversion. Nuclei were again recovered by centrifugation, resuspended in 1 ml of lysis buffer 3 (10 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% sodium deoxycholate, 0.5% Sarkosyl, 1× protease inhibitors), and sonicated in an ice-water bath using a Misonix microtip-equipped sonicator at setting 6 (∼6 W [root mean square] output power) for 12 cycles of 15 s of sonication followed by a 1-min cooling interval. The sonicated chromatin was adjusted to 1% Triton X-100 from a 10% stock solution, and debris was removed by centrifugation at 20,000 × g at 4°C for 20 min. The protein concentration of solubilized chromatin was determined by BCA assay, and approximately 300 μg of chromatin was immunoprecipitated overnight at 4°C with the indicated antibodies. Protein G Dynabeads (20 to 30 μl) were added, and immunoprecipitations were continued for an additional 2 h. Beads were washed four times with 1 ml of ChIP-RIPA wash buffer (50 mM HEPES-KOH [pH 7.6], 500 mM LiCl, 1 mM EDTA, 1.0% IGEPAL-CA630, 0.7% sodium deoxycholate) and once with TE containing 50 mM NaCl.
Following the final wash, DNA was recovered as described by Nelson et al. (29). Briefly, Dynabeads and precipitated input chromatin were resuspended in 100 μl of 10% Chelex resin (Bio-Rad) and incubated for 10 min at 100°C. Samples were cooled to room temperature and then digested with proteinase K (0.2 mg/ml) for 30 min at 55°C. Samples were again boiled for 10 min to inactivate proteinase K and centrifuged at 20,000 × g for 3 min to pellet the Chelex-Dynabead mixture. Supernatants (80 μl) containing the immunoprecipitated DNA were transferred to clean 1.5-ml tubes, and the Chelex-Dynabead resins were resuspended in an additional 120 μl of water, vortexed, and centrifuged as before. Supernatants were combined, yielding 200 μl of immunoprecipitated DNA.
Determination of relative enrichment was performed by qPCR using an ABI PRISM 7900HT 384-well real-time PCR machine with SYBR green PCR master mix (Applied Biosystems) and primers designed as described below. Threshold cycle (CT) values of ChIP-enriched DNA were exponentiated and expressed as percent recovery relative to the input DNA analyzed in parallel.
Primer pairs for qPCR were designed using Primer3Plus against human genome sequence (NCBI36/hg18) retrieved using the UCSC Genome Browser (19). The primer sequences and relative positions of the ChIP amplicons are provided in the supplemental material. The antibodies (amounts used and sources [antibody names]) used for ChIP were against THAP11 (5 μl, custom affinity purification), HCF-1 (1 μg, Bethyl Laboratories [A301-399A]), RNAPII (4 μg, Santa Cruz [SC-899X]), RNAPII-S5P (2 μg, Bethyl Laboratories [A300-655A]), RNAPII-S2P (2 μg, Bethyl Laboratories [A300-654A]), histone H3 (4 μg, Abcam [ab1791]), histone H3 acetyl K27 (2 μg, Abcam [ab4729]), histone H3 trimethyl K4 (4 μg, Abcam [ab8580]), histone H3 acetyl K9 (4 μg, Abcam [ab4441]), and FLAG-M2 (4 μg, Sigma [F1804]).
Sequential ChIP.
For sequential ChIP, first-round ChIPs were performed as described above except that following the final wash, beads were resuspended in 10 mM Tris-HCl (pH 7.6)–1 mM EDTA–2% SDS–20 mM DTT and the precipitated complexes were eluted by incubation at 37°C for 30 min. The 50-μl eluate was divided into two 20-μl aliquots with 10 μl reserved for qPCR analysis. Eluates were diluted 20-fold with 10 mM Tris-HCl (pH 7.6)–100 mM NaCl–1 mM EDTA–1% Triton X-100 and adjusted to 1 μg/μl BSA. THAP11 ChIP eluates were subjected to ChIP again (reChIP) with 1 μg of IgG or HCF-1, and HCF-1 ChIP eluates were subjected to reChIP with 1 μg of IgG or 5 μl of THAP11 antibodies overnight at 4°C. The resulting reChIP products were collected using protein G Dynabeads, washed, and eluted as described above for conventional ChIP. Enrichment was determined by qPCR and expressed as percent recovery relative to the input of the first-round ChIP.
alamarBlue cell proliferation assay.
Proliferation of SW620 knockdown cells was determined using the alamarBlue cell viability reagent according to the manufacturer's instructions (Invitrogen). Puromycin-selected SW620 or control THAP11 knockdown cells were seeded at 5,000 per well (eight wells per condition) in black-wall 96-well tissue culture plates in 100 μl of DMEM–1% FBS. Five identical 96-well plates were seeded for determination of cell viability on 5 consecutive days. Twenty-four hours after seeding, 10 μl of alamarBlue reagent was added per well to one 96-well plate and this plate was incubated for 2 h at 37°C in a cell culture incubator. Fluorescence measurements were then performed with a BioTek Synergy HT multidetection microplate reader equipped with 540-nm excitation and 590-nm emission filters. Average background fluorescence was calculated from wells (n = 16) containing medium alone and subtracted from the fluorescence of wells containing cells. This process was repeated every 24 h for 5 consecutive days using one replicate 96-well plate per day.
Crystal violet cell proliferation assay.
SW620 knockdown cells were plated at 3 × 105 per well in six-well tissue culture plates and grown in DMEM–1% FBS. At the indicated time points, cells were fixed with 4% paraformaldehyde for 15 min at room temperature, rinsed with PBS, and stained with 0.2% crystal violet. Cells were washed extensively with water and air dried. Bound crystal violet was eluted with 10% acetic acid and measured by absorbance at 595 nm in a BioTek Synergy HT multidetection microplate reader.
RESULTS
THAP11 expression in colon cancer.
THAP domain proteins remain poorly characterized, and their role in human diseases, including cancer, is largely unknown. In an effort to further characterize the physiological role of THAP domain-containing proteins, we performed a systematic survey of publicly available gene expression data sets to identify conditions of differential THAP protein expression. Using this approach, we identified increased THAP11 mRNA expression in a microarray data set originally designed to elucidate gene expression differences in the SW480/SW620 cell culture model of colon cancer progression (16, 31).
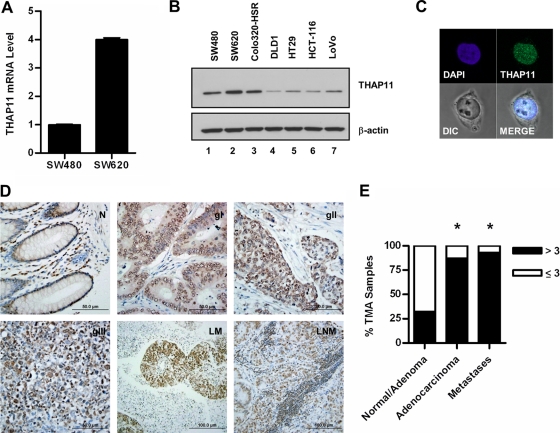
To corroborate the microarray finding, we prepared total RNA and whole-cell extracts from SW480 and SW620 cells and determined the relative amounts of THAP11 mRNA and protein by quantitative RT-PCR and immunoblotting, respectively. Both cell types express THAP11 mRNA, but metastasis-derived SW620 cells were found to express approximately 4 times as much THAP11 mRNA as isogenic, primary-tumor-derived SW480 cells (Fig. 1A). SW620 cells also expressed more THAP11 protein than SW480 cells (Fig. 1B, compare lanes 1 and 2), but SW480 cells contain greater amounts of THAP11 than primary-tumor-derived colon cancer cell lines HCT-116 and HT29 or the metastasis-derived LoVo cell line (Fig. 1B, compare lanes 1 and 4 to 7). The neuroendocrine colon cancer cell line Colo320-HSR contains nearly as much THAP11 as SW620 cells, suggesting that elevated THAP11 expression is not restricted to the SW480/SW620 model system (Fig. 1B, compare lanes 2 and 3). Immunofluorescence analysis revealed endogenous THAP11 to be located almost exclusively within the nuclei of SW620 (Fig. 1C) and SW480 cells (data not shown), consistent with a putative function in chromatin-dependent processes, including transcriptional regulation. These results suggest that gain of THAP11 expression may play a role in colon cancer cell function.
