Abstract
Human norovirus infections are the most common cause of acute nonbacterial gastroenteritis in humans worldwide, and glycan binding plays an important role in the susceptibility to these infections. However, due to the lack of an efficient cell culture system or small animal model for human noroviruses, little is known about the biological role of glycan binding during infection. Murine noroviruses (MNV) are also enteric viruses that bind to cell surface glycans, but in contrast to their human counterparts, they can be grown in tissue culture and a small animal host. In this study, we determined glycan-binding specificities of the MNV strains MNV-1 and CR3 in vitro, identified molecular determinants of glycan binding, and analyzed infection in vivo. We showed that unlike MNV-1, CR3 binding to murine macrophages was resistant to neuraminidase treatment and glycosphingolipid depletion. Both strains depended on N-linked glycoproteins for binding, while only MNV-1 attachment to macrophages was sensitive to O-linked glycoprotein depletion. In vivo, CR3 showed differences in tissue tropism compared to MNV-1 by replicating in the large intestine. Mapping of a glycan-binding site in the MNV-1 capsid by reverse genetics identified a region topologically similar to the histo-blood group antigen (HBGA)-binding sites of the human norovirus strain VA387. The recombinant virus showed distinct changes in tissue tropism compared to wild-type virus. Taken together, our data demonstrate that MNV strains evolved multiple strategies to bind different glycan receptors on the surface of murine macrophages and that glycan binding contributes to tissue tropism in vivo.
INTRODUCTION
Noroviruses are small nonenveloped, plus-sense RNA viruses in the calicivirus family. Infection with human norovirus is the most common cause of nonbacterial gastroenteritis worldwide (19, 20). Although noroviruses infect a broad range of mammalian hosts, individual strains are species specific (reviewed in references 20 and 30). Most human noroviruses belong to genogroup I or II (GI and GII), while murine norovirus (MNV) strains cluster in the fifth genogroup (GV) (20). MNV is the only norovirus that efficiently grows in tissue culture (in macrophages and dendritic cells) and in a small animal host (31, 61, 62). Many biological features, including fecal-oral transmission, replication in the intestine, and fecal shedding, are shared between murine and human noroviruses (30, 62).
MNV is highly abundant in research mice (e.g., see references 26, 32, and 39). MNV-1 was originally isolated from immunocompromised mice (31) but was later shown to infect wild-type mice (9, 41). MNV-1 infections in immunocompetent mice typically show no visible signs of disease, but mild gastroenteritis and inflammation have been described (41), while MNV-1 infections can be lethal in mice with defects in the innate immune system, such as STAT-1−/− mice (31). Different clonal isolates of the original MNV-1 strain are available (58). Of importance for this study are the MNV-1.CW3 and MNV-1.CW1 isolates, the latter being the basis for a reverse genetics system (60). Compared to MNV-1.CW3, MNV-1.CW1 contains three mutations in open reading frame 1 (ORF1) and a mutation (K296E) in ORF2, the major capsid protein, which is linked to an attenuated lethality phenotype in STAT1−/− mice (4, 61). Additional strains, including CR3, have been isolated from feces from multiple wild-type or genetically modified mice (58). Currently there are 56 fully sequenced MNV strains with ∼70% nucleotide identity deposited in GenBank (NCBI), and all belong to GV. CR3 and other fecally isolated strains persist in wild-type mice over 35 days postinfection (dpi), while the MNV-1 strain is not detected in stool or tissues after 7 dpi (25, 58).
