Abstract
Neurotropic flaviviruses can efficiently replicate in the developing and mature central nervous systems (CNS) of mice causing lethal encephalitis. Insertion of a single copy of a target for brain-expressed microRNAs (miRNAs) in the 3′ noncoding region (3′NCR) of the flavivirus genome (chimeric tick-borne encephalitis virus/dengue virus) abolished virus neurovirulence in the mature mouse CNS. However, in the developing CNS of highly permissive suckling mice, the miRNA-targeted viruses can revert to a neurovirulent phenotype by accumulating deletions or mutations within the miRNA target sequence. Virus escape from miRNA-mediated suppression in the developing CNS was markedly diminished by increasing the number of miRNA target sites and by extending the distance between these sites in the virus genome. Insertion of multiple miRNA targets into the 3′NCR altered virus neuroinvasiveness, decreased neurovirulence and neuroinflammatory responses, and prevented neurodegeneration without loss of immunogenicity. Although the onset of encephalitis was delayed, a small number of suckling mice still succumbed to lethal intracerebral infection with the miRNA-targeted viruses. Sequence analysis of brain isolates from moribund mice revealed that the viruses escaped from miRNA-mediated suppression exclusively through the deletion of miRNA targets and viral genome sequence located between the two miRNA targets separated by the greatest distance. These findings offer a general strategy to control the reversion of virus to a virulent phenotype: a simultaneous miRNA targeting of the viral genome at many different functionally important regions could prevent virus escape from miRNA-based attenuation, since a deletion of the targeted genomic sequences located between the inserted miRNA binding sites would be lethal for the virus.
INTRODUCTION
Cellular small noncoding microRNAs (miRNAs) regulate diverse processes in many plant and animal species through the assembly of the miRNA-induced silencing complex (RISC), which binds the complementary target sequences in mRNA and subsequently cleaves or posttranscriptionally represses the targeted mRNA (5, 10, 14). Many miRNAs have a distinctive cell- and tissue-specific pattern of expression (6, 10, 21). These distinctively expressed miRNAs are now being exploited to modulate the tissue tropism of a variety of viruses and to limit viral pathogenesis by engineering virus genomes that contain the target sequences recognized by cellular miRNA-preprogrammed RISCs (4, 16–18, 35). Recently, we demonstrated that the inclusion of a single copy of a target for a brain-expressed miRNA into the genome of neurotropic chimeric tick-borne encephalitis virus/dengue virus 4 (TBEV/DEN4) flavivirus (28) was sufficient to prevent the development of otherwise lethal encephalitis in adult mice infected intracerebrally (i.c.) with a large dose of virus (15). Viruses bearing a complementary binding sequence for mir-9, mir-124a, or let-7c miRNA had limited access to the central nervous systems (CNS) of immunocompromised mice lacking T and B cell responses. These findings indicate that miRNA targeting is an effective strategy for controlling flavivirus neurotropism and pathogenesis. However, even a single-nucleotide mutation in the miRNA target sequence led to a base pairing mismatch with the corresponding miRNA and restored the ability of the virus to cause paralysis and death in mice (15). This potential virus reversion to the neurovirulent phenotype by accumulation of mutations within the miRNA target sequence prompted us to more closely analyze the genetic stability of miRNA-targeted flaviviruses in the relevant animal models.
Previously, we demonstrated that neurovirulent escape mutants did not emerge in the mature CNS of immunocompetent adult mice inoculated directly into the brain with miRNA-targeted TBEV/DEN4 viruses (15); this might be due to both the miRNA-mediated suppression of virus replication in the mature CNS and the virus clearance by the fully developed antiviral immune responses. In the present study, we evaluated the neuropathogenesis and genetic stability of miRNA-targeted TBEV/DEN4 viruses in the developing CNS of suckling mice, a more sensitive animal model for assessment of flavivirus neurovirulence (29, 30, 33). Since the central nervous and immune systems of suckling mice are still immature compared to those of adult mice, we assumed that the less potent antiviral response in suckling mice might allow for prolonged replication of miRNA-targeted viruses and emergence of virulent escape mutants. Following intracerebral inoculation of suckling mice with TBEV/DEN4 virus carrying a single target for mir-124a miRNA, all virus isolates from the brains of mice that developed encephalitis contained partially or completely deleted mir-124a target sequences. The risk of virus escape from miRNA-mediated suppression in the CNS of suckling mice was minimized by increasing the number of targets in the viral genome for broadly brain-expressed miRNAs (mir-9 and/or mir-124a). We demonstrate that the location of miRNA target sites in the 3′ noncoding region (3′NCR) of the TBEV/DEN4 genome and their spacing affect the level of virus suppression. Analysis of the escape mutants derived from the brains of moribund mice shows that resistant viruses overcame miRNA-mediated suppression exclusively by acquiring large deletions encompassing the introduced miRNA-targeted sequences and the genome sequence located between the two miRNA targets separated by the greatest distance. Our findings indicate that the increase in the number of miRNA targets in the 3′NCR and in the distance between the targets results in significant attenuation of flavivirus neurovirulence even in the developing mouse CNS.
MATERIALS AND METHODS
Viruses.
Chimeric TBEV/DEN4 cDNA (GenBank accession no. FJ28986) (28) that contains the prM and E protein genes of Far Eastern subtype TBEV strain Sofjin with the remaining sequence derived from recombinant DEN4 virus was used previously to generate recombinant viruses carrying a single target for let-7c, mir-9, mir-124a, mir-128a, or mir-218 miRNA inserted immediately after the TAA stop codon into the 3′NCR of the TBEV/DEN4 genome between nucleotides (nt) 10280 and 10281 (15). The recovered viruses derived from simian Vero or mosquito C6/36 cells were designated let-7cT, mir-9T, mir-124aT, mir-128aT, and mir-218T.
To generate the 3′NCR deletion mutant virus, the BstBI-KpnI DNA fragment (from nt 10551 to 10664) was amplified by PCR and used for replacement of the corresponding DNA fragment sequence in the full-length cDNA genome of TBEV/DEN4Δ243 (15). The resulting cDNA clone (TBEV/DEN4Δ270) differed from parental TBEV/DEN4 cDNA by the insertion of the BstBI cleavage site located immediately after the TAA stop codon and the deletion of 270 nt of the TBEV/DEN4 genome (from nt 10281 to 10550).
To generate TBEV/DEN4 viruses for simultaneous multiple targeting for homologous (mir-124a) or two different (mir-9 and mir-124a) brain-expressed miRNAs, five sites for miRNA target insertions (see Fig. 3 and 4) were selected in the 3′NCR, and these sites were located between the following nucleotide positions: 10280 and 10281 (site 1), 10292 and 10293 (site 2), 10307 and 10308 (site 3), 10469 and 10470 (site 4), and 10554 and 10555 (site 5). Each BstBI-KpnI DNA fragment that encoded the 3′NCR (from nt 10281 to 10664) with inserted miRNA target sequences was synthesized by Blue Heron Biotechnology and cloned into the full-length TBEV/DEN4 cDNA genome. The sequences of the 3′NCRs of eight engineered TBEV/DEN4 cDNA genomes carrying 2, 3, or 4 miRNA targets are shown in Fig. S1 in the supplemental material. RNA transcripts derived from these modified TBEV/DEN4 cDNA clones were generated by transcription with SP6 polymerase and used to transfect Vero or C6/36 cells in the presence of Lipofectamine as described previously (9, 33). Eight viruses [designated 2x mir-124aT(1,2), mir-9T-124aT(1,2), mir-9T-124aT-124aT(1,2,3), 3x mir-124aT(1,2,3), 3x mir-124aT(1,2,4), 3x mir-124aT(1,2,5), 4x mir-124aT(1,2,3,4), and 4x mir-124aT(1,2,3,5)] carrying 2, 3, or 4 copies of targets (T stands for target) for mir-124a and/or mir-9 at the insertion sites (the site numbers are shown in the parentheses) were recovered from Vero cells, biologically cloned by two or three terminal dilutions, and amplified by two passages in Vero cells. Virus titer was determined in Vero cells using a plaque-forming assay (PFA) as described previously (9, 33). To verify the presence of the introduced miRNA target insertions in the virus genome, RNA for each virus was isolated, and the consensus sequence of the genome was determined.