Fig 1.
THAP11 expression in human colon cancer cell lines and tumors. (A) THAP11 mRNA levels in SW480 and SW620 cells determined by quantitative RT-PCR and expressed relative to the level in SW480 cells. Values represent the mean ± standard deviation of triplicate quantitative RT-PCRs from a representative experiment performed at least three times. (B) Immunoblot assays of whole-cell extracts from the indicated colon cancer cell lines. (C) Immunofluorescence localization of endogenous THAP11 in SW620 cells. Nuclei were identified by counterstaining with DAPI, and cells were visualized by differential interference contrast (DIC) microscopy. MERGE is an overlay of the THAP11, DAPI, and DIC images. (D) Representative images of THAP11 immunohistochemical staining in normal colon epithelium (N); grade I (gI), grade II (gII), and grade III (gIII) adenocarcinomas; liver metastasis (LM); and lymph node metastasis (LNM). (E) Quantitative analysis of THAP11 expression in TMA samples. THAP11 immunoreactivity was scored as described in Materials and Methods, and samples were placed into either the high/strong (>3) or the low/weak (≤3) THAP11 expression group. An asterisk denotes a statistically significant difference from normal/benign adenomas, as measured by chi-square analysis (P < 0.001).
To further explore this possibility, we next examined a large cohort of human colon cancer samples to determine if THAP11 is overexpressed in colon cancer and determine a potential link between THAP11 expression and disease progression. Immunohistochemical analysis of THAP11 expression in human colon cancer TMAs revealed that colon cancer has significantly higher THAP11 expression than normal colonic mucosa (Fig. 1D). The majority of samples from normal colonic epithelium (n = 33) and benign adenomas (n = 7) stained both weakly and infrequently for THAP11 (Fig. 1D, part N), while increased THAP11 staining frequency and intensity were observed in primary colon adenocarcinomas (n = 133) (Fig. 1D, compare parts gI, gII, and gIII with part N), as well as both liver (n = 3) and lymph node metastases (n = 37) (Fig. 1D, parts LM and LNM, respectively). A quantitative assessment of THAP11 immunoreactivity scored as a combination of the intensity and the percentage of staining revealed significantly higher THAP11 expression in primary colon tumors and metastases than in normal tissues/benign adenomas (P < 0.001) (Fig. 1E). Taken together, these data demonstrate that increased THAP11 expression occurs in multiple stages of colon cancer and suggest that SW620 cells may represent a tractable model to evaluate the molecular function of THAP11 in colon cancer.
Profiling of gene expression in SW620-THAP11 knockdown cells.
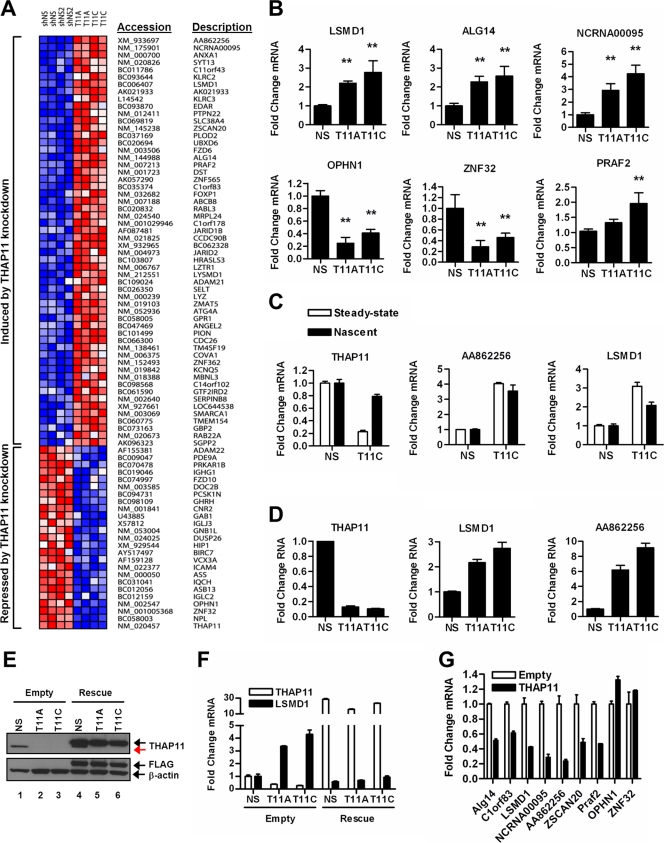
Previous work in our laboratory and others has suggested that THAP proteins can both activate and repress transcription (9, 24–26). Initial experiments performed using a Gal4 DNA-binding domain fusion of THAP11 in luciferase reporter assays suggested that THAP11 functions as a transcriptional repressor (data not shown). However, considering the artificial nature of the Gal4 fusion-based luciferase reporter assay and to independently evaluate whether endogenous THAP11 has transcriptional regulatory properties, we performed profiling of gene expression in SW620 cells depleted of THAP11 via retrovirally expressed shRNA. THAP11-specific shRNA constructs shT11A and shT11C significantly diminished THAP11 mRNA and protein, as determined by quantitative RT-PCR and immunoblotting (see Fig. S1 in the supplemental material). Two independent pools of SW620 cells expressing either control (shNS, shNS2) or THAP11 (shT11A, shT11C) shRNAs were analyzed for global gene expression changes using oligonucleotide microarrays. Genes displaying a 1.5-fold change and a P value of <0.01 (Student's t test) between control (shNS, shNS2) and THAP11 (shT11A, shT11C) shRNA-expressing cells were defined as differentially expressed. This gene set was further processed to remove predicted genes which have been subsequently “discontinued” by the NCBI and additionally lack supporting mRNA or expressed sequence tag sequences. This stringent analysis identified only 80 transcripts (excluding THAP11) as differentially expressed between THAP11 and control knockdown groups (Fig. 2A). Of these differentially expressed RNAs, 70% (56/80) showed increased expression with THAP11 knockdown while 30% of the genes were downregulated, suggesting that THAP11 possesses transcriptional regulatory activity. Independent verification of microarray-determined gene expression changes by quantitative RT-PCR of activated and repressed genes recapitulated the majority of these findings (Fig. 2B; data not shown), suggesting that the data set as a whole likely represents a THAP11-dependent gene expression program.
Fig 2.
THAP11 regulates transcription in colon cancer cells. (A) Heat map depicting microarray-based profiling of gene expression in SW620 cells expressing either control (NS and NS2) or THAP11 (T11A and T11C) shRNAs. (B) Validation of microarray-determined gene expression changes by quantitative RT-PCR. Values represent the mean ± standard deviation of four independent experiments. Double asterisks denote a statistically significant difference (P < 0.01) from control (NS) shRNA, as determined by Dunnett's posttest following analysis of variance. (C) Steady-state and nascent mRNA levels of putative THAP11 gene targets in control (NS) or THAP11 knockdown (T11C) SW620 cells. (D) Gene expression determined by quantitative RT-PCR in Colo320-HSR cells expressing control (NS) or THAP11 (T11A or T11C) shRNA. (E, top) THAP11 immunoblotting with SW620 cells expressing either control (Empty) or THAP11-Rescue-3xFLAG (Rescue) and the indicated shRNA. Endogenous THAP11 is indicated by the red arrow. The black arrow indicates THAP11-Rescue-3xFLAG. (E, bottom) Blots were stripped and reprobed with anti-FLAG and anti-β-actin antibodies. (F) Quantitative RT-PCR of THAP11 and LSMD1 expression in SW620 cells expressing either control (Empty) or THAP11-Rescue-3xFLAG (Rescue) and the indicated shRNA. RNA levels are normalized to β-actin and expressed as n-fold changes relative to SW620 cells expressing control shRNA (NS) and rescue (Empty) constructs. (G) Quantitative RT-PCR analysis of THAP11 target genes in HCT-116 cells transduced with either control (Empty) or THAP11-overexpressing retrovirus. In panels C, D, F, and G, values represent means ± standard deviations of triplicate quantitative RT-PCRs of a representative experiment performed at least three times.