Noroviruses recognize specific glycans present on proteins and lipids during host cell attachment. MNV-1 uses terminal sialic acids (SA) on the ganglioside GD1a (56), while human norovirus strains have been shown to bind to different histo-blood group antigens (HBGA) (reviewed in references 15 and 53) and the proteoglycan heparan sulfate (50). Glycans are synthesized by the enzymatic addition of monosaccharides, including fucose or SA, onto glycolipids and glycoproteins (1). The majority of surface proteins are glycosylated, and most of them are N-glycosylated (18). Because glycans are abundantly expressed on gastrointestinal and respiratory epithelial cells, they are exploited as receptors by many viruses, bacteria, and toxins (e.g., see references 2, 36, 43, 49, 54, 63). Glycan composition differs greatly between species, tissues, and cell types and subsequently contributes to host specificity, tropism, and pathogenicity of many different pathogens (reviewed in references 5, 43, and 54). In the case of human noroviruses, successful infection is determined by the compatibility of individual strains with the specific HBGA expressed by the human host (reviewed in references 3, 7, 23). Therefore, virus infection is restricted to a subset of susceptible individuals in the population (37, 38, 46). Crystal structure analysis of human norovirus-HBGA binding revealed that glycan-binding sites are located in distinct conformational epitopes within the protruding (P) domain of the viral capsid protein VP1, but the location, conformation, and amino acid composition of the sites differ between the two major genotypes GI and GII (8, 11, 22, 44, 47).
Despite the great importance of glycan binding in host susceptibility and evolution of human norovirus, the molecular mechanisms occurring during infection are poorly understood. Therefore, the comprehensive tools available for MNV, including tissue culture system (61), small animal host (31), and multiple reverse genetics systems (10, 60, 64), lend themselves to the study of the biological role of glycan binding during norovirus infection. We previously showed that the MNV strains MNV-1, WU11, and S99 bind terminal SA on the ganglioside GD1a on the macrophage surface in vitro (56). We have now extended this analysis to another MNV strain, CR3, and additional glycan structures, mapped a functional glycan-binding site in the viral capsid, and studied the role of glycan binding in vivo.
MATERIALS AND METHODS
Cell culture and mice.
HEK293T (293T) and RAW 264.7 (RAW) cells were purchased from ATCC and maintained as previously described (55, 61). Wild-type C57BL/6 mice were purchased from Jackson Laboratory and housed at the University of Michigan animal facilities in accordance with federal and university guidelines. All mice used in this study were initially seronegative for anti-MNV antibodies, as determined by enzyme-linked immunosorbent assay (ELISA) (61).
Virus.
MNV strains MNV-1 (GV/MNV1/2002/USA), plaque isolate CW3, and CR3 (GV/CR3/2005/USA) were used at passage 6. Recombinant viruses MNV-1.CW1 and MNV-1.RVSS were used at passage 2.
Reverse genetics.
Mutagenesis of MNV-1.CW1 was based on the reverse genetics system by Ward and colleagues (60). Site-directed mutagenesis was performed using the Quikchange XL kit (Stratagene) as described previously (57). Mutations were introduced into the pMNV* vector containing the MNV-1.CW1 genome and confirmed by sequencing the capsid region. In accordance with previous literature (60), the asterisk refers to a silent mutation in ORF1 introduced in the reverse genetics system in all recombinant viruses. Mutations I439V and T441S in pMNV-1* were introduced with primers KY-134 and KY-135, generating pMNV*.VSS. The K345R mutation was introduced with the primers KY-131 and KY-132, generating pMNV*.K345R. The combined mutant pMNV*.RVSS was generated sequentially, introducing K345R into pMNV*.VSS using KY-131 and KY-132. The mutagenesis primers are shown in Table 1. Recombinant viruses MNV-1.CW1, MNV-1.K345R, MNV-1.VSS, and MNV-1.RVSS were recovered by transfecting their respective plasmids into 293T cells as previously described (57). Recombinant viruses were plaque purified three times and amplified in RAW cells. The presence of the mutation in the final working stock was confirmed by sequencing viral cDNA. Genome titers (genome equivalents [eq]) and infectious units (PFU) were determined for each stock as described below.
Table 1.
Mutagenesis primers used in this study
| Primer | Sequence (5′→3′)a |
|---|---|
| KY-131 | GGTCCAGACCGAGACCACCAgaACTGGAGACAAGCTCAAGG |
| KY-132 | CCTTGAGCTTGTCTCCAGTtcTGGTGGTCTCGGTCTGGACC |
| KY-134 | CCGGACCTACATGCGTCAGgTtGACAgtagcGACGCCGCAGCAGAGGCGATAG |
| KY-135 | CTATCGCCTCTGCTGCGGCGTCgctacTGTCaAcCTGACGCATGTAGGTCCGG |
Nucleotide changes compared with the sequence of CW1 are indicated in lowercase.
