Abstract
The mechanism of hepatitis E virus (HEV) replication remains largely unknown. Here we demonstrate that HEV replication requires an active ubiquitin-proteasome system and that proteasome inhibitors affect HEV replication, possibly by inhibition of viral transcription or/and translation without a significant effect on cellular translation. Overexpression of ubiquitin in inhibitor-treated cells partially reverses the inhibitor effect on HEV replication. The results suggest that HEV replication requires interactions with proteasome machinery, which could be a potential therapeutic target against HEV.
TEXT
The cellular ubiquitin-proteasome system (UPS) is important for intracellular protein degradation in eukaryotic cells (40, 46). The UPS is composed of ubiquitination and substrate-degrading machinery. Ubiquitination is the conjugation of proteins with ubiquitin and occurs through the sequential enzymatic reactions of an E1 activating enzyme, E2 conjugation enzyme, and E3 ligase (17). Viruses manipulate the infrastructure and metabolism of their host cell to effect their own survival. UPS has been implicated in the infection cycle and virus-host interplay of several viruses (3, 9, 28, 36, 51). Bortezomib is an FDA-approved proteasome inhibitor that has demonstrated clinical efficacy in the treatment of multiple myeloma (7, 10, 27). Therefore, in this study we examined the role of UPS in the replication of hepatitis E virus (HEV) and evaluated the potential use of UPS inhibitors as therapeutic agents against HEV infection.
HEV, a nonenveloped single-strand positive-sense RNA virus in the family Hepeviridae (30, 32), is an important but understudied human pathogen (2, 11, 31, 33). The genome of HEV is ∼7.2 kb and contains a 5′-m7G cap (20) and three open reading frames (ORFs) (39). The ORF1 protein possesses domains for replicase enzymes (25) and among these, functional activities of RdRp (1), Hel (21, 22), and MetT (29) have been experimentally verified. ORF2 encodes the viral capsid protein (16, 50). ORF3 encodes a small multifunctional protein that interacts with various signaling pathways (5, 6, 13, 24, 26, 33–35, 43–45, 48, 49). The ORF2 and ORF3 proteins are translated from a 2.2-kb subgenomic RNA (15, 18). The expression of the ORF3 protein is not required for virus replication, virion assembly, or infection in vitro (12, 14).
It has been reported that proteasome inhibitors affect the replication of herpesviruses (9), vaccinia virus (36), influenza virus (47), human immunodeficiency virus (38), and cytomegaloviruses (42). Many viruses encode proteins that can modify the host's ubiquitin machinery, (19). Recently, a papain-like cysteine protease has been described as a deubiquitinating enzyme in HEV (23), indicating that a ubiquitin system may influence the life cycle of HEV.
For all experiments, a subclone of a human hepatocellular carcinoma cell line, Huh7-S10-3, which is permissive for HEV replication, was used, and the cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum under a 5% CO2 atmosphere at 37°C. Transfected cells were maintained under the same conditions except at 34.5°C.
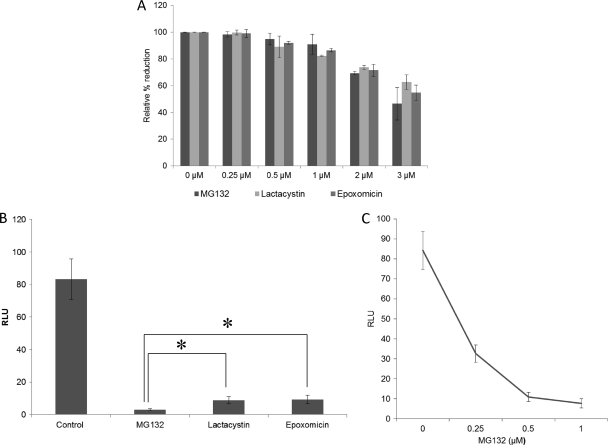
First, to determine whether proteasome activity is required for HEV replication, we tested the effects of proteasome inhibitors MG132, lactacystin, and epoxomicin (Sigma) on HEV replication. The toxicities of the inhibitors were tested by the alamarBlue assay (Invitrogen), and we showed that there was >80% cell survival when concentrations of inhibitors were less than or equal to 1 μM (Fig. 1A). Thus, for all further experiments in this study, the concentration of inhibitors we used was 1 μM or less.