Gene expression measurements by oligonucleotide microarray and quantitative RT-PCR reflect steady-state mRNA levels and by themselves are incapable of assessing whether regulation occurs at the transcriptional or posttranscriptional level. To determine if the gene expression changes observed in THAP11 knockdown cells were attributable to increased transcription, we performed a modified nuclear run-on assay with SW620 cells expressing either control or THAP11 shRNAs (8). Run-on transcription from isolated nuclei was performed using BrU to label nascent RNA transcripts in the presence of 0.5% Sarkosyl. Inclusion of Sarkosyl in run-on reactions prevents reinitiation of transcription, thus allowing only the completion of transcripts actively engaged by RNA polymerase II (RNAPII) at cell lysis (8). BrU-labeled nascent transcripts were then immunoprecipitated, and transcript levels were determined by quantitative RT-PCR. In THAP11 knockdown cells, we found nascent transcript levels for LSMD1, NCRNA00095, AA862256, and ANXA1 (annexin A1) upregulated in a manner qualitatively similar to that of their steady-state mRNAs (Fig. 2C; data not shown). Importantly, THAP11 steady-state but not nascent transcript levels were depleted in SW620 cells expressing THAP11 shRNAs (Fig. 2C). This expected discrepancy is in agreement with the proposed mechanism of RNA interference as a posttranscriptional gene silencing event and provides an important verification of the specificity of the nuclear run-on assay to detect nascent rather than mature transcripts (27). Taking these findings together, we conclude that the increased RNA levels observed in THAP11 knockdown cells for LSMD1, NCRNA00095, AA862256, and ANXA1; decreased levels of ZNF32 and OPHN1 genes; and perhaps the microarray data set as a whole likely reflect THAP11-mediated changes in the transcription of these target genes. However, we cannot exclude the possibility that posttranscriptional regulatory mechanisms also contribute to the gene expression profile of THAP11 knockdown cells.
To determine if THAP11 knockdown similarly changes gene expression in other colon cancer cell lines, we expressed THAP11 shRNAs in Colo320-HSR cells, which also have elevated THAP11 expression (Fig. 1B). Knockdown of THAP11 in Colo320-HSR cells derepressed the putative THAP11 target genes ALG14, LSMD1, AA862256, NCRNA00095, and AK021933, similar to its knockdown in SW620 cells (Fig. 2D; data not shown). Interestingly, annexin A1 was not derepressed in Colo320-HSR cells upon THAP11 knockdown (data not shown), suggesting that some gene targets may also be cell type specific.
Transcripts induced by THAP11 knockdown may represent an authentic cellular response to diminished THAP11 or, alternatively, may arise from nonspecific shRNA events despite our use of multiple control and THAP11-targeted shRNAs. To discriminate between these possibilities, we performed rescue experiments using a THAP11 expression construct rendered nonsilenceable by mutation of three consecutive codons in each of the shRNA targeting sequences. SW620 cells were first transduced with either control or THAP11 shRNA and then with either control (empty) or THAP11-Rescue-3xFLAG retroviruses. Cells expressing both constructs were selected by puromycin resistance (shRNA) and FACS (THAP11 rescue). Immunoblotting with THAP11 antibody revealed robust expression of rescue but not endogenous THAP11 in cells transduced with THAP11-Rescue-3xFLAG but not control (empty) virus (Fig. 2E). The identity of the THAP11 bands in rescue-expressing cells was further validated by immunoblotting with monoclonal anti-FLAG antibody (Fig. 2E).
If the gene expression profile observed in THAP11 knockdown cells is attributable to specific depletion of THAP11, then restoration of THAP11 status in THAP11-Rescue-3xFLAG cells should reverse this effect. Indeed, quantitative RT-PCR analysis of putative THAP11 gene targets revealed that expression of nonsilenceable THAP11 prevented the differential gene expression previously observed in THAP11 knockdown cells (Fig. 2F; see Table S1 in the supplemental material). Importantly, this rescue effect was functional irrespective of the magnitude of target gene induction. Modestly (1.2- to 1.6-fold) induced genes such as SMARCA1 and ATG4A and robustly (3- to 8-fold) induced genes, including LSMD1 and AA862256, were rescued by nonsilenceable THAP11. The expression of THAP11-Rescue-3xFLAG rerepressed putative THAP11 gene targets below the level observed in cells expressing endogenous amounts of THAP11 (shNS and empty), likely due to rescue construct overexpression. Next, we asked whether a gain in THAP11 expression is sufficient to alter transcription at these target genes in colon cancer cells that express small amounts of endogenous THAP11. Retrovirus-mediated overexpression of THAP11 in HCT-116 cells also resulted in repression (ALG14, C1orf83, LSMD1, NCRNA00095, AA862256, ZSCAN20, PRAF2) or activation (OPHN1, ZNF32) of genes previously shown to be derepressed or repressed by THAP11 knockdown, respectively (Fig. 2G). Taken together, these results indicate that human THAP11 functions as a transcriptional regulator of these genes in colon cancer cells.
Identification of direct THAP11 gene targets in SW620 cells.
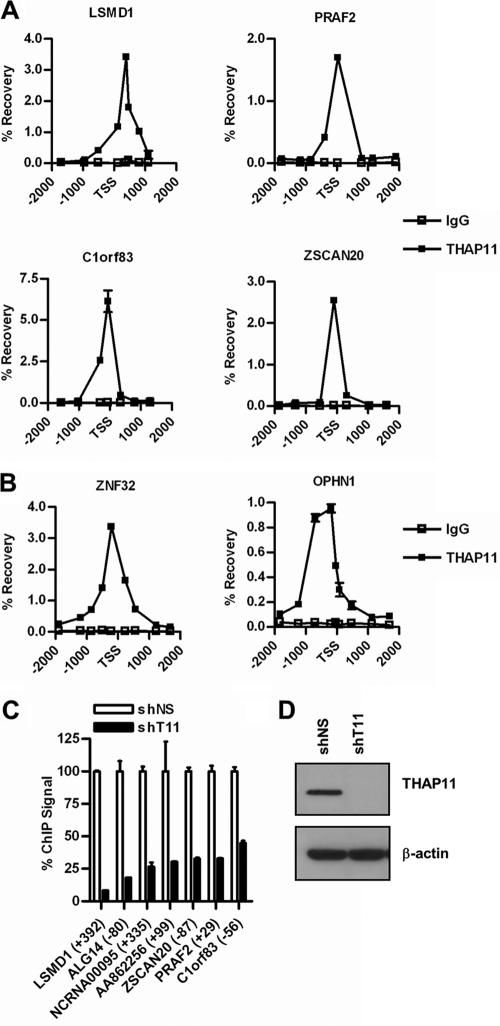
THAP11-dependent changes in gene expression may reflect a direct role for THAP11 in transcriptional regulation. To identify direct THAP11 gene targets, we performed ChIP with normal SW620 cells using our custom anti-THAP11 antibody and monitored the enrichment of immunoprecipitated chromatin by qPCR using amplicons spaced approximately 300 to 600 bp apart and spanning at least 1 kb on either side of the transcriptional start sites of both repressed (Fig. 3A; see Fig. S2 in the supplemental material) and activated (Fig. 3B) putative THAP11 target genes, including PRAF2, LSMD1, AA862256, ALG14, NCRNA00095, C1orf83, ZSCAN20, ZNF32, and OPHN1. This ChIP scanning approach identified endogenous THAP11 binding within 500 bp of the annotated transcriptional start site at each putative THAP11 target gene examined. Similar patterns of enrichment were observed when ChIP was performed using anti-FLAG monoclonal antibody with SW620 cells depleted of endogenous THAP11 but expressing THAP11-Rescue-3xFLAG (data not shown). Together, these results show that THAP11 directly binds to both activated and repressed genes.