Fig 3.
Construction of recombinant TBEV/DEN4 viruses bearing multiple copies of miRNA targets. Proposed secondary structure for the DEN4 3′NCR part of the TBEV/DEN4 genome (GenBank accession number FJ828986) (28) and schematic diagram of five miRNA target insertion sites (shown by red arrows) at the positions between the nucleotides that are indicated in parentheses. The conserved sequence CS1, CS2, and RCS motifs are shown in blue. The designations for engineered recombinant TBEV/DEN4 viruses carrying 2, 3, or 4 copies of complementary targets (T) for mir-124a and/or mir-9 that were introduced in the insertion sites [site(s) 1, 2, 3, 4, or 5] specified in the parentheses are shown in the gray box.
Fig 4.
Localization and size of deletions in the 3′NCR of escape mutants detected in the brains of moribund mice infected i.c. with miRNA-targeted viruses. (A) Schematic representation of miRNA target insertion sites 1 to 5 in the 3′NCR as specified in the legend to Fig. 3. (B) The position and number of mir-9 and/or mir-124a target sites in the 3′NCR of the virus genome are shown by the black or white boxes, respectively. The size of the deletions (number of nucleotides deleted [Δnts]), their localization in the 3′NCR (arrows), and the day of virus isolation from the moribund mouse are indicated for each escape mutant derived from a mouse brain. The exact sequence boundaries for each deletion are shown in Fig. S3 in the supplemental material. Each Δ58 or Δ53 nucleotide deletion was found in virus progenies derived from the brains of two mice (indicated by an asterisk).
RNA isolation, reverse transcription, sequence analysis, and quantitation.
Viral RNA from cell culture medium or mouse brain homogenates was isolated using the QIAamp viral RNA minikit (Qiagen), and one-step reverse transcription-PCR (RT-PCR) was performed using the Superscript One-Step kit (Invitrogen) with DEN4- or TBEV-specific primers. The nucleotide consensus sequences of the virus genomes were determined through direct sequence analysis of the PCR fragments on a 3730 Genetic Analyzer using TBEV or DEN4 virus-specific primers in BigDye Terminator cycle sequencing reactions (Applied Biosystems) and were analyzed using Sequencher 4.7 software (Gene Codes Corporation).
Evaluation of viruses in mice.
All animal experiments were done at the animal biosafety level 3 (ABSL-3) facilities of the National Institute of Allergy and Infectious Diseases (NIAID), in compliance with the guidelines of the NIAID/NIH Institutional Animal Care and Use Committee.
The neurovirulence of parental and miRNA target TBEV/DEN4 viruses was evaluated in 3-day-old Swiss Webster mice (Taconic Farms) by intracerebral (i.c.) inoculation. Suckling mice in litters of 9 or 10 were inoculated with 1, 10, 102, or 103 PFU of virus and monitored for morbidity and mortality up to 21 days postinoculation (p.i.). The 50% lethal dose (LD50) was determined by the Reed and Muench method (32). Moribund (paralyzed) mice were humanely euthanized, and their brain homogenates were prepared to assess the level of virus replication in the brain as described previously (9, 30). Also, viral RNA from brain homogenates was extracted, and the sequence of the genomic region containing the engineered miRNA target was determined. For study of virus replication in the mouse brain, 5-day-old mice were inoculated i.c. with 103 PFU of TBEV/DEN4 or its derivative viruses, and the brains of three mice in each group were harvested every other day from day 1 through day 19 p.i. Virus titers in brain suspensions were quantitated by titration in Vero cells. Virus RNA was extracted from homogenates and used for sequence analysis to investigate the stability of the engineered miRNA target insertions after virus replication in the brain.
To investigate the neuroinvasiveness of TBEV/DEN4 and its derivatives, 3-week-old SCID mice (ICRSC-M; Taconic Farms) in groups of 5 were inoculated intraperitoneally (i.p.) with 105 PFU of virus and observed for 49 days for signs of morbidity typical of CNS involvement, including paralysis. Moribund mice were humanely euthanized.
Immunogenicity and protective efficacy of parental and miRNA target viruses were assessed in 3-week-old Swiss Webster mice that were inoculated i.p. with 105 PFU of virus. Survivors were bled on day 28 p.i. to evaluate antibody response, challenged i.c. on day 33 with 100 i.c. LD50s of unmodified TBEV/DEN4 virus, and observed for morbidity and mortality for an additional 3 weeks.
Histopathological and immunohistochemical analyses.
Five-day-old Swiss Webster mice were inoculated i.c. with 103 PFU of the TBEV/DEN4, mir-124aT, or 4x mir-124aT(1,2,3,5) virus. Control mice were mock inoculated with Leibovitz's L-15 medium (Invitrogen). The brains of three mice were collected on each predetermined time point or when the mice became moribund. The brains were collected on days 3 and 6 p.i. (moribund) for TBEV/DEN4-infected mice and on days 3, 6, 8, and 13 p.i. (moribund) for mir-124aT-infected mice. Mice inoculated with 4x mir-124aT(1,2,3,5) remained asymptomatic during the 20-day observation period, and their brains were collected on days 3, 6, 8, 13, and 20 p.i.
The brains were fixed with perfusion fixative (4% paraformaldehyde, 4% sucrose, 1.4% sodium cacodylate; Electron Microscopy Sciences) for 72 h, cryopreserved in 30% sucrose, and treated overnight with 20% glycerol and 2% dimethyl sulfoxide. Multiple brains were embedded in gelatin matrix blocks using MultiBrain Technology (NeuroScience Associates) and sectioned coronally at 30 μm beginning at the olfactory bulbs and proceeding caudally. Serial sections were stored in the antigen preserve solution (50% phosphate-buffered saline [PBS] [pH 7.0], 50% ethylene glycol, 1% polyvinylpyrrolidone) at −20°C until staining. A set of 14 serial sections was used for each staining.
Hematoxylin and eosin staining was performed according to a standard procedure and used for general assessment of virus-induced histopathology. For detailed analysis of viral antigen distribution, microglial activation, reactive astrocytosis, and the integrity of somatodendritic neuronal compartments, we performed immunohistochemistry on free-floating sections following a standard immunoperoxidase method. The following primary rabbit polyclonal antibodies were used: anti-TBEV (1:30,000) to reveal viral antigen distribution; anti-Iba1 (1:15,000; Wako Chemicals USA) to reveal immunoreactivity (IR) for ionized calcium binding adapter molecule and to evaluate microglial activation; anti-GFAP (1:20,000; Dako) to reveal IR for glial fibrillary acidic protein (GFAP) and to evaluate reactive astrocytosis; and anti-MAP2 (1:500; Millipore) to reveal IR for microtubule-associated protein 2 (MAP2) and to evaluate the integrity of somatodendritic neuronal compartments. Primary antibody staining was visualized using appropriate secondary antibodies and Vectastain Elite ABC kit (Vector Labs) with diaminobenzidine (Sigma-Aldrich). The sections were then mounted on 2- by 3-in. (50.8- by 76.2-mm) slides, dehydrated in alcohol, cleared in xylene, and covered with coverslips. All stained brain sections were converted into the digital whole-slide images using Aperio GL scanner and analyzed using ImageScope software (Aperio).
RESULTS
Neurovirulence of TBEV/DEN4 viruses bearing a single copy of a miRNA target.