Surface glycan depletion.
Surface glycans were depleted from RAW cells by treatment with either Vibrio cholerae neuraminidase (N7885; Sigma-Aldrich) for 2 h or d-threo-1-ethylenedioxyphenyl-2-palmitoyl-3-pyrrolidino-propanol (d-threo-P4) (45) for 72 h, as described previously (56), and tunicamycin from Streptomyces sp. (T7765; Sigma-Aldrich) for 24 h, benzyl 2-acetamido-2-deoxy-α-d-galactopyranoside (benzylGalNAc) (B4894; Sigma-Aldrich) for 48 h, or peptide:N-glycosidase F (PNGase F) endoglycosidase (P0704; Invitrogen) for 18 h. Cell viability was monitored for all conditions with WST-1 reagent (Roche) as described previously (56). Prior to neuraminidase and PNGase F treatments, 2 × 106 cells/well were plated, while prior to tunicamycin treatment, 2 × 105 cells/well were plated in 12-well dishes. For benzylGalNAc and d-threo-P4 treatments, 2 × 107 cells were batch treated in a T25 cell culture flask for 48 h and 72 h, respectively, and cells were split into individual tubes with 2 × 106 cells each before MNV infection. Pretreatment with d-threo-P4 and PNGase F was performed in the absence of fetal calf serum and by replacing Dulbecco's modified Eagle's medium (DMEM) with VP-SFM medium (Gibco).
To confirm a reduction in cell surface glycans, treated cells were incubated with 2 μg/ml Alexa Fluor 647-labeled wheat germ agglutinin (WGA) (Invitrogen) in 1× phosphate-buffered saline (PBS) plus 20 mM sodium azide for 1 h on ice. The unbound WGA was washed off three times with ice-cold PBS, and the cells were fixed in 1× PBS plus 2% paraformaldehyde plus 1% fetal calf serum plus 20 mM sodium azide for 10 min. The cells were washed in PBS and resuspended in 500 μl PBS plus 1% fetal calf serum for analysis by flow cytometry using FACScanto (Becton, Dickinson), and WGA binding was quantified by calculating the mean fluorescence intensity (MFI) of treated over untreated cells using FlowJo software (Tree Star Inc.).
Binding assays.
Binding assays are based on previously reported binding studies (56), but normalizing the inoculum to genome equivalents (eq). After the depletion of surface glycans, cells were washed twice with 2 ml PBS per well and incubated with 200 genome eq per cell for 60 min on ice in a volume of 0.5 ml. Unbound virus was removed after 1 h by washing three times with 1 ml ice-cold PBS. Cells were resuspended in 140 μl PBS and frozen at −80°C before RNA extraction and quantification by quantitative reverse transcription-PCR (qRT-PCR).
Quantification of MNV titers.
To measure MNV genome titers in tissue culture, total RNA was extracted using the QIAamp viral RNA minikit (Qiagen) according to the manufacturer's recommendations and resuspended in 50 μl AVE buffer (Qiagen). Viral cDNA was reverse transcribed using a 5-μl RNA suspension in a reaction volume of 35 μl as previously described (56) and used for quantitative PCR (TaqMan qPCR) on an Applied Biosystems 7500 fast real-time PCR system in a reaction volume of 20 μl using previously described primers, probe, and qPCR conditions (56). Genome eq were determined based on the MNV-1 cDNA standard.
To measure genome titers in mouse tissue or feces, samples were submitted to the Research Animal Diagnostic Services (Charles River Laboratories, Wilmington, MA) for qRT-PCR and analyzed as described previously (58).
To measure infectious virus, plaque assays were performed on duplicate samples in the case of tissue culture experiments or from homogenized tissue samples as previously described (24, 56, 61). Titers are presented as plaque-forming units (PFU) per ml.
Statistics.