Fig 3.
ChIP analysis of endogenous THAP11. THAP11 ChIP at THAP11-repressed genes (A) and THAP11-activated genes (B) in SW620 cells using anti-THAP11 antibody or control IgG. (C) Control (shNS) or THAP11 (shT11) knockdown SW620 cells were analyzed by ChIP as described for panel A. Values represent the mean ± standard deviation of duplicate qPCRs from a representative experiment performed three times with similar results. (D) Immunoblotting of SW620 cells from panel C expressing control (shNS) or THAP11 (shT11) shRNAs.
To further confirm the specificity of the THAP11 ChIP assay, we repeated the experiment with THAP11 knockdown SW620 cells. As expected, cells expressing THAP11 shRNA exhibited reduced, albeit detectable, levels of chromatin-bound THAP11. Residual binding in THAP11 knockdown cells ranged from 18% at LSMD1 to 45% at C1orf83 relative to that observed with control knockdown cells (Fig. 3C), despite undetectable levels of THAP11 in whole-cell extracts (Fig. 3D), suggesting that knockdown was substantial but incomplete. Because each of the aforementioned genes was effectively regulated by and contained chromatin-bound THAP11 near the transcriptional start site, we conclude that these genes are likely direct targets of THAP11-mediated transcriptional regulation.
Decreased RNAPII occupancy at THAP11-repressed genes.
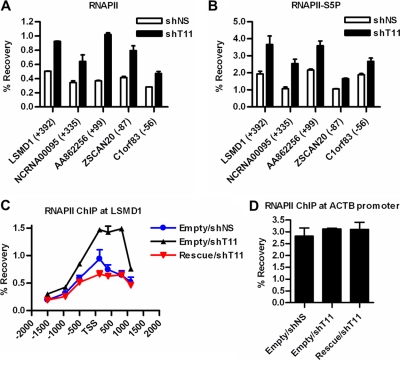
Since 70% of differentially expressed genes were derepressed in THAP11 knockdown cells, we focused our attention on further characterizing the mechanism of THAP11-mediated transcriptional repression. We examined RNAPII occupancy at THAP11 target genes by ChIP using an antibody that recognizes the N-terminal domain of RNAPII and found increased total RNAPII binding near the transcriptional start sites of genes induced by THAP11 knockdown (Fig. 4A; data not shown). The increase in total RNAPII was paralleled by increases in C-terminal domain (CTD)-phosphorylated and transcriptionally active RNAPII, as revealed by ChIP using S2P and S5P CTD-specific antibodies (Fig. 4B; data not shown). The elevated RNAPII occupancy observed at LSMD1 in THAP11 knockdown cells was reversed in SW620 rescue cells that simultaneously express nonsilenceable THAP11, thus confirming that increased RNAPII occupancy reflects diminished THAP11 levels (Fig. 4C). Importantly, no change in RNAPII occupancy was observed at the β-actin promoter upon either THAP11 knockdown or nonsilenceable THAP11 rescue expression, demonstrating the specificity of the assay for THAP11 target genes (Fig. 4D). These results suggest that THAP11 may repress transcription by limiting or destabilizing RNAPII at THAP11 target genes.
Fig 4.
THAP11 regulates RNAPII occupancy at repressed genes. Total RNAPII (A) and RNAPII-S5P (B) occupancy as determined by ChIP assay of SW620 cells expressing the indicated shRNAs. (C) ChIP analysis of RNAPII occupancy at LSMD1 in SW620 cells expressing either control (shNS) or THAP11 (shT11) shRNA and either the control (Empty) or THAP11-Rescue-3xFLAG (Rescue). (D) ChIP analysis of RNAPII occupancy at the ACTB (β-actin) promoter from the indicated SW620 cells as in panel C. Values represent the mean ± standard deviation of duplicate qPCRs from a representative experiment performed three times.
Analysis of histone modifications at THAP11-repressed genes.
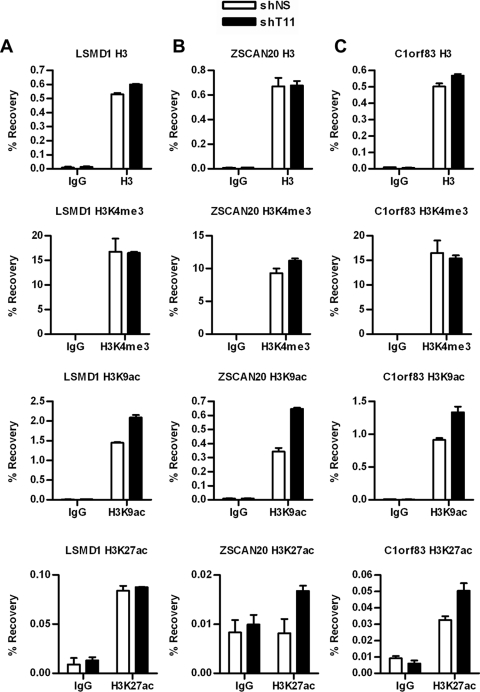
To determine whether THAP11 occupancy modulates histone acetylation and methylation patterns associated with transcriptional activation, we analyzed levels of histone H3 lysine 4 trimethylation (H3K4me3), lysine 9 acetylation (H3K9ac), and lysine 27 acetylation (H3K27ac) in SW620 THAP11 knockdown cells using ChIP assays (Fig. 5). Our results show that while the level of total histone H3 and H3K4me3 did not change at any of the target genes analyzed, H3K9ac was upregulated at the LSMD1 (Fig. 5A), ZSCAN20 (Fig. 5B), and C1orf83 (Fig. 5C) genes upon THAP11 knockdown. Interestingly, however, while enhanced acetylation of lysine 27 was observed for the ZSCAN20 and C1orf83 genes, no such changes were noticeable in LSMD1 under similar conditions. These results suggest that histone H3 hyperacetylation of target genes may contribute to their upregulation in THAP11-depleted cells.
Fig 5.
THAP11 regulates histone acetylation at repressed genes. Histone H3 modifications in SW620 cells expressing either control (shNS) or THAP11 (shT11) shRNAs as determined by ChIP using histone H3, H3K4me3, H3K9ac, and H3K27ac antibodies. Values represent the mean ± standard deviation of duplicate qPCRs from a representative experiment performed three times.
HCF-1 associates with THAP11 and regulates THAP11 target gene expression.