Fig 1.
The proteasome inhibitors significantly reduced the levels of HEV replication. (A) Toxicity of inhibitors to Huh7 S10 cells. The results shown are from an alamarBlue assay for Huh7 S10 cells treated with the inhibitors MG132, lactacystin, and epoxomicin. The assay was performed on the fourth day after treatment. The concentrations of inhibitors are indicated. Mean values from three independent experiments are plotted. (B) HEV replication is reduced by treatment with proteasome inhibitors. Relative luciferase activities are shown for Huh7 S10 cells transfected with capped RNA transcript of the pSK-HEV2RLuc clone. Inhibitor treatment started 1 day posttransfection, and the concentration of inhibitors was 1 μM. A luciferase assay was performed on the fifth day posttransfection. Mean values from six independent experiments are plotted. Statistical analysis was performed using analysis of variance followed by Dunnett's procedure, and significance was set at a P level of <0.05 (indicated with an asterisk). Data analysis was performed using JMP9 software. (C) Effect of MG132 on HEV replication. Relative luciferase activities are shown for Huh7 S10 cells transfected with capped RNA transcript of the pSK-HEV2RLuc clone. Inhibitor treatment started 1 day posttransfection, and the concentration of MG132 used is indicated. A luciferase assay was performed on the fifth day posttransfection. Mean values from three independent experiments are plotted.
HEV replicon expressing the Renilla luciferase (Rluc) gene system (designated pSK-HEV-2RLuc) was developed previously using the genotype 1 human HEV infectious clone pSK-HEV-2 (4). The capped RNA transcript of the pSK-HEV-2RLuc clone was transfected into Huh7-S10-3 cells by using the DMRIE-C reagent (Invitrogen). UPS inhibitors were added to culture medium at 24 h posttransfection. The luciferase activities were measured with a dual luciferase reporter assay system (Promega) at 5 days posttransfection. Firefly luciferase RNA was cotransfected with HEV Rluc replicon RNA to normalize the Renilla luciferase signal. All the inhibitors tested in this study caused a significant reduction in the level of virus replication, suggesting that the UPS is important for HEV replication. The MG132 inhibitor had a more pronounced effect on virus replication than other inhibitors (Fig. 1B). Furthermore, we found that this inhibition of HEV replication was concentration dependent (Fig. 1C).
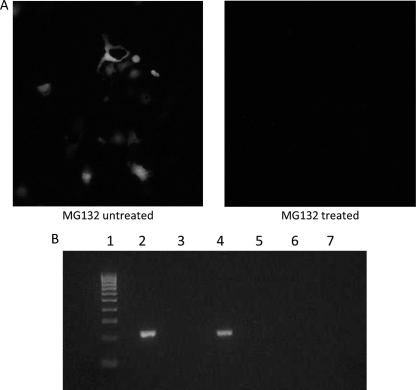
To investigate which specific step(s) of the HEV replication cycle might be affected by lack of proteasome activity, we performed an immunofluorescence assay (IFA) to detect viral capsid protein synthesis, and we performed negative-strand-specific reverse transcription-PCR (RT-PCR) to detect replicative negative-strand viral RNA. Briefly, Huh7 cells were transfected with the full-length capped RNA transcripts of the pSK-HEV2 infectious clone, and capsid protein synthesis was monitored by IFA (8). When 1 μM MG132 inhibitor was added to culture medium at 24 h posttransfection, no capsid protein synthesis was detected by IFA (Fig. 2A), further confirming that HEV replication requires an active ubiquitin proteasome system.
Fig 2.