TBEV/DEN4 viruses bearing a complementary target for brain-expressed miRNA (let-7c, mir-9, mir-124a, mir-128a, or mir-218) were greatly restricted for neurovirulence in adult mice (15). Since suckling mice have a less developed CNS and an immature immune system, they represent a highly sensitive animal model for measuring the pathogenic potential of flaviviruses including the assessment of residual neurovirulence of live attenuated vaccine viruses (29, 30, 33). To determine whether the miRNA target insertions into the TBEV/DEN4 genome could attenuate viral neurovirulence in suckling mice, we evaluated the 50% lethal dose (LD50) of viruses in a classical dose escalation study. Three-day-old Swiss mice were inoculated by the intracerebral (i.c.) route with a virus dose ranging from 1 to 100 PFU. With the exception of the let-7cT virus-infected mice, animals in other groups developed encephalitis, and the morbidity/mortality rate in these groups of mice was virus dose dependent (Fig. 1). mir-128aT and mir-218T viruses did not show any substantial decrease in the level of neurovirulence compared to the parental TBEV/DEN4 virus. However, introduction of a target for mir-9, mir-124a, or let-7c miRNA into the TBEV/DEN4 genome increased the LD50 of the resulting viruses by 6-, 66-, or >125-fold and also extended the average survival time (AST) of moribund mice from 7.6 days to 14.5, 13, or >21 days, respectively (Table 1). The increase in the i.c. LD50 for the miRNA-targeted viruses over that of the TBEV/DEN4 parent suggested that their replication in the developing CNS was restricted. However, the neurovirulence of these viruses was significantly higher for suckling mice compared to the adult mice (15), despite similar (let-7 and mir-124a) or slightly higher (mir-9) levels of miRNA expression in the CNS of suckling mice (miRNA array analysis; see Fig. S2 and Table S1 in the supplemental material). These results suggest that most likely, other factors such as the level of immune system maturation and/or specific differences in the cellular patterns of miRNA expression in the CNS may play a role in the observed age-dependent susceptibility of mice to these miRNA-targeted viruses.
Fig 1.
Survival curves of suckling mice infected with recombinant TBEV/DEN4 viruses carrying a single copy of miRNA target. Litters of 10 3-day-old Swiss Webster mice were inoculated i.c. with 1 PFU (A), 10 PFU (B), or 100 PFU (C) of the indicated viruses and then monitored daily for morbidity for 21 days. The intracerebral LD50 for each virus was determined by the Reed and Muench method (32), and the values are shown in Table 1.
Table 1.
Relative neurovirulence of the parental and miRNA target TBEV/DEN4 viruses in suckling mice
| Virus | i.c. LD50 (PFU) | Fold reductiona | ASTb (no. of days) |
|---|---|---|---|
| TBEV/DEN4 | 0.8 | 7.6 | |
| let-7cT | >100 | >125 | >21 |
| mir-9T | 5 | 6 | 14.5 |
| mir-124aT | 53 | 66 | 13 |
| mir-128aT | 1 | 9.7 | |
| mir-218T | <1 | 8.8 |
Fold reduction in neurovirulence for each miRNA target virus compared to the neurovirulence of the TBEV/DEN4 parent.
Average survival times (AST) of moribund mice following inoculation with 10 PFU of virus.
Since the mir-124aT virus bearing a single target for a brain-specific and broadly expressed mir-124a (2, 20, 34) exhibited the most attenuated neurovirulence, it was selected for further evaluation of the replication level and genetic stability in the developing CNS. Following i.c. inoculation of suckling mice with 103 PFU of virus, TBEV/DEN4 replicated efficiently in the brain, attained a mean peak virus titer of 10.2 log10 PFU/g by day 5, and caused paralysis and death on day 7 (Fig. 2A). At the same inoculation dose, the peak level of mir-124aT replication in the brain was 500-fold lower than the parental TBEV/DEN4 virus but similar to that of the DEN4 virus. In addition, the mir-124aT virus replicated at a lower rate, and mice became moribund at later time points after inoculation, one mouse became paralyzed on day 7, and all the remaining mice (n = 18) developed symptoms of encephalitis between days 9 and 13 p.i. Sequence analysis of mir-124aT progenies derived from the brains of moribund mice identified single- or double-nucleotide substitutions in the central part of the mir-124a target sequence in four of the 10 escape mutants (Fig. 2B). These data are consistent with our previous observations (15) demonstrating that a single-nucleotide mutation in the inserted miRNA target (such as the let-7c target) can restore the virus neurovirulent phenotype. Interestingly, the predominant (60%) mutation in the brain-derived mir-124aT mutants was a partial or complete deletion of the miRNA target sequence (Fig. 2B). Thus, the insertion of a single copy of mir-124a target into the genome was not sufficient to prevent the reversion of virus to the neurovirulent phenotype in the developing CNS. The development of encephalitis was associated with the accumulation of mutations or deletions within the miRNA target sequences of brain-derived viral progenies. This suggests that perfect base pairing between a target and its corresponding miRNA is critical for miRNA target recognition and effective inhibition of virus replication in the CNS.
Fig 2.
Replication and genetic stability of the mir-124aT virus in the brains of suckling mice infected i.c. with a lethal dose. (A) Replication kinetics of parental TBEV/DEN4 and DEN4 viruses and their mir-124aT derivative bearing complementary sequence for mir-124a in the mouse brains. Suckling mice were inoculated i.c. with 103 PFU of the TBEV/DEN4 or mir-124aT virus. The brains were removed on odd days postinfection from three mice in each group, and virus titer was determined by plaque assay in Vero cells. The limit of detection (1.7 log10 PFU/g) is indicated by the broken line. The lines terminate for TBEV/DEN4 and mir-124aT viruses after all mice had succumbed to infection. The values for DEN4 virus are historical data included for comparison (30). (B) Mutations that accumulated in the miRNA target sequences of mir-124aT progenies isolated from the mouse brain (MB) on the indicated day postinoculation (MB1, mouse brain 1 from group). The miRNA target sequence that is flanked with BstBI and XhoI sites (underlined) inserted between positions 10280 and 10281 of the TBEV/DEN4 genome is shown on a gray background. The ORF stop codon is shown boxed. Deletions are shown by dashed arrows. Nucleotides that differ from those in the mir-124aT virus sequence are indicated in bold type.
Generation of TBEV/DEN4 viruses carrying multiple miRNA target sites.
We next explored whether the introduction of multiple miRNA binding sites for two CNS-abundant miRNAs (mir-9 and mir-124a) into the 3′NCR of the viral genome would reduce the emergence of virulent escape mutants in the developing CNS of mice. From our previous studies, we learned that the TBEV/DEN4 genome can tolerate a large deletion of 243 nt in the 3′NCR (Fig. 3; the nucleotides from positions 10281 to 10523) without a significant effect on viral replication in vitro and on neurovirulence in suckling mice (15). In the present study, the first four sites on the proposed secondary structure of the TBEV/DEN4 3′NCR (Fig. 3) were utilized for the insertions of 2, 3, or 4 targets that contained a perfect complementary sequence to mir-9 and/or mir-124a miRNAs. These miRNA targets were inserted in the predicted accessible, A/U nucleotide-rich loop structures of the 3′NCR (31), since A/U-rich nucleotide composition near the miRNA binding site increases the miRNA target site accessibility for the RISC complex and can promote effective repression of RNA translation (6, 10, 12). In order to target the longest portion of the 3′NCR of the viral genome, we first identified the minimum 3′NCR terminal sequence required for virus viability and replication in vitro. We deleted 270 nt from the TBEV/DEN4 genome beginning with nt 10281 (following the TAA stop codon of the open reading frame [ORF]) and extending into the core element of the 3′NCR (nucleotide position 10550). Compared to the shorter deletion (243 nt) mentioned above, extension of the deletion into the core element of the 3′NCR resulted in a failure of the TBEV/DEN4Δ270 virus to be recovered in Vero cells, suggesting that the sequence between nucleotide positions 10523 and 10550 is necessary for virus viability. Therefore, in order to position a potential target site in the TBEV/DEN4 genome in a sequence region that is important for virus replication, site 5 (nt 10554 to 10555) (Fig. 3) in the 3′NCR was used for miRNA target insertion. Six viruses [designated 2x mir-124aT(1,2), 3x mir-124aT(1,2,3), 3x mir-124aT(1,2,4), 3x mir-124aT(1,2,5), 4x mir-124aT(1,2,3,4), and 4x mir-124aT(1,2,3,5)] (Fig. 3) containing 2, 3, or 4 copies of mir-124a target sequence were recovered from the engineered full-length cDNA clones in Vero cells as described previously (15). For simultaneous targeting of the viral genome with two different brain-expressed miRNAs, two additional viruses [designated mir-9T-124aT(1,2) and mir-9T-124aT-124aT(1,2,3)] (Fig. 3) were constructed. Each of these viruses carried one target for mir-9 and one or two targets for mir-124a using the insertion sites 1 and 2 or sites 1, 2, and 3, respectively. Following successful recovery in Vero cells, all recombinant viruses were amplified by one additional passage, biologically cloned by two or three subsequent terminal dilutions, and finally amplified in Vero cells by two serial passages. All miRNA-targeted TBEV/DEN4 viruses grew well in Vero cells, achieving titers greater than 106 PFU/ml. Sequence analysis revealed that all recombinant miRNA-targeted TBEV/DEN4 viruses were genetically stable in Vero cells and contained the desired miRNA target insertions (3′NCR sequences are shown in Fig. S1 in the supplemental material).