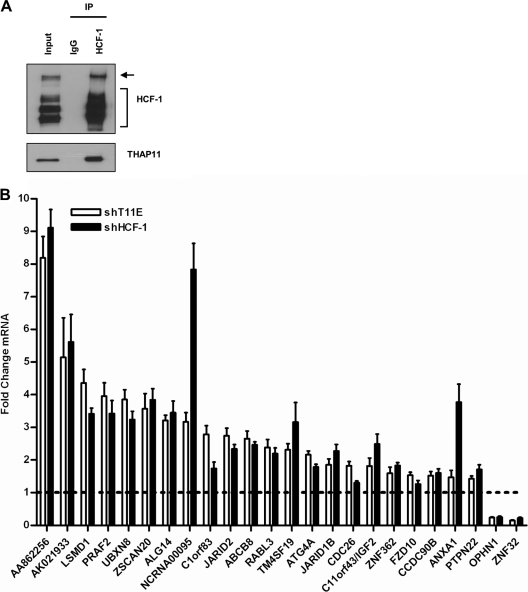
THAP11 and most other THAP proteins have recently been shown to contain a functional HBM, a four-amino-acid signature (E/DHxY) that mediates the interaction of DNA- and chromatin-associated proteins with the N-terminal Kelch domain of the transcriptional coregulator HCF-1 (10, 26). These observations inspired us to ask whether THAP11 associates with HCF-1 in colon cancer cells and if this interaction contributes to THAP11-mediated transcriptional regulation. To address this question, we immunoprecipitated HCF-1 from SW620 nuclear extract and probed the immunoprecipitate for endogenous THAP11. As shown in Fig. 6A, a significant fraction of THAP11 was found to specifically coprecipitate with HCF-1. The reciprocal experiment revealed that a small but detectable fraction of HCF-1 also coprecipitated with THAP11 (data not shown). This observation was also extended to additional colon cancer cell lines where THAP11 was found to coprecipitate with HCF-1 from nuclear extracts prepared from Colo320-HSR, SW480, and HCT-116 cells (see Fig. S3 in the supplemental material). Since THAP11 associated with HCF-1 in cells, we next analyzed the effect of HCF-1 knockdown on THAP11 target gene expression. As shown in Fig. 6B, knockdown of HCF-1 alone was sufficient to regulate transcription at THAP11 target genes in a manner qualitatively similar to that of THAP11 knockdown.
Fig 6.
HCF-1 associates with THAP11 and coregulates transcription. (A) SW620 nuclear extract was immunoprecipitated with the indicated antibody, and the immunoprecipitates (IP) were immunoblotted for HCF-1 and THAP11. The input corresponds to 10% (50 μg) of the starting material. The HCF-1 precursor and the HCF-1C subunit polypeptides are indicated by the arrow and the bracket, respectively. (B) Gene expression in SW620 cells expressing the indicated shRNAs. Values represent the mean ± standard deviation of four independent experiments. The dashed line indicates the relative expression of control shRNA-expressing cells.
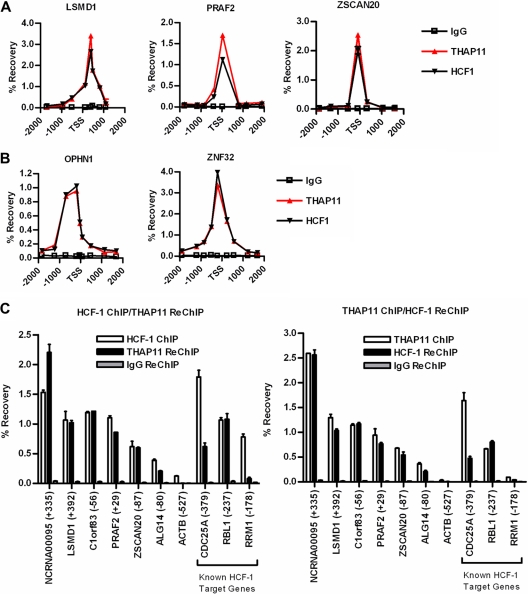
Because endogenous THAP11 and HCF-1 physically associate and coordinately regulate gene expression, we next wished to determine if HCF-1 is recruited to promoter regions of THAP11 target genes. To address this question, we used ChIP to determine the chromatin occupancy profile of HCF-1 in SW620 cells at THAP11-repressed target genes LSMD1, ALG14, NCRNA00095, AA862256, ZSCAN20, PRAF2, and C1orf83 (Fig. 7A; see Fig. S3B in the supplemental material) and THAP11-activated target genes ZNF32 and OPHN1 (Fig. 7B). Compared with the chromatin occupancy profile previously determined for THAP11 (Fig. 3), we found that the distribution of chromatin-bound HCF-1 was strikingly similar in each genomic region analyzed (Fig. 7A and B; see Fig. S3B in the supplemental material), suggesting that HCF-1 is recruited to THAP11-bound promoters. Sequential ChIP, or ChIP-reChIP, experiments subsequently confirmed that THAP11 and HCF-1 simultaneously co-occupy promoters of THAP11-regulated genes. Chromatin that was immunoprecipitated with anti-HCF-1 antibody was effectively subjected to reChIP with anti-THAP11 antibody but only at THAP11-bound target genes and not the control ACTB (β-actin) promoter or the THAP11–HCF-1-regulated RRM1 promoter (Fig. 7C, left half) (26). Similar results were observed when the order of antibodies was reversed, i.e., ChIP with anti-THAP11 and reChIP with anti-HCF-1 (Fig. 7C right half). Interestingly, we also find that THAP11 co-occupies previously identified HCF-1- and E2F-responsive promoters CDC25A and RBL1(p107) (Fig. 7C), suggesting that THAP11, in addition to E2F transcription factors, may contribute to HCF-1 binding at these genes (39).
Fig 7.
THAP11 and HCF-1 co-occupy chromatin in colon cancer cells. ChIP assay at THAP11-repressed (A) and -activated (B) genes in SW620 cells. THAP11 ChIP data from Fig. 3 are replotted here for comparison. (C) Sequential ChIP assays of SW620 cells at THAP11 and previously identified HCF-1 target genes. (Left half) HCF-1 ChIP followed by control IgG or THAP11 reChIP. (Right half) THAP11 ChIP followed by control IgG or HCF-1 reChIP. Enrichment was analyzed by qPCR and expressed as percent recovery relative to the input from the first ChIP. Values are means ± standard deviations of duplicate PCRs from a single experiment performed at least three times with similar results.
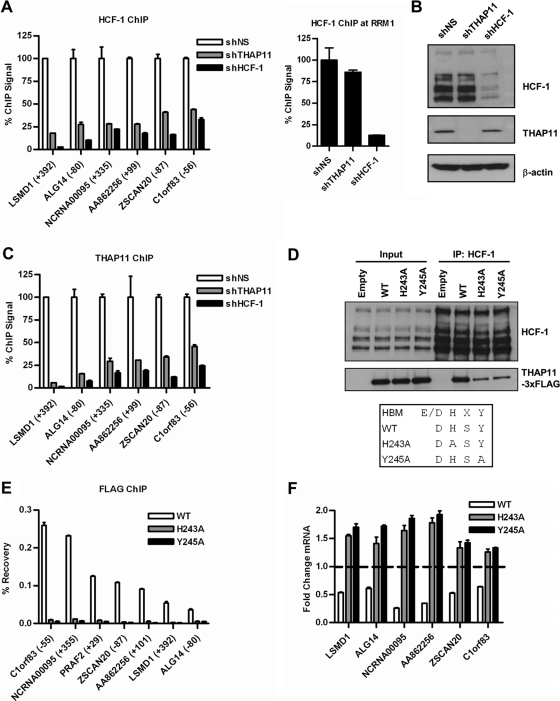
HCF-1 is not known to possess intrinsic DNA-binding activity, and its association with chromatin is thought to require interaction with DNA-bound factors (43). Accordingly, sequence-specific binding by THAP11 in a targeted genomic region may result in HCF-1 recruitment by virtue of their physical interaction. To test this directly, we performed ChIP assays for HCF-1 and THAP11 in SW620 cells depleted of either THAP11 or HCF-1 by shRNA. THAP11 knockdown results in a marked reduction in HCF-1 occupancy in each THAP11-bound genomic region examined (Fig. 8A, left half) but not the THAP1 target RRM1 (Fig. 8A, right half), suggesting that THAP11 specifically recruits HCF-1 to its target promoters. Importantly, HCF-1 protein levels were unaltered in THAP11 knockdown cells (Fig. 8B), indicating that the differential recruitment of HCF-1 observed at THAP11 target promoters does not result from a reduction in total HCF-1 protein. The reciprocal experiment, ChIP assay for THAP11 in HCF-1 knockdown cells (Fig. 8C), unexpectedly revealed that THAP11 binding to chromatin was HCF-1 dependent, suggesting that a functional THAP11–HCF-1 complex is necessary for chromatin association of both factors. However, we note that in multiple repeat experiments, HCF-1 knockdown resulted in a modest but reproducible decrease in total THAP11 protein expression (Fig. 8B; data not shown). To demonstrate that physical association with HCF-1 is indeed necessary for THAP11 targeting to chromatin, we generated THAP11 HBM mutants and determined their abilities to bind chromatin and repress transcription in HCT-116 cells, which express small amounts of endogenous THAP11. The FLAG-tagged wild-type and THAP11H243A and THAP11Y245A HBM mutant forms were stably expressed in HCT-116 cells at similar levels, but the HBM mutant forms showed strongly diminished HCF-1 interaction, as determined by coimmunoprecipitation (Fig. 8D). A ChIP assay performed with anti-FLAG monoclonal antibody to detect only ectopic THAP11 revealed a similarly strong reduction in chromatin binding of HBM mutant but not wild-type THAP11 (Fig. 8E). Consistent with their reduced chromatin association, we found that overexpressed THAP11H243A and THAP11Y245A not only failed to repress transcription but instead stimulated gene expression relative to that in empty-vector-expressing cells (Fig. 8F), suggesting that HBM mutants may contain weak dominant negative activity. From these data, we conclude that THAP11–HCF-1 complex formation is necessary for the targeting of both factors to THAP11 target promoters and subsequent transcriptional regulation.