MG132 inhibits viral transcription and/or translation. (A) Immunofluorescent staining of a subclone of Huh7 cells transfected with similar amounts of capped full-length RNA transcripts. (Left) MG132 untreated; (right) MG132 treated. Inhibitor treatment started 1 day posttransfection, and cells were stained for HEV ORF2 protein by using chimpanzee 1313 anti-HEV immune serum. (B) Detection of HEV replication by strand-specific anchored RT-PCR. A subclone of Huh7 cells was transfected with similar amounts of capped full-length RNA transcript, 1 μM MG132 treatment started 1 day posttransfection, and cells were harvested on the fifth day posttransfection. For detection of replicative negative-sense viral RNA, a strand-specific anchored RT-PCR was carried out. Lane 1, 100-bp marker; lane 2, RT-PCR results for positive-control negative-strand RNA transcript generated by in vitro transcription; lane 3, PCR performed with positive-control negative-strand RNA transcript generated by in vitro transcription (reaction without RT); lane 4, RT-PCR performed with RNA isolated from full-length capped RNA transfected cells; lane 5, PCR performed with RNA isolated from full-length capped RNA transfected cells (reaction without RT); lane 6, RT-PCR performed with RNA isolated from full-length capped RNA-transfected cells and treated with 1 μM MG132; lane 7, RT-PCR performed with mock-transfected cells.
For the detection of negative-strand replicative viral RNA, a strand-specific anchored RT-PCR was carried out essentially as described previously (41). RNA was reverse transcribed with a forward primer (5′-GGGGGGGGGGGGCCGCGCCCATACTTTCGATGA-3′), and both the first and second amplifications were carried out using the forward poly(G) primer (5′-GGGGGGGGGGGGGGGGG-3′) and reverse primer (5′-CAGGGAGCGCGGAACGGAACGCAG-3′). As for a positive control, a negative-strand HEV RNA was prepared by in vitro transcription of a PCR DNA template amplified with a forward primer (5′-CCAGCAGTATTCAAAGACC-3′) and a reverse primer (5′-GATCATCTCCCTATAGTGAGTCGTATTATTTCAGGGAGCGCGAAACGC-3′; T7 polymerase promoter sequence, underlined). Huh7 cells were transfected with the capped full-length RNA transcripts of the pSK-HEV2 infectious clone, and cells were treated with 1 μM MG132 inhibitor at 1 day posttransfection and harvested on the fifth day posttransfection. No negative-strand viral RNA was detected when cells were treated with MG132 inhibitor (Fig. 2B), suggesting that the proteasome activity is needed for the replication of the HEV genome, possibly by inhibition of viral transcription or translation or both. We believe that inhibition of early or multiple stages of virus replication will result in little or no synthesis of negative-strand RNA, thus explaining our observation of the absolute negative result on the detection of the negative-strand RNA.
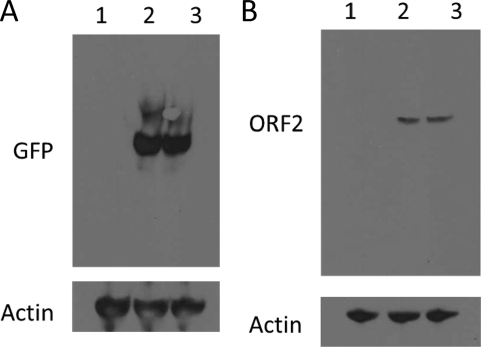
Due to the long duration of treatment, we tested the cytotoxicities of the inhibitors in cell culture to determine the concentration ranges of inhibitors. It is possible that inhibitors may impair cellular translation and thus could attribute to the inhibition of viral replication. Therefore, we subsequently tested the effects of the inhibitor drug treatments on green fluorescent protein (GFP) synthesis. In addition, in another set of experiments we expressed part of the viral capsid protein that is known to form virus-like particles (VLPs) in the presence and absence of MG132. Huh7 cells were transfected, inhibitor treatment was the same as described above, and pAcGFP N1 and pTrix-neo-ORF2 (with deletion of amino acids 1 to 111 [Δ1-111]) were transfected to Huh7 cells. Immunoblotting was performed with anti-GFP rabbit polyclonal antibody (1:500), anti-HEV chimpanzee polyclonal serum (1:200), and anti-actin goat polyclonal antibody (1:200) (all from Santa Cruz Biotechnology) and with appropriate secondary antibody. Comparable levels of GFP, capsid protein, and actin were observed in drug-treated and untreated cells (Fig. 3A and B). Also, when the full-length RNA genome of HEV was transfected into Huh7 cells in the presence of MG123, no capsid protein was detected. These results strongly suggest that there is inhibition of viral replication without a significant effect on cellular translation.