Statistical analysis of the binding assays was performed using a two-tailed unpaired t test on log10-transformed titers. Differences are shown as percentages relative to the control treatment and presented as means ± standard errors of the means (SEM) of duplicate samples from at least three independent experiments. Statistical analysis of the virus load in mouse tissues between strains was performed using the nonparametric Mann-Whitney rank-sum test. All statistical analyses were carried out using the Prism software package (GraphPad Software).
RESULTS
MNV binds glycolipids and glycoproteins on murine macrophages in a strain-dependent manner.
We previously demonstrated that three MNV strains (MNV-1.CW3, WU11, and S99) bind to primary and cultured murine macrophages in a terminal SA- and ganglioside-dependent manner (56). To expand upon the strain specificities and glycan species, we compared attachment of MNV-1.CW3 and CR3 to RAW cells after removal of terminal SA or depletion of glycosphingolipids and O- or N-linked surface glycans (Fig. 1).
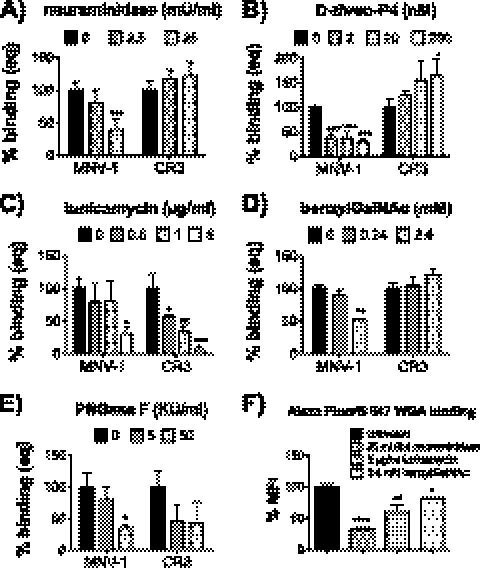
Fig 1.
MNV attachment to murine macrophages is strain dependent. Prior to incubation with MNV, surface glycans on RAW 264.7 cells were depleted with Vibrio cholerae neuraminidase (A), d-threo-P4 (B), tunicamycin (C), benzylGalNAc (D), or PNGase F (E). Cell viability was monitored using WST-1 reagent and remained above 85% during all treatments (data not shown). Cells were infected with 200 genome equivalents of MNV-1.CW3 (MNV-1) and CR3 per cell for 1 h on ice. The number of bound genomes was determined by qRT-PCR. Results are shown as percent binding normalized to the mock treatment. (F) Wheat germ agglutinin (WGA) binding to glycan-depleted RAW 264.7 cells. Binding of Alexa Fluor 647-labeled WGA to treated RAW 264.7 cells was determined by flow cytometry. WGA binding was quantified by calculating the mean fluorescence intensity (MFI) of treated over untreated cells. Results of all assays are presented as means ± SEM from three to four independent experiments. Statistical analysis was performed using the two-tailed unpaired t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
CR3 is a field isolate that shares over 95% amino acid identity within the capsid region of MNV-1.CW1, MNV-1.CW3, WU11, or S99 and has similar growth kinetics in vitro (data not shown). Interestingly, CR3 produces smaller plaques (58), and virus stocks consistently had an increased genome/PFU ratio compared to MNV-1.CW3 or MNV-1.CW1 in RAW cells (data not shown). To avoid compounding effects in our binding analysis due to the difference in noninfectious particles, binding assays were performed with equal genome equivalents of each virus, using 200 genome equivalents per cell. For all studies, mitochondrial dehydrogenase activity as a measure of cell viability was monitored using WST-1 (Roche), and viability remained above 85% in all experiments (data not shown). We furthermore validated that the respective treatments resulted in changes in cell surface glycans by measuring the reduction of cell surface SA by using Alexa Fluor 647-labeled wheat germ agglutinin (WGA; Invitrogen) with flow cytometry.