Multiple miRNA target insertions altered virus neuroinvasiveness without negative impact on immunogenicity and protective efficacy.
The parental TBEV/DEN4 and eight recombinant viruses carrying 2, 3, or 4 miRNA targets were assessed in immunodeficient SCID mice. To determine the ability of miRNA-targeted viruses to replicate in the periphery, and possibly invade the CNS and cause fatal encephalitis, SCID mice were inoculated intraperitoneally (i.p.) with a 105 PFU dose of virus. The TBEV/DEN4 parent is a neuroinvasive virus in SCID mice with an i.p. LD50 of 25,000 PFU (33), and all mice infected with TBEV/DEN4 developed paralysis between days 15 and 21 p.i. Unlike TBEV/DEN4, miRNA-targeted viruses failed to induce CNS disease during the 35-day observation period (data not shown). Thus, the introduction of multiple binding sites for brain-expressed mir-9 and/or mir-124a reduced the neuroinvasive potential of TBEV/DEN4 in immunodeficient mice.
Immunogenicity and protective efficacy of parental and miRNA-targeted viruses were evaluated in immunocompetent adult Swiss mice at a dose of 105 PFU. The parental and miRNA-targeted viruses induced a moderate titer of TBEV-specific neutralizing antibodies as measured on day 28 p.i. (Table 2). On day 33, mice were challenged by the i.c. route of inoculation with an i.c. 100 LD50s (600 PFU) of virulent unmodified TBEV/DEN4 virus; the i.c. LD50 of TBEV/DEN4 for adult mice was previously determined to be 6 PFU (15). All mock-inoculated mice developed encephalitis between days 7 and 10 postchallenge. However, mice immunized with a single dose of any of the miRNA-targeted viruses were partially or completely protected against severe i.c. challenge during the 21-day period of observation. Moreover, there was no difference in the level of protection induced in mice by parental TBEV/DEN4 (67%) or miRNA-targeted viruses (50 to 100%).
Table 2.
Immunogenicity and protective efficacy of TBEV/DEN4 miRNA-targeted viruses in adult Swiss mice
| Virus | No. of mice immunized | Serum neutralizing antibody response against TBEVa |
Survival (%) after i.c. challenge with 100 i.c. LD50s of TBEV/DEN4d | ASTe (no. of days) | |
|---|---|---|---|---|---|
| No. (%) of seropositive miceb | GMT (PRNT50%)c | ||||
| mir-9T | 5 | 4 (80) | 55 | 3 (60) | 7 |
| mir-124aT | 5 | 4 (80) | 55 | 3 (60) | 11.5 |
| 2x mir-124aT(1,2) | 10 | 6 (60) | 18 | 5 (50) | 9.5 |
| mir-9T-124aT(1,2) | 10 | 5 (50) | 14 | 6 (60) | 9 |
| 3x mir-124aT(1,2,3) | 10 | 4 (40) | 17 | 8 (80) | 10 |
| mir-9T-124aT-124aT(1,2,3) | 10 | 8 (80) | 29 | 8 (80) | 13 |
| 3x mir-124aT(1,2,4) | 10 | 9 (90) | 48 | 9 (90) | 10 |
| 3x mir-124aT(1,2,5) | 10 | 7 (70) | 33 | 7 (70) | 10 |
| 4x mir-124aT(1,2,3,4) | 10 | 6 (60) | 39 | 5 (50) | 12.3 |
| 4x mir-124aT(1,2,3,5) | 10 | 10 (100) | 60 | 10 (100) | >21 |
| TBEV/DEN4 | 6 | 5 (83) | 197 | 4 (67) | 13 |
| None (mock infected) | 5 | 0 | <5 | 0 (0) | 8 |
Three-week-old Swiss mice were inoculated i.p. with 105 PFU of virus, and serum for neutralization assay was collected on day 28.
Number (percent) of seropositive mice with plaque reduction (50%) neutralizing antibody titers (PRNT50%) of ≥5. PRNT50% (reciprocal) was determined against the unmodified TBEV/DEN4 virus.
GMT, geometric mean titers (reciprocal) are calculated for mice that were seropositive.
Groups of mice that had been immunized with the designated virus on day 0 were challenged by an i.c. route with 100 LD50s (600 PFU) of TBEV/DEN4 administered i.c. on day 33.
AST, average survival times of moribund mice following the TBEV/DEN4 challenge.
Multiple site miRNA targeting within the 3′NCR strongly attenuates virus neurovirulence in suckling mice.
The effect of multiple miRNA targeting on viral neurovirulence was evaluated in suckling mice. Although an increase in the number of miRNA binding sites in targeted RNAs is usually associated with an enhancement of mRNA translational inhibition or virus miRNA-mediated suppression (4, 5, 7, 16), introduction of two tandem targets for mir-124a did not diminish the neurovirulence of the 2x mir-124aT(1,2) virus compared to that of mir-124aT carrying just a single copy (Tables 1 and 3). The i.c. LD50 was 53 PFU for both viruses (Tables 1 and 3). On the other hand, targeting the viral genome at sites 1 and 2 for two different brain-expressed miRNAs (mir-124a and mir-9) was effective, since the i.c. LD50 of the constructed mir-9T-124aT(1,2) virus increased by 11- or 112-fold compared to that of mir-124aT or mir-9T, respectively. However, addition of a third mir-124a target into the mir-9T-124aT(1,2) genome at insertion site 3 had no additional effect on the neurovirulence of the mir-9T-124aT-124aT(1,2,3) virus (Table 3). A moderate decrease in the neurovirulence was observed for the 3x mir-124aT(1,2,3) virus carrying three tandem repeats of mir-124a target at sites 1, 2, and 3 compared to the 2x mir-124aT(1,2) virus (i.c. LD50s of 214 and 53 PFU, respectively). In addition, a more substantial attenuation of virus neurovirulence (greater than 1,250-fold increase in the i.c. LD50 compared to parental TBEV/DEN4) was achieved when a third mir-124a target was inserted at the more distant position, site 4 or 5, in the 3′NCR of the 3x mir-124aT(1,2,4) or 3x mir-124aT(1,2,5) genome, respectively. In total, only 5 mice from three groups of animals inoculated with 10, 100, and 1,000 PFU of 3x mir-124aT(1,2,4) or 3x mir-124aT(1,2,5) succumbed to infection after a significant delay in the onset of encephalitis compared to that of parental TBEV/DEN4 virus (Table 3). Furthermore, addition of a fourth mir-124a target copy into the 3x mir-124aT(1,2,3) virus at site 4 or 5 led to further attenuation of the resulting 4x mir-124aT(1,2 3,4) or 4x mir-124aT(1,2,3,5) virus, respectively. Only 1 of 30 mice infected with 4x mir-124aT(1,2,3,5) developed neurological signs (on day 20 p.i.). These results indicate that the increase in both the number of mir-124a targets and the length of viral genome sequence between the target sites had an additive attenuating effect on virus neurovirulence as demonstrated by a delay in the onset of encephalitis and an increase in mouse survival.