Fig 3.
The inhibitory effect of MG132 on HEV replication does not result from the inhibition of translation. (A) Effect of MG132 treatment on GFP synthesis. A subclone of Huh7 cells was transfected with a similar amount of pAcGFP N1 plasmid in six-well plates. Inhibitor treatment started 1 day posttransfection, cells were harvested on the fifth day posttransfection, and immunoblotting was performed with anti-GFP polyclonal serum produced in a rabbit. Lane 1, mock-transfected cells; lane 2, pAcGFP N1-transfected cells; lane 3, pAcGFP N1-transfected cells with 1 μM MG132 treatment. (B) Effect of MG132 treatment on ORF2 protein synthesis. A subclone of Huh7 cells were transfected with a similar amount of pTrix-neo-ORF2 (Δ1–111) plasmid in six-well plates. Inhibitor treatment started 1 day posttransfection, cells were harvested on the fifth day posttransfection, and immunoblotting was performed with chimpanzee 1313 anti-HEV immune serum. Lane 1, mock-transfected cells; lane 2, pTrix-neo-ORF2 (Δ1–111)-transfected cells; lane 3, pTrix-neo-ORF2 (Δ1–111)-transfected cells treated with 1 μM MG132.
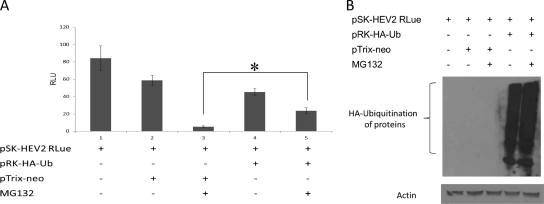
It has been shown that MG132 reduces the pool of free ubiquitin in cells (28). MG132 inhibits budding of human parainfluenza virus 5 by depletion of free ubiquitin in cells by blocking the 26S proteasomal degradation of polyubiquitinated proteins (37). Therefore, to determine whether the observed inhibition of HEV replication by MG132 was due to depletion of free ubiquitin, we cotransfected plasmid pRK5-HA-ubiquitin (kindly provided by Ted Dawson [Addgene plasmid]) with capped viral RNA transcript, and the effect on viral replication was monitored. An increase in viral replication was observed when the cells were cotransfected with capped HEV RNA transcript with pRK5-HA-ubiquitin compared to cells cotransfected with capped HEV RNA transcript with the pTrix neo plasmid (Fig. 4A). Immunoblotting was performed to detect the expression of hemagglutinin (HA)-ubiquitin in the transfected cells by using an anti-HA tag monoclonal antibody produced in mice (Sigma) (Fig. 4B). In the case of overexpression of HA-ubiquitin and MG132 treatment, recycling of ubiquitin molecules may be affected; in this case, the pool of free HA-ubiquitin must be large enough to cause depletion of free HA-ubiquitin but sufficient to restore virus replication. Therefore, we believe that the reason that there was no difference between lanes 4 (without MG132) and 5 (with MG132) is likely due to the overexpression of HA-ubiquitin. Viral replication was not completely restored, and this may have been due to cotransfection efficiency. Nevertheless, the results suggest that depletion of free ubiquitin may be important for inhibition of viral replication.
Fig 4.
Overexpression of HA-ubiquitin partially restores virus replication. (A) Effect of HA-ubiquitin overexpression on HEV replication in the context of MG132 treatment. In six-well plates, cotransfection of capped RNA transcripts of the pSK-HEV2RLuc clone and pRK-HA-Ub/pTrix-neo was carried out. MG132 treatment started 1 day posttransfection. A luciferase assay was performed on the fifth day posttransfection. Mean relative light unit (RLU) values from three independent experiments are plotted. Statistical analysis was performed using analysis of variance followed by contrast procedure, and significance was set at a P level of <0.05 (indicated with an asterisk). Data analysis was performed using JMP9 software. (B) Representative results from an immunoblot assay performed with the anti-HA tag monoclonal antibody of the experiment shown in panel A.