To measure the effect of terminal SA depletion on MNV attachment to macrophages, RAW cells were pretreated with neuraminidase and incubated with MNV-1.CW3 or CR3. Neuraminidase treatment reduced binding of WGA by 68% ± 6% at the highest concentration of neuraminidase (25 mU/ml), indicating that our treatment effectively removed SA-containing side chains (Fig. 1F). At the same concentration, MNV-1.CW3 binding was significantly reduced by 61% ± 8%, while binding of CR3 was increased by 22% ± 12% (Fig. 1A). This confirmed that MNV-1.CW3 binding to murine macrophages is terminal SA dependent (56) and suggested that CR3 binds other cell surface moieties. Similarly, pretreatment with d-threo-P4, an inhibitor of glycosphingolipid synthase (35, 45), reduced MNV-1.CW3 binding by 70% ± 5% but significantly increased CR3 binding by 65% ± 35% at 200 nM (Fig. 1B). Our previous work demonstrated that under the same conditions, d-threo-P4 treatment of RAW cells reduces GD1a ganglioside levels by 93% (56). Thus, the observed increase in CR3 binding may indicate that the removal of glycosphingolipids from the cell surface exposed previously inaccessible CR3 binding sites. Overall, these results confirmed our previous findings that MNV-1.CW3 requires terminal SA and gangliosides for macrophage binding (56) and determined that CR3 binding was independent of gangliosides and terminal SA.
Because internal SA are resistant to neuraminidase treatment (28) and competition with lectins specific for α2,6-linked SA affected MNV-1 infection of murine macrophages (56), glycoproteins were analyzed as alternative attachment receptors. RAW cells were treated with inhibitors of N-linked glycosylation, tunicamycin (14, 59), or PNGase F (34), as well as an inhibitor of O-linked glycosylation, benzyl-N-acetyl-α-d-galactosaminide (benzylGalNAc) (6, 33), before incubation with MNV-1.CW3 or CR3. Treatments with 5 μg/ml tunicamycin and 2.4 mM benzylGalNAc reduced binding of our control, WGA, by 40% ± 10% and 18% ± 8%, respectively (Fig. 1F). The different degree of sialic acid removal by each treatment reflects the relative abundance of N-linked versus O-linked surface glycoproteins (18). With regard to MNV, tunicamycin treatment reduced CR3 binding to murine macrophages in a dose-dependent manner, while MNV-1.CW3 was only affected at the highest concentration (Fig. 1C). At the highest tunicamycin concentration (5 μg/ml), binding of MNV-1.CW3 and CR3 was reduced by 70% ± 11% and 94% ± 5%, respectively, indicating that both strains bind to N-linked glycoproteins on the macrophage surface. On the other hand, only MNV-1.CW3 binding but not CR3 binding was reduced by 47% ± 1% at the highest concentration of benzylGalNAc (2.4 mM).
To independently verify the importance of N-linked glycans for MNV-1 and CR3 binding and to rule out secondary effects of tunicamycin treatment on global alterations in cell surface protein makeup, RAW cells were treated with the endoglycosidase PNGase F, which removes high-mannose N-linked glycans from glycoproteins (40) (Fig. 1E). At the highest concentration of PNGase F (50,000 U/ml), MNV-1 binding was reduced by 98% ± 1% and CR3 binding was reduced by 58% ± 10%, confirming that both MNV strains depended on N-linked glycans for macrophage cell surface attachment. Unfortunately, we were unable to perform such independent verification in the case of O-linked glycans, as to the best of our knowledge there are no drugs that exclusively remove O-linked glycans.
Overall, these data show that MNV-1.CW3 and CR3 require different glycans for attachment in vitro and indicate that glycan binding for MNV is strain dependent (Table 2).
Table 2.
Summary of glycan dependency of the wild-type and recombinant MNV strains
| Characteristic | Presence of characteristic in strain: |
|||
|---|---|---|---|---|
| MNV-1.CW3 | CR3 | MNV-1.CW1 | MNV-1.RVSS | |
| Terminal SA dependent | + | − | + | − |
| Ganglioside dependent | + | − | + | + |
| N-linked GP dependent | + | + | + | + |
| O-linked GP dependent | + | − | + | − |
CR3 displays an altered infectivity profile in vivo compared to MNV-1.