Fig 8.
Interaction with HCF-1 is necessary for THAP11 chromatin association. (A) HCF-1 ChIP with SW620 cells expressing the indicated shRNAs. (B) Immunoblotting of THAP11 and HCF-1 in SW620 cells from panels A and C. (C) THAP11 ChIP with SW620 cells expressing the indicated shRNAs. (D) Coimmunoprecipitation of THAP11 by HCF-1 in HCT-116 cells expressing FLAG-tagged wild-type (WT) THAP11 or HCF-1 binding domain H243A and Y245A mutant proteins. Coprecipitating THAP11 was detected by FLAG immunoblotting. IP, immunoprecipitate. (E) ChIP assay of HCT-116 cells from panel D using anti-FLAG antibody. (F) Quantitative RT-PCR of THAP11 target genes in HCT-116 cells from panel D. The dashed line indicates the relative expression of empty-vector-expressing cells.
Cell growth suppression in THAP11 knockdown SW620 cells.
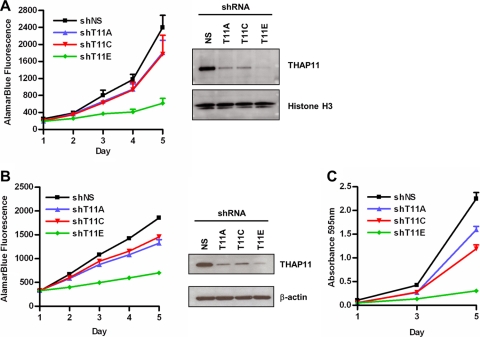
We have shown that THAP11 expression is upregulated in primary and metastatic colon cancer tumors and cell lines (Fig. 1), suggesting that increased THAP11 expression may confer a growth and/or survival advantage on expressing cells. To explore the possibility that THAP11 knockdown affects cell proliferation, we performed an alamarBlue cell enumeration assay, which detects the metabolic conversion of nonfluorescent resazurin to fluorescent resorufin in viable cells. SW620 cells were transduced with control (shNS) or THAP11 shRNA (shT11A, shT11C, or shT11E), at 2 days postransduction, selected with puromycin for an additional 2 days, and then seeded into 96-well plates for the alamarBlue assay. As shown in Fig. 9A, THAP11 knockdown resulted in a significant decrease in the number of viable cells over time with each THAP11-specific shRNA examined. Knockdown with shRNA T11E was found to be substantially more effective at reducing cell proliferation than shRNAs T11A or T11C, and this difference correlates with the extent of knockdown, as determined by immunoblotting of nuclear extracts (Fig. 9A, right half). Similar findings were obtained when the alamarBlue assay was performed with Colo320-HSR cells (Fig. 9B) or when cell proliferation was measured by crystal violet staining of SW620 cells (Fig. 9C).
Fig 9.
Knockdown of THAP11 decreases cell proliferation. Shown are results of alamarBlue cell proliferation assays of SW620 cells (A, left half) or Colo320-HSR cells (B, left half) expressing the indicated shRNAs. Values represent the mean ± standard deviation (n = 8 wells) from a representative experiment performed at least three times with similar results. Also shown are immunoblot assays of extracts from SW620 cells (A, right half) or Colo320-HSR cells (B, right half) expressing the indicated shRNAs. NS, control shRNA. (C) Crystal violet cell proliferation assay with SW620 cells expressing either control or THAP11 shRNAs. shNS, control shRNA.
DISCUSSION
In this work, we investigated the role of THAP11 in regulating the transcription and proliferation of colon cancer cells. We identified a novel set of genes that have previously not been linked to colon cancer cell function. Our work also demonstrates that HCF-1 is an obligatory partner for stable chromatin association and target gene regulation by THAP11. Based on these and additional data presented here, we propose that THAP11 plays a role in colon cancer cell function, at least in part, by regulating a subset of coding and noncoding RNAs (ncRNAs).
Several lines of evidence presented here suggest that THAP11, in complex with HCF-1, functions as a transcriptional regulator in colon cancer cells. Profiling of gene expression in THAP11 knockdown cells in conjunction with ChIP assays revealed direct THAP11-mediated transcriptional regulation. The majority of differentially expressed genes were derepressed upon THAP11 knockdown, suggesting that the THAP11–HCF-1 complex may function predominantly as a transcriptional repressor. Nonetheless, we find that 30% of the genes are repressed upon depletion of endogenous THAP11 and several are indeed direct THAP11–HCF-1 target genes. This finding indicates that THAP11–HCF-1 can both activate and repress transcription in colon cancer cells and is in good agreement with a recent genome-wide analysis of THAP11-dependent transcription in mouse ES cells which has identified both directly activated and repressed genes (10). While our work extensively characterizes the role of THAP11–HCF-1 in transcriptional regulation, the precise molecular mechanism by which THAP11–HCF-1-mediated activation and repression occur remains to be determined. As a transcriptional coregulator, HCF-1 has been alternately linked to both activation and repression of transcription (39, 42). In the context of transcriptional repression, HCF-1 is known to associate with the SIN3/HDAC histone deacetylase complex, O-linked glycosyltransferase, and protein phosphatase 1 (1, 42). Consistent with the above observations, we find that THAP11 depletion at repressed genes increases histone acetylation concomitant with increased RNAPII binding. The identity of HCF-1-associated activities that may contribute to THAP11-mediated repression and/or activation is an unanswered but pertinent question. Our studies, together with previous work, suggest that the newly discovered THAP family of proteins functions in part by regulating the transcription of their target genes (6, 10, 24–26).