In summary, in this study we demonstrated an important role of UPS in the life cycle of HEV. Proteasome inhibitors affected viral replication, possibly by inhibition of viral transcription or/and translation. The results strongly suggested that an active proteasome system is essential for HEV replication, and therefore proteasome inhibitors could be useful as therapeutics against HEV infection.
ACKNOWLEDGMENTS
This study was supported by grants from the U.S. National Institutes of Health (R01 AI074667 and R01 AI050611).
We thank Laura Cordoba, Scott Kenny, Dianjun Cao, and Barbara Dryman for their support. We also thank Suzanne U. Emerson and Robert H. Purcell for kindly providing the chimpanzee 1313 anti-HEV antisera, the pSK-HEV-2 infectious clone, and the Huh7-S10-3 subclone of the hepatocellular carcinoma cell line.
Footnotes
Published ahead of print 21 March 2012
REFERENCES
- 1. Agrawal S, Gupta D, Panda SK. 2001. The 3′ end of hepatitis E virus (HEV) genome binds specifically to the viral RNA polymerase (RdRp). Virology 282:87–101 [DOI] [PubMed] [Google Scholar]
- 2. Arankalle VA, et al. 1994. Seroepidemiology of water-borne hepatitis in India and evidence for a third enterically-transmitted hepatitis agent. Proc. Natl. Acad. Sci. U. S. A. 91:3428–3432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Burch AD, Weller SK. 2004. Nuclear sequestration of cellular chaperone and proteasomal machinery during herpes simplex virus type 1 infection. J. Virol. 78:7175–7185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cao D, Huang YW, Meng XJ. 2010. The nucleotides on the stem-loop RNA structure in the junction region of the hepatitis E virus genome are critical for virus replication. J. Virol. 84:13040–13044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chandra V, Kar-Roy A, Kumari S, Mayor S, Jameel S. 2008. The hepatitis E virus ORF3 protein modulates epidermal growth factor receptor trafficking, STAT3 translocation, and the acute-phase response. J. Virol. 82:7100–7110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chandra V, Kalia M, Hajela K, Jameel S. 2010. The ORF3 protein of hepatitis E virus delays degradation of activated growth factor receptors by interacting with CIN85 and blocking formation of the Cbl-CIN85 complex. J. Virol. 84:3857–3867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen D, Frezza M, Schmitt S, Kanwar J, Dou QP. 2011. Bortezomib as the first proteasome inhibitor anticancer drug: current status and future prospective. Current Cancer Drug Target 11:23953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cordoba L, et al. 2011. Three amino acid mutations (F51L, T59A, and S390L) in the capsid protein of the hepatitis E virus collectively contribute to virus attenuation. J. Virol. 85:5338–5349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Delboy MG, Roller DG, Nicola AV. 2008. Cellular proteasome activity facilitates herpes simplex virus entry at a postpenetration step. J. Virol. 82:3381–3390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Edelmann MJ, Benjamin N, Kessler BM. 2011. Pharmacological targets in the ubiquitin system offer new ways of treating cancer, neurodegenerative disorders and infectious diseases. Expert Rev. Mol. Med. 13:e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Emerson SU, Purcell RH. 2003. Hepatitis E virus. Rev. Med. Virol. 13:145–154 [DOI] [PubMed] [Google Scholar]
- 12. Emerson SU, et al. 2004. In vitro replication of hepatitis E virus (HEV) genomes and of an HEV replicon expressing green fluorescent protein. J. Virol. 78:4838–4846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Emerson SU, Nguyen H, Torian U, Purcell RH. 2006. ORF3 protein of hepatitis E virus is not required for replication, virion assembly, or infection of hepatoma cells in vitro. J. Virol. 80:10457–10464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Emerson SU, et al. 2010. Release of genotype 1 hepatitis E virus from cultured hepatoma and polarized intestinal cells depends on open reading frame 3 protein and requires an intact PXXP motif. J. Virol. 