To investigate whether MNV strains with different in vitro glycan-binding phenotypes differ in their in vivo tissue tropisms, 7-week-old C57BL/6 mice were infected with 1 × 106 PFU/ml of MNV-1.CW1 or CR3. Tissues were harvested from multiple regions of the intestine, mesenteric lymph nodes (MLN), liver (LI), lung (LU), and spleen (SP) at 2 or 35 dpi and measured by plaque assay or qRT-PCR, respectively (Fig. 2A).
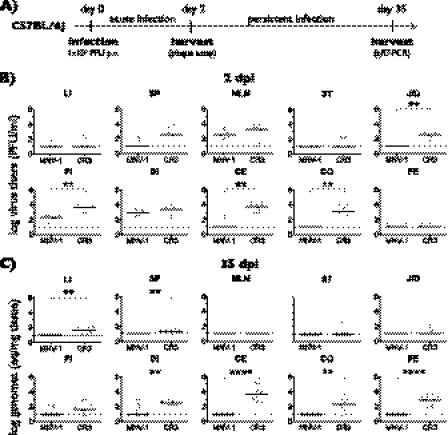
Fig 2.
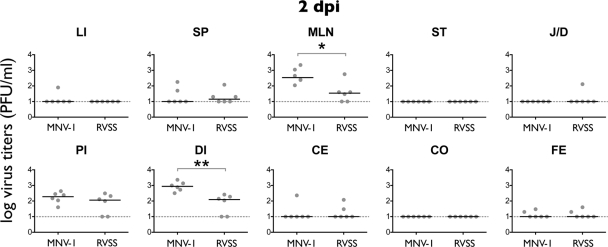
Comparison of the tissue tropism of MNV-1 and CR3. (A) The outline of the experimental design is shown. Seven-week-old C57BL/6 mice were infected orally (per os [p.o.]) with 1 × 106 PFU of MNV-1.CW1 (MNV-1) or CR3. Three sets of two mice were sacrificed during acute infection at 2 days postinfection (dpi), and three sets of three mice were sacrificed during persistent infection at 35 dpi and analyzed by plaque assay or qRT-PCR, respectively. (B) Tissues were harvested 2 dpi, and viral titers were determined by plaque assay from liver (LI), spleen (SP), mesenteric lymph nodes (MLN), stomach (ST), jejunum/duodenum (J/D), proximal ileum (PI), distal ileum (DI), cecum (CE), and colon (CO) and single fecal pellets from the colon (FE). (C) Viral loads at 35 dpi were determined by qRT-PCR. For all data, the limit of detection is represented by the dashed line. Titers from each mouse are represented by a circle, and medium titers are indicated by a horizontal line. Significant differences in titers between CR3 and MNV-1 were determined using the nonparametric Mann-Whitney test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
At 2 dpi, MNV-1.CW1 and CR3 were consistently detected in the proximal ileum (PI), distal ileum (DI), and MLN and sporadically detected in LU, LI, and SP (Fig. 2B) (data not shown for LU). In addition, CR3 was only sporadically detected in the stomach (ST), while MNV-1 was sporadically detected in the cecum (CE) and was absent from ST, jejunum/duodenum (J/D), and colon (CO). CR3 viral loads were consistently detectable and significantly higher in the J/D, PI, CE, and CO compared to MNV-1.CW1 viral loads. Infectious particles in the feces were only sporadically detected for both strains, suggesting shedding was limited at this time point.
CR3 persistently infects C57BL/6 mice for at least 35 days (58). Therefore, we extended our comparison to this late time point and used the more sensitive qRT-PCR assay instead of the plaque assay to detect viral genomes (Fig. 2C) (data not shown for LU). MNV-1.CW1 genomes were sporadically detected in PI and DI at 35 dpi in individual mice, suggesting that MNV-1.CW1 may not always be completely cleared from C57BL/6 mice. Significantly higher levels of CR3 were consistently detected in DI, CE, CO, and feces and sporadically detected in ST, J/D, LI, and SP, while being absent from MLN and LU. The presence of CR3 but not MNV-1.CW1 genomes in feces confirmed that CR3 is persistently shed for weeks. These data demonstrated that MNV-1.CW1 and CR3 differ significantly in their tissue tropisms, raising the intriguing possibility that differential glycan usage contributes to organ distribution and persistence in the gastrointestinal tract.