THAP11 expression was found to positively correlate with disease progression in human primary tumor specimens. The increase in THAP11 expression in colon cancer tumors and cell lines suggests that THAP11-dependent transcriptional regulation may contribute to the pathogenesis of colon cancer. Consistent with this hypothesis, we find that knockdown of THAP11 in SW620 colon cancer cells results in a significant decrease in cell proliferation. Interestingly, however, the THAP11-dependent gene expression profile is largely devoid of genes with previously known functions in tumorigenesis or cell proliferation. Exceptions to this generalization are annexin A1, a member of the annexin family of calcium-dependent phospholipid-binding proteins, and PRAF2, a novel proapoptotic Bcl-xL/Bcl2-interacting protein (40). The role of annexin A1 in cancer cell function is complex, but several reports have suggested that annexin A1 can function in an antiproliferative capacity (15, 17, 20). We speculate that increased PRAF2 and/or annexin A1 gene expression resulting from THAP11 knockdown may contribute to the cell proliferation defect observed in these cells. Alternatively, the proliferation defect resulting from THAP11 knockdown may arise from the cumulative effect of multiple dysregulated genes. In this context, we also note that, in addition to protein coding genes, several annotated or putative long ncRNAs were also identified as direct targets of THAP11-mediated transcriptional repression, including NCRNA00095 and AA862256. The fortuitous discovery of these ncRNAs as THAP11-regulated transcripts by microarray-based gene expression profiling likely reflects their previous but erroneous annotation as protein coding genes with subsequent inclusion in the microarray design. Since many long ncRNAs have been discovered to be regulated by the same transcriptional control mechanisms utilized by protein coding mRNAs (14), we speculate that additional long ncRNAs are likely regulated by THAP11. It is now well established that long ncRNAs contribute to a diverse array of biological functions, including transcriptional regulation, cell growth, and apoptosis, but the vast majority of long ncRNAs remain uncharacterized (37). We were surprised to observe such a small THAP11 target gene set in our microarray analysis. One possible explanation for this is that THAP11 target genes are individually sensitive to a given level of THAP11 in cells, while knockdown by THAP11 A and C constructs decreases the level of THAP11 to a point where only the gene set identified in this microarray are uncovered. The use of a more robust knockdown construct or THAP11 knockout cells may uncover additional and novel THAP11 targets. Additional studies are required to identify the full complement of THAP11-regulated transcripts, as well as the protein coding or noncoding gene(s) downstream of THAP11 that contributes to cell proliferation.
THAP11 associates with and recruits HCF-1 to promoters, and all of the THAP11 target genes analyzed here contain one or more putative THAP11-binding sites near their respective transcription start sites. The stable association of the THAP11–HCF-1 complex on chromatin was unexpectedly found to require both proteins; depletion of either protein was sufficient to disrupt the binding of both factors at THAP11 target genes. This mutual interdependence of HCF-1 and a sequence-specific transcription factor has not been described previously and may represent a unique feature of THAP11. Recent structural analyses of prototypical THAP domains bound to their respective DNA elements have revealed that THAP proteins bind DNA in a bipartite manner, making simultaneous major- and minor-groove DNA contacts (5, 35). Nonspecific contacts with the sugar-phosphate backbone of the minor groove are critical for THAP domain binding to DNA and are mediated primarily by basic amino acids present in a flexible loop structure positioned between the zinc-coordinating residues and the conserved AVPTIF box. However, the length of this loop structure is dramatically shortened in THAP11 and has been suggested to result in reduced minor-groove contacts and a diminished affinity for DNA (35). Since HCF-1 does not bind DNA directly, it appears unlikely that HCF-1 is functioning as a true DNA-binding heterodimeric partner of THAP11. Alternatively, HCF-1 may function as a non-DNA-binding heterodimeric partner or instead as a bridge between THAP11 and another transcription factor. Indeed, sequence-based analysis of the THAP11-bound promoters described here has identified several high-probability Sp1-binding sites close to THAP11-bound regions. Sp1 has been previously shown to associate with HCF-1 but in a region separate from the THAP11-interacting Kelch domain (13, 42), suggesting that the binding of Sp1 and that of THAP11 to HCF-1 are not likely to be mutually exclusive. We hypothesize that THAP11 guides HCF-1 to promoters harboring a THAP11 response element but that stable association of the THAP11–HCF-1 complex requires additional interaction between HCF-1 and factors like Sp1.
The role of HCF-1 in cell proliferation and cell cycle progression has been attributed largely to HCF-1-dependent transactivation of E2F target genes during G1/S-phase progression (18, 39, 43). Our results suggest that THAP11-dependent gene regulation is a novel and previously unknown component of HCF-1-dependent cell proliferation. Consistent with this interpretation, we note that other non-E2F factors have also recently been linked to HCF-1-dependent cell proliferation. HCF-1 has been shown to link the transcriptional coactivator and deubiquitinating enzyme BAP1 with YY1 to regulate cell proliferation and growth control genes (44), while Mazars et al. have shown that THAP1, rather than E2F proteins, recruits HCF-1 to activate the cell cycle-regulated gene RRM1 in proliferating endothelial cells (26). Interestingly, we find that, in addition to the novel gene set described here, THAP11 and HCF-1 are also corecruited at the cell cycle- and E2F-regulated genes CDC25A and RBL1. This finding raises the intriguing possibility that THAP11 complements or cooperates with E2F proteins for the recruitment of HCF-1 at some cell cycle-regulated genes. These findings further underscore the notion that THAP proteins represent a large but poorly characterized family of HCF-1-associated transcription factors with potential roles in cell proliferation, development, and apoptosis.
Our findings contrast with a previous report that suggests that THAP11 is downregulated in several human cancers and functions as a cell growth suppressor through direct transcriptional repression of MYC (45). These conflicting results may represent a tissue-specific role for THAP11 in human colon cancer versus other types of cancer. However, we note that the assertion by Zhu et al. that THAP11 is largely repressed in human cancers is based solely on mRNA expression data and may not accurately reflect THAP11 protein status. Nonetheless, close inspection of the THAP11 expression data from their multiple-tissue Northern blot array (Fig. 1 in reference 45) suggests elevated THAP11 mRNA in several tumor versus normal colon tissues, consistent with our immunohistochemistry results. We were unable to detect endogenous THAP11 or HCF-1 at the MYC promoter in SW620 cells (data not shown), and knockdown of either factor similarly failed to increase MYC gene expression (data not shown), suggesting that MYC is not a THAP11 target gene in colon cancer cells and providing a potential explanation for the contrasting results regarding THAP11-dependent cell proliferation.
THAP11 has recently been identified as a critical factor in the maintenance of ES cell pluripotency and proliferation (9). Interestingly, small-interfering-RNA-mediated knockdown of THAP11 was reported not to affect ES cell proliferation significantly, despite an approximately 85% reduction in THAP11 expression (9). These findings are in good agreement with our results indicating that the significant but incomplete knockdown observed with shRNAs T11A and T11C results in a modest cell proliferation defect while nearly complete depletion of THAP11 with shRNA T11E yields a markedly enhanced reduction of cell proliferation.
In summary, our studies show for the first time that THAP11 expression increases dramatically in colon cancer and plays an important role in colon cancer cell proliferation. We show that THAP11 functions as a transcriptional regulator that requires HCF-1 as an obligatory partner in DNA binding and target gene regulation. Finally our study identifies a novel set of genes that previously have not been linked to cancer cell function. Future studies will investigate the roles of these novel protein coding and ncRNA target genes, identify the full complement of THAP11 and THAP11–HCF-1 target genes, and determine whether THAP11 has a role in other human cancers.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH/NCI grant R01 CA133755 (D.C.). J.B.P. is supported by NIH/NCI Institutional NRSA Training Program in Signal Transduction and Cancer grant T32 CA070085. We gratefully acknowledge the Northwestern University Cell Imaging Facility, Flow Cytometry Facility, and Genomics Core Facility, which are supported by Cancer Center Support grant NCI CA060553.
We have no conflict of interest to declare.