84:9059–9069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Graff J, Torian U, Nguyen H, Emerson SU. 2006. A bicistronic subgenomic mRNA encodes both the ORF2 and ORF3 proteins of hepatitis E virus. J. Virol. 80:5919–5926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Graff J, et al. 2008. Mutations within potential glycosylation sites in the capsid protein of hepatitis E virus prevent the formation of infectious virus particles. J. Virol. 82:1185–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hershko A, Ciechanover A. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425–479 [DOI] [PubMed] [Google Scholar]
- 18. Ichiyama K, et al. 2009. Determination of the 5′-terminal sequence of subgenomic RNA of hepatitis E virus strains in cultured cells. Arch. Virol. 154:1945–1951 [DOI] [PubMed] [Google Scholar]
- 19. Isaacson MK, Ploegh HL. 2009. Ubiquitination, ubiquitin-like modifiers, and deubiquitination in viral infection. Cell Host Microbe 5:559–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kabrane-Lazizi Y, Meng XJ, Purcell RH, Emerson SU. 1999. Evidence that the genomic RNA of hepatitis E virus is capped. J. Virol. 73:8848–8850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Karpe YA, Lole KS. 2010. NTPase and 5′ to 3′ RNA duplex-unwinding activities of hepatitis E virus helicase domain. J. Virol. 84:3595–3602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Karpe YA, Lole KS. 2010. RNA 5′ triphosphatase activity associated with hepatitis E virus helicase domain. J. Virol. 84:9637–9641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Karpe YA, Lole KS. 2011. Deubiquitinating activity associated with hepatitis E virus putative papain like cysteine protease. J. Gen. Virol. 92:2088–2092 [DOI] [PubMed] [Google Scholar]
- 24. Kar-Roy A, Korkaya H, Oberoi R, Lal SK, Jameel S. 2004. The hepatitis E virus open reading frame 3 protein activates ERK through binding and inhibition of the MAPK phosphatase. J. Biol. Chem. 279:28345–28357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Koonin EV, et al. 1992. Computer-assisted assignment of functional domains in the nonstructural polyprotein of hepatitis E virus: delineation of an additional group of positive-strand RNA plant and animal viruses. Proc. Natl. Acad. Sci. U. S. A. 89:8259–8263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Korkaya H, et al. 2001. The ORF3 protein of hepatitis E virus binds to Src homology 3 domains and activates MAPK. J. Biol. Chem. 276:42389–42400 [DOI] [PubMed] [Google Scholar]
- 27. Kouroukis CT, et al. 2011. A phase II study of bortezomib and gemeitabine in relapsed mantle cell lymphoma from the National Cancer Institute of Canada clinical trial group (IND 172). Leuk. Lymphoma 52:394–399 [DOI] [PubMed] [Google Scholar]
- 28. Lopez T, Silvia-Ayala D, Lopez S, Arias CA. 2011. Replication of the rotavirus genome requires an active ubiquitin-proteasome system. J. Virol. 85:11964–11971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Magden J, et al. 2001. Virus-specific mRNA capping enzyme encoded by hepatitis E virus. J. Virol. 75:6249–6255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Meng XJ. 2010. Recent advances in hepatitis E virus. J. Viral Hepat. 17:153–161 [DOI] [PubMed] [Google Scholar]
- 31. Meng XJ. 2011. From barnyard to food table: the omnipresence of hepatitis E virus and risk for zoonotic infection and food safety. Virus Res. 161:23–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Meng XJ, et al. 2011. Hepeviridae, p 1021–1028 In King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. (ed), Virus taxonomy, 9th report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, London, England [Google Scholar]
- 33. Moin SM, Panteva M, Jameel S. 2007. The hepatitis E virus ORF3 protein protects cells from mitochondrial depolarization and death. J. Biol. Chem. 282:21124–21133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Moin SM, Chandra V, Arya R, Jameel S. 2009. The hepatitis E virus ORF3 protein stabilizes HIF-1α and enhances HIF-1-mediated transcriptional activity through p300/CBP. Cell. Microbiol. 11:1409–1421 [DOI] [PubMed] [Google Scholar]