Table 3.
Neurovirulence of multiple miRNA target viruses in 3-day-old Swiss mice inoculated i.c.a
| Virus and group | Dose (PFU) | No. of moribund mice on the following day p.i.: |
Morbidity (%) | LD50 (PFU) | Fold reductionb | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | |||||
| 2x mir-124aT (1,2) | 53 | 66 | |||||||||||||||||
| Group 1 | 10 | 2 | 3 | 1 | 60 | ||||||||||||||
| Group 2 | 102 | 2 | 2 | 1 | 1 | 60 | |||||||||||||
| Group 3 | 103 | 2 | 3 | 50 | |||||||||||||||
| mir-9T-124aT (1,2) | 562 | 703 | |||||||||||||||||
| Group 4 | 10 | 0 | |||||||||||||||||
| Group 5 | 102 | 1 | 1 | 1 | 1 | 40 | |||||||||||||
| Group 6 | 103 | 1 | 1 | 1 | 1 | 1 | 50 | ||||||||||||
| 3x mir-124aT(1,2,3) | 214 | 268 | |||||||||||||||||
| Group 7 | 10 | 2 | 20 | ||||||||||||||||
| Group 8 | 102 | 2 | 2 | 4 | 80 | ||||||||||||||
| Group 9 | 103 | 1 | 2 | 1 | 2 | 60 | |||||||||||||
| mir-9T-124aT-124aT (1,2,3) | 490 | 613 | |||||||||||||||||
| Group 10 | 10 | 1 | 10 | ||||||||||||||||
| Group 11 | 102 | 1 | 1 | 20 | |||||||||||||||
| Group 12 | 103 | 1 | 4 | 1 | 60 | ||||||||||||||
| 3x mir-124aT (1,2,4) | >1,000 | >1,250 | |||||||||||||||||
| Group 13 | 10 | 1 | 10 | ||||||||||||||||
| Group 14 | 102 | 1 | 11 | ||||||||||||||||
| Group 15 | 103 | 1 | 1 | 1 | 30 | ||||||||||||||
| 3x mir-124aT (1,2,5) | >1,000 | >1,250 | |||||||||||||||||
| Group 16 | 10 | 0 | |||||||||||||||||
| Group 17 | 102 | 1 | 1 | 2 | 40 | ||||||||||||||
| Group 18 | 103 | 1 | 10 | ||||||||||||||||
| 4x mir-124aT (1,2,3,4) | >1,000 | >1,250 | |||||||||||||||||
| Group 19 | 10 | 0 | |||||||||||||||||
| Group 20 | 102 | 1 | 1 | 1 | 33 | ||||||||||||||
| Group 21 | 103 | 1 | 1 | 20 | |||||||||||||||
| 4x mir-124aT (1,2,3,5) | >1,000 | >1,250 | |||||||||||||||||
| Group 22 | 10 | 0 | |||||||||||||||||
| Group 23 | 102 | 1 | 10 | ||||||||||||||||
| Group 24 | 103 | 0 | |||||||||||||||||
| TBEV/DEN4, group 25 | 10 | 9 | 1 | 100 | 0.8c | ||||||||||||||
Groups of 9 or 10 mice were inoculated i.c. with the indicated dose of virus and monitored for signs of encephalitis for 21 days. The brains of moribund mice were harvested on the indicated day, and virus RNA was isolated from brain homogenate. To determine the stability of the miRNA target insertions in the genome, the 3′NCRs of brain-derived viruses from each group of mice were directly sequenced from brain homogenates. The results of sequence analysis of 29 escape mutant viruses are shown in Fig. S3 in the supplemental material.
Fold reduction in neurovirulence for each miRNA-targeted virus compared to the neurovirulence of the parental TBEV/DEN4 virus.
The estimated LD50 of TBEV/DEN4 from previous studies (Table 1).
Genetic stability of miRNA-targeted viruses in the developing CNS.
Because of the high mutation rate of RNA viruses due to the low fidelity of the RNA polymerase (8), we examined whether the delayed onset of viral encephalitis observed in suckling mice was a result of emerging mutations within the miRNA target insertions. We sequenced the 3′NCRs of 29 viruses that were isolated from individual brains of mice that succumbed to the i.c. infection with miRNA-targeted viruses (Table 3). The results of this sequence analysis are shown in Fig. S3 in the supplemental material. Surprisingly, all brain-derived viruses contained large deletions (from 53 to 367 nt) that began close to the TAA stop codon of the ORF and extended into the 3′NCR as shown schematically in Fig. 4; the exact boundaries of the deletions are shown in Fig. S3. The majority of deletion mutations in the recovered viruses included partially or completely deleted sequences of the two most distant miRNA binding sites as well as the viral genome sequence located between them. The largest deletion, 367 nt, was found in the 3′NCR of the Δ367 escape mutant of the 4x mir-124aT(1,2,3,5) virus. The 3′NCR of the Δ367 virus retained 135 nt of which 110 nt represented the 3′-terminal sequence of the genome and the other 25 nt were the remains of the two most distantly inserted sequences at sites 1 and 5 (14 and 11 nt, respectively). Sequence analysis of the Δ367 genome also revealed a single G2274 → A nucleotide mutation in the envelope E protein gene that resulted in a Gly443 → Asp substitution. Interestingly, the Δ367 virus was infectious for Vero cells and replicated efficiently at 37°C with virus titer reaching 6 × 106 PFU/ml on day 7 p.i. Sequence analysis revealed that a Gly443 → Asp substitution is an unique adventitious mutation for this escape mutant and was not present in the genomes of four 3x mir-124aT(1,2,5) escape mutants having large deletions between the same two most distantly inserted sequences at sites 1 and 5 (Δ335, Δ336, Δ337, and Δ347 deletion mutants; Fig. 4; see also Fig. S3 in the supplemental material). Insertion of this point mutation into the E protein gene of TBEV/DEN4Δ270 cDNA was not able to support virus recovery and replication in Vero cells. However, in the absence of a G2274 → A mutation in the E protein gene, the parental TBEV/DEN4 virus, carrying the 3′NCR sequence derived from the Δ367 genome, replicated very slowly in Vero cells and reached a titer of less than 103 PFU/ml during the 14 days of incubation, which was ∼104-fold lower than that attained by parental TBEV/DEN4 or Δ367 virus. In addition, this mutant virus produced small plaques with a size of <0.1 mm, whereas both TBEV/DEN4 and Δ367 produced plaques averaging 2 mm. Thus, these findings suggest that a single G2274 → A mutation (Gly443 → Asp) in E protein may contribute to efficient replication of the Δ367 virus in Vero cells and possibly, in the mouse brain.
To verify the ability of the Δ367 virus isolated from the mouse brain or Vero cells to cause CNS disease, suckling mice were inoculated by the i.c. route with 10-fold dilutions ranging from 1 to 103 PFU of virus. Similar to the unmodified TBEV/DEN4 parent, the Δ367 virus derived either from the brain homogenate or Vero cells was highly neurovirulent in mice, with a calculated i.c. LD50 of 4 or 1 PFU, respectively, and attained a virus titer in the brains of moribund mice between 1.3 × 108 and 3.2 × 108 PFU/g of brain tissue.