Footnotes
Published ahead of print 27 February 2012
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Ajuh PM, et al. 2000. Association of a protein phosphatase 1 activity with the human factor C1 (HCF) complex. Nucleic Acids Res. 28:678–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Balakrishnan MP, et al. 2011. THAP5 is a DNA-binding transcriptional repressor that is regulated in melanoma cells during DNA damage-induced cell death. Biochem. Biophys. Res. Commun. 404:195–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Balakrishnan MP, et al. 2009. THAP5 is a human cardiac-specific inhibitor of cell cycle that is cleaved by the proapoptotic Omi/HtrA2 protease during cell death. Am. J. Physiol. Heart Circ. Physiol. 297:H643–H653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bessière D, et al. 2008. Structure-function analysis of the THAP zinc finger of THAP1, a large C2CH DNA-binding module linked to Rb/E2F pathways. J. Biol. Chem. 283:4352–4363 [DOI] [PubMed] [Google Scholar]
- 5. Campagne S, Saurel O, Gervais V, Milon A. 2010. Structural determinants of specific DNA-recognition by the THAP zinc finger. Nucleic Acids Res. 38:3466–3476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cayrol C, et al. 2007. The THAP-zinc finger protein THAP1 regulates endothelial cell proliferation through modulation of pRB/E2F cell-cycle target genes. Blood 109:584–594 [DOI] [PubMed] [Google Scholar]
- 7. Clouaire T, et al. 2005. The THAP domain of THAP1 is a large C2CH module with zinc-dependent sequence-specific DNA-binding activity. Proc. Natl. Acad. Sci. U. S. A. 102:6907–6912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Core LJ, Waterfall JJ, Lis JT. 2008. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322:1845–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dejosez M, et al. 2008. Ronin is essential for embryogenesis and the pluripotency of mouse embryonic stem cells. Cell 133:1162–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dejosez M, et al. 2010. Ronin/Hcf-1 binds to a hyperconserved enhancer element and regulates genes involved in the growth of embryonic stem cells. Genes Dev. 24:1479–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Freiman RN, Herr W. 1997. Viral mimicry: common mode of association with HCF by VP16 and the cellular protein LZIP. Genes Dev. 11:3122–3127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fuchs T, et al. 2009. Mutations in the THAP1 gene are responsible for DYT6 primary torsion dystonia. Nat. Genet. 41:286–288 [DOI] [PubMed] [Google Scholar]
- 13. Gunther M, Laithier M, Brison O. 2000. A set of proteins interacting with transcription factor Sp1 identified in a two-hybrid screening. Mol. Cell. Biochem. 210:131–142 [DOI] [PubMed] [Google Scholar]
- 14. Guttman M, et al. 2009. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458:223–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Guzmán-Aránguez A, et al. 2005. Differentiation of human colon adenocarcinoma cells alters the expression and intracellular localization of annexins A1, A2, and A5. J. Cell. Biochem. 94:178–193 [DOI] [PubMed] [Google Scholar]
- 16. Hewitt RE, et al. 2000. Validation of a model of colon cancer progression. J. Pathol. 192:446–454 [DOI] [PubMed] [Google Scholar]
- 17. Hsiang CH, Tunoda T, Whang YE, Tyson DR, Ornstein DK. 2006. The impact of altered annexin I protein levels on apoptosis and signal transduction pathways in prostate cancer cells. Prostate 66:1413–1424 [DOI] [PubMed] [Google Scholar]
- 18. Julien E, Herr W. 2003. Proteolytic processing is necessary to separate and ensure proper cell growth and cytokinesis functions of HCF-1. EMBO J. 22:2360–2369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kent WJ, et al. 2002. The human genome browser at UCSC. Genome Res. 12:996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lecona E, et al. 2008. Upregulation of annexin A1 expression by butyrate in human colon adenocarcinoma cells: role of p53, NF-Y, and p38 mitogen-activated protein kinase. Mol. Cell. Biol. 28:4665–4674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee TI, Johnstone SE, Young RA. 2006. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat. Protoc. 1:729–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lu R, Misra V. 2000. Zhangfei: a second cellular protein interacts with herpes simplex virus accessory factor HCF in a manner similar to Luman and VP16. Nucleic Acids Res. 28:2446–2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lu R, Yang P, Padmakumar S, Misra V. 1998. The herpesvirus transactivator VP16 mimics a human basic domain leucine zipper protein, luman, in its interaction with HCF. J. Virol. 72:6291–6297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Macfarlan T, et al. 2005. Human THAP7 is a chromatin-associated, histone tail-binding protein that represses transcription via recruitment of HDAC3 and nuclear hormone receptor corepressor. J. Biol. Chem. 280:7346–7358 [DOI] [PubMed] [Google Scholar]
- 25. Macfarlan T, Parker JB, Nagata K, Chakravarti D. 2006. Thanatos-associated protein 7 associates with template activating factor-Ibeta and inhibits histone acetylation to repress transcription. Mol. Endocrinol. 20:335–347 [DOI] [PubMed] [Google Scholar]
- 26. Mazars R, et al. 2010. The thap-zinc finger protein thap1 associates with coactivator HCF-1 and O-GLcNAc transferase: a link between DYT6 and DYT3 dystonias. J. Biol. Chem. 285:13364–13371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Meister G, Tuschl T. 2004. Mechanisms of gene silencing by double-stranded RNA. Nature 431:343–349 [DOI] [PubMed] [Google Scholar]
- 28. Morgenstern JP, Land H. 1990. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 18:3587–3596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nelson JD, Denisenko O, Bomsztyk K. 2006. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat. Protoc. 1:179–185 [DOI] [PubMed] [Google Scholar]
- 30. Perissi V, Jepsen K, Glass CK, Rosenfeld MG. 2010. Deconstructing repression: evolving models of co-repressor action. Nat. Rev. Genet. 11:109–123 [DOI] [PubMed] [Google Scholar]
- 31. Provenzani A, et al. 2006. Global alterations in mRNA polysomal recruitment in a cell model of colorectal cancer progression to metastasis. Carcinogenesis 27:1323–1333 [DOI] [PubMed] [Google Scholar]
- 32. Reynolds A, et al. 2004. Rational siRNA design for RNA interference. Nat. Biotechnol. 22:326–330 [DOI] [PubMed] [Google Scholar]
- 33. Roussigne M, Cayrol C, Clouaire T, Amalric F, Girard JP. 2003. THAP1 is a nuclear proapoptotic factor that links prostate-apoptosis-response-4 (Par-4) to PML nuclear bodies. Oncogene 22:2432–2442 [DOI] [PubMed] [Google Scholar]
- 34. Roussigne M, et al. 2003. The THAP domain: a novel protein motif with similarity to the DNA-binding domain of P element transposase. Trends Biochem. Sci. 28:66–69 [DOI] [PubMed] [Google Scholar]
- 35. Sabogal A, Lyubimov AY, Corn JE, Berger JM, Rio DC. 2010. THAP proteins target specific DNA sites through bipartite recognition of adjacent major and minor grooves. Nat. Struct. Mol. Biol. 17:117–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 37. Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. 2010. Non-coding RNAs: regulators of disease. J. Pathol. 220:126–139 [DOI] [PubMed] [Google Scholar]
- 38. Tamiya G. 2009. Transcriptional dysregulation: a cause of dystonia? Lancet Neurol. 8:416–418 [DOI] [PubMed] [Google Scholar]
- 39. Tyagi S, Chabes AL, Wysocka J, Herr W. 2007. E2F activation of S phase promoters via association with HCF-1 and the MLL family of histone H3K4 methyltransferases. Mol. Cell 27:107–119 [DOI] [PubMed] [Google Scholar]
- 40. Vento MT, et al. 2010. Praf2 is a novel Bcl-xL/Bcl-2 interacting protein with the ability to modulate survival of cancer cells. PLoS One 5:e15636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wei JJ, et al. 2006. Ethnic differences in expression of the dysregulated proteins in uterine leiomyomata. Hum. Reprod. 21:57–67 [DOI] [PubMed] [Google Scholar]
- 42. Wysocka J, Myers MP, Laherty CD, Eisenman RN, Herr W. 2003. Human Sin3 deacetylase and trithorax-related Set1/Ash2 histone H3-K4 methyltransferase are tethered together selectively by the cell-proliferation factor HCF-1. Genes Dev. 17:896–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wysocka J, Reilly PT, Herr W. 2001. Loss of HCF-1-chromatin association precedes temperature-induced growth arrest of tsBN67 cells. Mol. Cell. Biol. 21:3820–3829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yu H, et al. 2010. The ubiquitin carboxyl hydrolase BAP1 forms a ternary complex with YY1 and HCF-1 and is a critical regulator of gene expression. Mol. Cell. Biol. 30:5071–5085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhu CY, et al. 2009. Cell growth suppression by thanatos-associated protein 11 (THAP11) is mediated by transcriptional downregulation of c-Myc. Cell Death Differ. 16:395–405 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.