- 35. Ratra R, Kar-Roy A, Lal SK. 2008. The ORF3 protein of hepatitis E virus interacts with hemopexin by means of its 26 amino acid N-terminal hydrophobic domain II. Biochemistry 47:1957–1969 [DOI] [PubMed] [Google Scholar]
- 36. Satheshkumar PS, Anton LC, Sanz P, Moss B. 2009. Inhibition of the ubiquitin-proteasome system prevents vaccinia virus DNA replication and expression of intermediate and late genes. J. Virol. 83:2469–2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schmitt AP, Leser GP, Morita E, Sundquist WI, Lamb RA. 2005. Evidence for a new viral late-domain core sequence, FPVI, necessary for budding of a paramyxovirus. J. Virol. 79:2988–2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schubert U, et al. 2000. Proteasome inhibition interferes with gag polyprotein processing, release, and maturation of HIV-1 and HIV-2. Proc. Natl. Acad. Sci. U. S. A. 97:13057–13062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tam AW, et al. 1991. Hepatitis E virus (HEV): molecular cloning and sequencing of the full-length viral genome. Virology 185:120–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tanahashi N, Kawahara H, Murakami Y, Tanaka K. 1999. The proteasome-dependent proteolytic system. Mol. Biol. Rep. 26:3–9 [DOI] [PubMed] [Google Scholar]
- 41. Thakral D, Nayak B, Rahman S, Durgapal H, Panda SK. 2005. Replication of recombinant hepatitis E virus genome tagged with reporter gene and generation of short-term cell line producing viral RNA and proteins. J. Gen. Virol. 86:1189–1200 [DOI] [PubMed] [Google Scholar]
- 42. Tran K, Mahr JA, Spector DH. 2010. Proteasome subunits relocalize during human cytomegalovirus infection, and proteasome activity is necessary for efficient viral gene transcription. J. Virol. 84:3079–3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tyagi S, Korkaya H, Zafrullah M, Jameel S, Lal SK. 2002. The phosphorylated form of the ORF3 protein of hepatitis E virus interacts with its non-glycosylated form of the major capsid protein, ORF2. J. Biol. Chem. 277:22759–22767 [DOI] [PubMed] [Google Scholar]
- 44. Tyagi S, Surjit M, Roy AK, Jameel S, Lal SK. 2004. The ORF3 protein of hepatitis E virus interacts with liver-specific α1-microglobulin and its precursor α1-microglobulin/bikunin precursor (AMBP) and expedites their export from the hepatocyte. J. Biol. Chem. 279:29308–29319 [DOI] [PubMed] [Google Scholar]
- 45. Tyagi S, Surjit M, Lal SK. 2005. The 41-amino-acid C-terminal region of the hepatitis E virus ORF3 protein interacts with bikunin, a kunitz-type serine protease inhibitor. J. Virol. 79:12081–12087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Voges D, Zwickl P, Baumeister W. 1999. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 68:1015–1068 [DOI] [PubMed] [Google Scholar]
- 47. Widjaja I, et al. 2010. Inhibition of the ubiquitin proteasome system affects influenza A virus infection at a postfusion step. J. Virol. 84:9625–9631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yamada K, et al. 2009. ORF3 protein of hepatitis E virus is essential for virion release from infected cells. J. Gen. Virol. 90:1880–1891 [DOI] [PubMed] [Google Scholar]
- 49. Zafrullah M, Ozdener MH, Panda SK, Jameel S. 1997. The ORF3 protein of hepatitis E virus is a phosphoprotein that associates with the cytoskeleton. J. Virol. 71:9045–9053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zafrullah M, Ozdener MH, Kumar R, Panda SK, Jameel S. 1999. Mutational analysis of glycosylation, membrane translocation, and cell surface expression of the hepatitis E virus ORF2 protein. J. Virol. 73:4074–4082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang Z, et al. 2000. Structural and functional characterization of interaction between hepatitis B virus X protein and the proteasome complex. J. Biol. Chem. 275:15157–15165 [DOI] [PubMed] [Google Scholar]