Sequence analysis data of brain isolates (29 viruses; Fig. 4; see also Fig. S3 in the supplemental material) indicated that deletion mutations predominantly encompassed the introduced miRNA target sequences, suggesting that these deletions emerged as a result of viral escape from miRNA-mediated suppression. Interestingly, eight escape mutants that were isolated from the brains of mice inoculated with TBEV/DEN4 viruses bearing two or three tandem miRNA targets at insertion sites 1 and 2 or 1, 2, and 3 contained nucleotide deletions extending from the miRNA target site 2 or 3 into the 3′ end of the 3′NCR (e.g., to nucleotide 10370, 10374, 10379, 10381, 10382, 10433, 10471, or 10529; Fig. 3 and Fig. 4; the exact boundaries of the deletions are shown in Fig. S3 in the supplemental material). These findings suggest that after an initial miRNA-mediated RISC cleavage of the 3′NCR at the target sites, the viral genome can be further degraded by cellular ribonucleases.
Introduction of four binding sites for mir-124a restricted virus replication and reduced neuroinflammation in the CNS.
Among all of the generated miRNA target viruses, the 4x mir-124aT(1,2,3,5) virus carrying four mir-124a targets demonstrated the most attenuated neurovirulent phenotype. For this reason, we analyzed the extent of its replication and neuroinflammatory responses induced in the CNS. Following i.c. inoculation of 30 suckling mice with a 103 PFU dose of virus, the brains of three mice were harvested every other day, up to day 19. The virus titer of each brain suspension was determined on Vero cells. In contrast to the efficient replication of TBEV/DEN4 or mir-124aT (Fig. 2A), the growth of 4x mir-124aT(1,2,3,5) virus was highly restricted. Replication of the 4x mir-124aT(1,2,3,5) virus in the brain was below the level of detection (<1.7 log10 PFU/g of brain) in the majority of mice and observed at a very low level (2 to 2.5 log10 PFU/g of brain) in two of the three mice on day 1 p.i. and in one mouse on day 7 p.i. There were no clinical signs of illness in any mice.
In a separate comparative study, histopathological and immunohistochemical analyses of the brain tissue were performed in three groups of suckling mice inoculated i.c. with the TBEV/DEN4, mir-124aT, or 4x mir-124aT(1,2,3,5) virus to evaluate the distribution of viral antigen, inflammatory cell infiltration, microgliosis, reactive astrocytosis, and presence of neuronal damage.
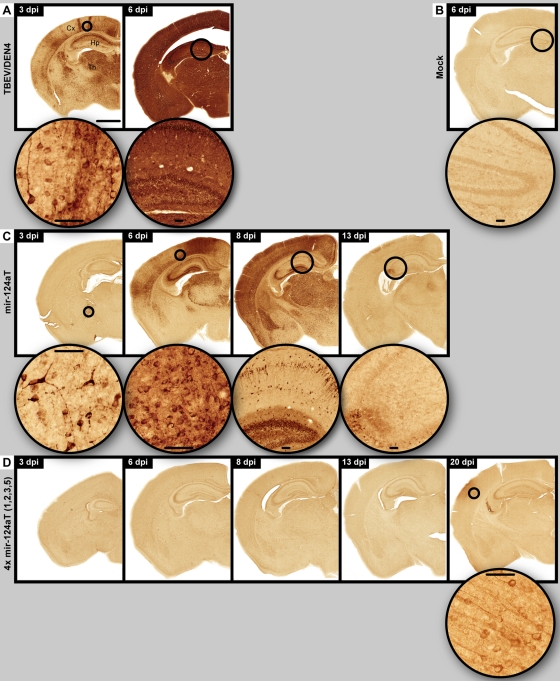
Extensive neuronal replication of the parental TBEV/DEN4 virus was evident by detection of viral antigens as early as day 3 p.i. and continued to increase to a very high level throughout the brain until day 6 p.i., when the mice became moribund (Fig. 5A). The mir-124aT virus replicated in neurons at a lower rate, apparently reaching a peak of replication on day 8 p.i. with a subsequent decrease by day 13 when all mice became moribund (Fig. 5C). In contrast, mice inoculated with the 4x mir-124aT(1,2,3,5) virus did not show any signs of the CNS infection or virus replication in neurons until day 20 p.i. when the viral antigens were detected in a small group of cortical neurons (Fig. 5D) of one of three mice.
Fig 5.
Spatiotemporal distribution of viral antigens in neurons. Immunohistochemistry for viral antigens was performed on CNS of mice infected with the parental TBEV/DEN4 (A), mir-124aT (C), or 4x mir-124aT(1,2,3,5) (D) virus on the indicated day p.i. (dpi). A mock control is shown in panel B. The bar (1,000 μm) (in panel A) applies to all panels showing a whole-mouse brain hemisphere. Round insets show circled areas of cortex (Cx), thalamus (Th), or hippocampus (Hp) at higher magnification (bars in insets, 50 μm).
The time course and magnitude of inflammatory responses (see Fig. S4 to S6 in the supplemental material) and neuronal degeneration (Fig. 6) in the CNS differed between the viruses and appeared to be an outcome of viral replication in neuronal cell populations. In mice infected with parental TBEV/DEN4 virus, profound neuroinflammatory changes with extensive microglial activation (Fig. S4B and S4F), reactive astrocytosis (Fig. S5B and S5F), inflammatory cell infiltration (Fig. S6B and S6F), and neuronal somatodendritic damage (Fig. 6B and F) were seen on day 6 p.i., the time when all mice became moribund.
Fig 6.
Neuronal somatodendritic damage. Somatodendritic neuronal compartments were revealed by specific IR for the microtubule-associated protein 2 (MAP2-IR) in the CNS of mice infected with the TBEV/DEN4 (B and F), mir-124aT (C, G, I, J, M, and N), or 4x mir-124aT(1,2,3,5) (D, H, K, L, O, and P) virus on indicated day p.i. Mock controls are shown in panels A and E. The boxed areas of the thalamus in panels A to D, I, and K or of the motor cortex in panels M and O are shown at higher magnification in the corresponding panels (E to H, J, L, N, and P). Bars, 1,000 μm (A to D, I, K, M, and O) and 50 μm (E to H, J, L, N, and P).
The mir-124aT virus carrying only one mir-124a target also induced neuronal somatodendritic damage (Fig. 6C, G, I, J, M, and N), microglial activation (see Fig. S4 in the supplemental material), reactive astrocytosis (Fig. S5), and widespread inflammatory cell infiltration (Fig. S6). However, there was a delay in these responses within the CNS, and all mice became moribund 1 week later (day 13 p.i.) compared to mice infected with TBEV/DEN4.
The brains of mice inoculated with the 4x mir-124aT(1,2,3,5) virus did not show signs of neuronal damage (Fig. 6) and had only minimal inflammatory reactions on days 6, 8 and 13 p.i. (see Fig. S4, S5, and S6 in the supplemental material). Nonetheless, at the end of the experiment (day 20 p.i.), the brain tissue of one of three mice with detectable viral antigens in cortical neurons (Fig. 5D) showed a moderate microglial activation (Fig. S4), reactive astrocytosis (Fig. S5), and inflammatory cell infiltration (Fig. S6). It should be noted that the somatodendritic neuronal compartments in the brain of this mouse were well preserved by the end of the experiment (not shown) despite the detection of viral antigens.
Taken together, these and previous (Table 3) data indicate that targeting of the 3′NCR with multiple mir-124a binding sites can markedly restrict virus replication in the developing CNS and prevent virus-induced neuroinflammation and neuronal degeneration.
DISCUSSION
The inclusion of a single copy of target sequences for miRNAs expressed in the CNS (mir-9, mir-124a, mir-128a, and mir-218) completely abolished the neurotropism of the TBEV/DEN4 virus in immunocompetent adult mice, but the effects of these miRNA target insertions on the viral neurovirulence in newborn mice and neuroinvasiveness in immunodeficient mice were less evident than those observed for the let-7c target (15). let-7c is a member of the large let-7 family of miRNAs which are ubiquitously expressed in a multitude of tissues and cells (2, 3, 21, 34), and the let-7c target inclusion into viral genome was most effective in terms of limiting TBEV/DEN4 neuropathogenesis in both newborn and adult mice. These findings suggest that a variety of factors including immune system maturation and levels and patterns of miRNA expression in the CNS and/or other tissues may play a role in age-dependent susceptibility of mice to miRNA-targeted virus infection. Sequence analysis data of viral isolates derived from the brains of moribund SCID (15) or suckling mice, which were infected with TBEV/DEN4 viruses carrying a single miRNA target, show that virus escape from the miRNA-mediated suppression can occur through deletions or mutations within the central region of the miRNA target sequence. These findings are in line with data obtained for other viruses (4, 11, 16, 22, 36, 37) indicating that the perfect complementarity between the target sequence and cellular miRNA is a critical factor for miRNA target recognition and catalytic cleavage of miRNA-targeted RNA by the RISC (6, 12). As we demonstrated previously (15), a single-nucleotide mutation within the central region of the let-7c target sequence was sufficient to restore the ability of the let-7cT mutant virus to cause lethal CNS disease in mice and to efficiently replicate in Vero cells or primary neurons expressing the let-7c miRNA.
Virus escape from the miRNA-mediated suppression in the developing mouse CNS was progressively reduced by increasing the number of miRNA target sites in the 3′NCR of the viral genome. The simultaneous tandem targeting of the TBEV/DEN4 genome for two different brain-expressed miRNAs (mir-9 and mir-124a) was more efficient in reducing virus neurovirulence than targeting of two or three tandem targets for mir-124a miRNA alone. In addition, we found that genomic targeting with a tandem repeat of three mir-124a targets (at sites 1, 2, and 3) was less effective in preventing the development of lethal encephalitis than targeting with the same number of mir-124a binding sites from which a third mir-124a target was inserted at a more distant position (at site 4 or 5).
Multiple miRNA target insertions (2, 3, or 4 copies) into the 3′NCR altered virus neuroinvasiveness in SCID mice and significantly attenuated its neurovirulence in immunocompetent mice without a negative impact on the immunogenicity and protective efficacy. Importantly, the miRNA targeting of a large portion of the 3′NCR with three or four miRNA binding sites greatly reduced the probability of virus to escape from the miRNA-mediated suppression even in the developing mouse CNS. However, the suppression of virus replication in the brain was not complete. Analysis of virus escape mutants derived from the brains of moribund mice showed that emerging viruses can overcome miRNA-mediated suppression by acquiring large deletions that included the inserted miRNA target sequences as well as the portion of viral genome located between the two most distant miRNA targets. It should be noted that this mode of virus escape in the CNS was a common trend among all viruses carrying two or more miRNA targets. This observation raises intriguing questions about the potential mechanism(s) involved in the generation of a large deletion in the viral RNA genome. Since each introduced miRNA target was fully complementary to the corresponding miRNA, the viral RNA should be cleaved by Argonaute-2 of the RISC at the miRNA binding sites (23). These cleavages would release 3′ and 5′ RNA fragments as well as the genomic RNA fragments located between a pair of miRNA targets that could serve as the RNA template segments for the subsequent formation of novel RNA genomes. Most likely, the mechanism involves the covalent joining of two noncontiguous RNA genome portions (the 5′ RNA fragment containing the sequences of the 5′NCR and polyprotein ORF and the 3′ RNA fragment containing the 3′-terminal part of the 3′NCR) by way of a viral polymerase-directed template-switching mechanism similar to that proposed for the generation of viral defective interfering RNAs (19, 25, 26). The precise mechanism of template switching by the RNA-dependent RNA polymerase (RdRp) is not well-known. The current model suggests that the viral RdRp together with the nascent RNA strand, which had been synthesized on the primary RNA template (the 3′ RNA fragment), jumps to the 5′ RNA fragment where it resumes RNA synthesis. Interestingly, among the nucleotide sequences of the 29 escape mutants analyzed in this study (see Fig. S3 in the supplemental material), the junction sites did not show nucleotide similarity but contained A/U-rich stretches, which are believed to promote RNA recombination by template switching (26). For each miRNA-targeted virus, the deleted 3′NCR sequences from virus isolates varied in size and localization and were always associated with and included the miRNA target elements (Fig. 4; see also Fig. S3 in the supplemental material). In the eight progenies of TBEV/DEN4 viruses bearing two or three tandem miRNA targets, deletions extended beyond the last miRNA target to the G/C-rich sequences of the 3′NCR (e.g., to the nucleotide 10370, 10374, 10379, 10381, 10382, 10433, 10471, or 10529; Fig. 3; see also Fig. S3) which can form hairpin structures (31) and promote viral RNA recombination (26). The presence of these large deletions in the viral escape mutants suggests that the cellular exo- and endoribonucleases may contribute to the further degradation of the initially RISC-generated RNA fragments prior to recombination.
The 3′NCR of the flavivirus genome is required for the formation of subgenomic RNAs (27) and is comprised of several conserved RNA structures that can modulate virus replication and pathogenicity (13, 24). Conserved sequence elements such as a tandem of the repeated conserved sequence 2 (RCS2) and conserved sequence 2 (CS2) in the 3′NCR are important for virus replication in mammalian cells. Indeed, we found that a 270-nt deletion between the stop codon and the last 3′-terminal 114 nt of the 3′NCR engineered into the recombinant TBEV/DEN4 Δ270 cDNA, including the RCS2 and CS2 sequences, was lethal, precluding virus recovery in simian Vero cells. Taking this into account, the recovery of brain-derived 3x mir-124aT(1,2,5) and 4x mir-124aT(1,2,3,5) escape mutants with large deletions in the 5′ part of the 3′NCR, including the conserved RCS2 and CS2 structures (Fig. 3 and 4), was unexpected. The 3′NCRs of five escape mutant viruses, one for 4x mir-124aT(1,2,3,5) and four for 3x mir-124aT(1,2,5), varied in length (127, 135, 137, 138, or 139 nt; see Fig. S3 in the supplemental material) but were always longer than the 3′NCR engineered in the TBEV/DEN4 Δ270 cDNA (120 nt) and retained the 110 proximal 3′-terminal nucleotides of the parental TBEV/DEN4 genome (nt 10555 to 10664) (Fig. 3). The spacer sequences between the stop codon and these last 3′-terminal 110 nt ranged from 17 to 29 nt in the length and were the remains of the sequence insertions at sites 1 and 5. Thus, the 5′ part of the 3′NCR between the stop codon and nucleotide 10555 could be substituted with short external sequences without loss of virus viability. These findings suggest that the size rather than the primary nucleotide sequence of the spacer region can affect the yield and replication of infectious virus. However, we also identified another unique mutation in the structural envelope E protein (a Gly443 → Asp substitution) of the Δ367 escape virus carrying a large deletion in the 3′NCR. This mutation occurred in an α-helix H2 element of E protein that is important for the prM-E dimer stability during virion assembly (1). The exact mechanism by which the selected escape mutant was able to regain the replicative capability and revert to a neurovirulent phenotype remains to be determined. In the absence of a Gly443 → Asp mutation in the E protein, the parental TBEV/DEN4 virus, carrying the 3′NCR sequence derived from the escape Δ367 virus, replicated very slowly in Vero cells and reached a titer 104-fold lower than that attained by parental TBEV/DEN4 or Δ367 virus. Thus, these findings suggest that a point mutation (Gly443 → Asp) in E protein may contribute to efficient replication of the Δ367 virus in Vero cells and possibly, in the mouse brain.
Taken together, our findings with multiple miRNA targeting of neurotropic flavivirus demonstrate that the degree of virus attenuation for the CNS can be modulated by increasing the number of miRNA target sites and targeting a large portion of the viral genome. Insertion of four target copies for the CNS-specific mir-124a in the 3′NCR [4x mir-124aT(1,2,3,5) virus] was the most effective approach for restricting viral replication, reducing inflammatory responses, and preventing neuronal damage even in the highly vulnerable developing CNS. With this approach, virus escape from miRNA-mediated suppression was very infrequent and observed only in the CNS of suckling mice. Sequence analysis of brain-derived progenies of the miRNA-targeted viruses has shown that virus escape occurs exclusively through the deletion of inserted miRNA targets and results in the loss of the viral genome sequence located between the two most distant miRNA targets. Importantly, deletions never extended upstream into the viral polyprotein ORF or downstream into the conserved 3′-terminal 110-nucleotide structure of the 3′NCR signifying the functionally important role of these regions for virus viability. These findings have prompted us to propose a rational vaccine design approach using multiple miRNA targets engineered into many different viral genome regions (5′NCR, ORF, and 3′NCR) that are crucial for virus viability in order to control virus neurotropism and prevent virus reversion to the neurovirulent phenotype. In such a design, the emergence of neurovirulent escape mutants under the miRNA-mediated suppression in the CNS would be unlikely due to the fact that multiple miRNA-induced cleavages would lead to deletions of critically important virus sequences located between inserted miRNA binding sites.
Supplementary Material
ACKNOWLEDGMENTS
We thank S. Whitehead (NIAID, NIH) for critically reading the manuscript.
This research was supported by the Intramural Research Program of the National Institutes of Health, National Institute of Allergy and Infectious Diseases. We do not have a conflict of financial or other interest.
Footnotes
Published ahead of print 14 March 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Allison SL, Stiasny K, Stadler K, Mandl CW, Heinz FX. 1999. Mapping of functional elements in the stem-anchor region of tick-borne encephalitis virus envelope protein E. J. Virol. 73:5605–5612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bak M, et al. 2008. MicroRNA expression in the adult mouse central nervous system. RNA 14:432–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barh D, Malhotra R, Ravi B, Sindhurani P. 2010. MicroRNA let-7: an emerging next-generation cancer therapeutic. Curr. Oncol. 17(1):70–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Barnes D, Kunitomi M, Vignuzzi M, Saksela K, Andino R. 2008. Harnessing endogenous miRNAs to control virus tissue tropism as a strategy for developing attenuated virus vaccines. Cell Host Microbe 4:239–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bartel DP. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297 [DOI] [PubMed] [Google Scholar]
- 6. Bartel DP. 2009. MicroRNAs: target recognition and regulatory functions. Cell 136:215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brown BD, Naldini L. 2009. Exploiting and antagonizing microRNA regulation for therapeutic and experimental applications. Nat. Rev. Genet. 10:578–585 [DOI] [PubMed] [Google Scholar]
- 8. Domingo E, et al. 1996. Basic concepts in RNA virus evolution. FASEB J. 10:859–864 [DOI] [PubMed] [Google Scholar]
- 9. Engel AR, et al. 2010. The neurovirulence and neuroinvasiveness of chimeric tick-borne encephalitis/dengue virus can be attenuated by introducing defined mutations into the envelope and NS5 protein genes and the 3′ non-coding region of the genome. Virology 405:243–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Filipowicz W, Bhattacharyya SN, Sonenberg N. 2008. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 9:102–114 [DOI] [PubMed] [Google Scholar]
- 11. Gitlin L, Stone JK, Andino R. 2005. Poliovirus escape from RNA interference: short interfering RNA-target recognition and implications for therapeutic approaches. J. Virol. 79:1027–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Grimson A, et al. 2007. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell 27:91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gritsun TS, Gould EA. 2007. Origin and evolution of 3′UTR of flaviviruses: long direct repeats as a basis for the formation of secondary structures and their significance for virus transmission. Adv. Virus Res. 69:203–248 [DOI] [PubMed] [Google Scholar]
- 14. He L, Hannon GJ. 2004. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Genet. 5:522–531 [DOI] [PubMed] [Google Scholar]
- 15. Heiss BL, Maximova OA, Pletnev AG. 2011. Insertion of microRNA targets into the flavivirus genome alters its highly neurovirulent phenotype. J. Virol. 85:1464–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kelly EJ, Hadac EM, Greiner S, Russell SJ. 2008. Engineering microRNA responsiveness to decrease virus pathogenicity. Nat. Med. 14:1278–1283 [DOI] [PubMed] [Google Scholar]
- 17. Kelly EJ, Russell SJ. 2009. MicroRNAs and regulation of vector tropism. Mol. Ther. 17:409–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kelly EJ, Nace R, Barber N, Russell SJ. 2010. Attenuation of vesicular stomatitis virus encephalitis through microRNA targeting. J. Virol. 84:1550–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kirkegaard K, Baltimore D. 1986. The mechanism of RNA recombination in poliovirus. Cell 47:433–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Krichevsky AM, King KS, Donahue CP, Khrapko K, Kosik KS. 2003. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA 9:1274–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lagos-Quintana M, et al. 2002. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 12:735–739 [DOI] [PubMed] [Google Scholar]
- 22. Lin SS, et al. 2009. Molecular evolution of a viral non-coding sequence under the selective pressure of amiRNA-mediated silencing. PLoS Pathog. 5(2):e1000312 doi:10.1371/journal.ppat.1000312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu J, et al. 2004. Argonaute2 is the catalytic engine of mammalian RNAi. Science 305:1437–1441 [DOI] [PubMed] [Google Scholar]
- 24. Markoff L. 2003. 5′- and 3′-noncoding regions in flavivirus RNA. Adv. Virus Res. 59:177–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nagy PD, Simon AE. 1997. New insights into the mechanisms of RNA recombination. Virology 235:1–9 [DOI] [PubMed] [Google Scholar]
- 26. Pathak KB, Nagy PD. 2009. Defective interfering RNAs: foes of viruses and friends of virologists. Viruses 1:895–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pijlman GP, et al. 2008. A highly structured, nuclease-resistant, noncoding RNA produced by flaviviruses is required for pathogenicity. Cell Host Microbe 4:579–591 [DOI] [PubMed] [Google Scholar]
- 28. Pletnev AG, Bray M, Huggins J, Lai CJ. 1992. Construction and characterization of chimeric tick-borne encephalitis/dengue type 4 viruses. Proc. Natl. Acad. Sci. U. S. A. 89:10532–110536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pletnev AG, Men R. 1998. Attenuation of the Langat tick-borne flavivirus by chimerization with mosquito-borne flavivirus dengue type 4. Proc. Natl. Acad. Sci. U. S. A. 95:1746–1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pletnev AG, Swayne DE, Speicher J, Rumyantsev AA, Murphy BR. 2006. Chimeric West Nile virus/dengue virus vaccine candidate: preclinical evaluation in mice, geese and monkeys for safety and immunogenicity. Vaccine 24:6392–6404 [DOI] [PubMed] [Google Scholar]
- 31. Proutski V, Gritsun TS, Gould EA, Holmes EC. 1999. Biological consequences of deletions within the 3′-untranslated region of flaviviruses may be due to rearrangements of RNA secondary structure. Virus Res. 64:107–123 [DOI] [PubMed] [Google Scholar]
- 32. Reed L, Muench H. 1938. A simple method of estimating fifty per cent endpoints. Am. J. Hyg. 27:493–497 [Google Scholar]
- 33. Rumyantsev AA, Chanock RM, Murphy BR, Pletnev AG. 2006. Comparison of live and inactivated tick-borne encephalitis virus vaccines for safety, immunogenicity and efficacy in rhesus monkeys. Vaccine 24:133–143 [DOI] [PubMed] [Google Scholar]
- 34. Sempere LF, et al. 2004. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 5:R13 http://genomebiology.com/content/5/3/R13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Skalsky RL, Cullen BR. 2010. Viruses, microRNAs, and host interactions. Annu. Rev. Microbiol. 64:123–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. von Eije KJ, ter Brake O, Berkhout B. 2008. Human immunodeficiency virus type 1 escape is restricted when conserved genome sequences are targeted by RNA interference. J. Virol. 82:2895–2903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yen LC, et al. 17 October 2011. Neurovirulent flavivirus can be attenuated in mice by incorporation of neuron-specific microRNA recognition elements into viral genome. Vaccine [Epub ahead of print.] doi:10.1016/j.vaccine.2011.09.102 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.