Mapping of a glycan-binding site in the MNV-1 P domain.
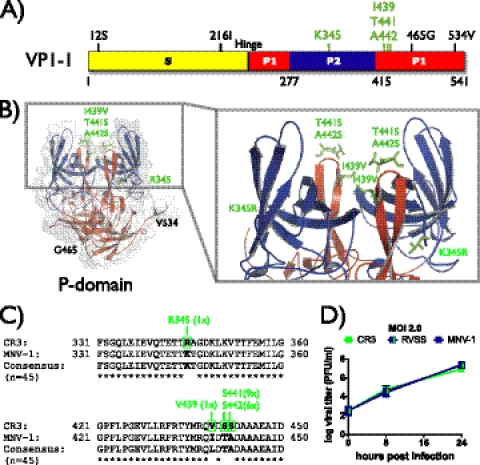
To test whether glycan binding contributes to tissue tropism, we first set out to map a glycan-binding site in the MNV-1 capsid. Because the human norovirus P domain contains carbohydrate-binding sites (e.g., see reference 51), we hypothesized that this may also be the case for MNV. In order to map a putative glycan-binding domain in MNV, the amino acid sequences of the capsid proteins of three neuraminidase-sensitive strains, MNV-1.CW3, WU11, and S99, were compared with those of the neuraminidase-resistant strain CR3. Based on the alignment, eight amino acids unique to CR3 were identified within the capsid protein that could account for neuraminidase resistance—two in the shell (S) domain and six in the P domain (Fig. 3A). The latter six amino acids were then located in our recently determined high-resolution crystal structure of the MNV-1.CW1 P domain (57) (Fig. 3B). Four amino acids, K345, I439, T441, and A442, were localized at the surface of the P domain and therefore hypothesized as the ones most likely to interact with host glycans. A comparison of these amino acids in an alignment of 45 known MNV capsid sequences showed that the exchanges K345R and I439V were exclusively found in CR3, while T441S and A442S sporadically appeared in other strains (Fig. 3C). To test whether amino acids 345, 439, 441, and 442 in the MNV capsid constitute a glycan-binding site of functional importance during host cell attachment, we used reverse genetics to exchange these amino acids in MNV-1.CW1, the background of the reverse genetics system (60), with the corresponding amino acids from CR3. Recombinant viruses MNV-1.K345R, MNV-1.VSS, and MNV-1.RVSS were successfully rescued and showed similar growth kinetics at low (0.05) and high (2.0) multiplicities of infection (MOI) to those of CR3, wild-type MNV-1.CW3, and recombinant MNV-1.CW1 in RAW cells (Fig. 3D) (data not shown). This demonstrated that the introduced mutations did not significantly alter the infection of murine macrophages in tissue culture and enabled further functional studies.
Fig 3.
Identification and mutagenesis of a putative glycan-binding site. (A) Schematic representation of the MNV-1 major capsid protein VP-1. The shell domain (S domain; amino acids [aa] 1 to 218) is shown in yellow, and the protruding domain (P domain) is shown in red (P1; aa 229 to 277 and aa 416 to 541) and blue (P2; aa 278 to 415). Amino acid positions that differ between neuraminidase-sensitive (MNV-1, S99, and WU11) and neuraminidase-resistant (CR3) viruses are labeled based on their position in MNV-1. Amino acids of the putative glycan-binding site K345, I439, T441, and A442 are emphasized in green. (B) Cartoon and surface reconstruction of the three-dimensional (3D) structure of the MNV-1 P domain dimer (Protein Data Bank [PDB] no. 3LQ6), showing the topology of the putative glycan-binding site. Colors correspond to regions shown in panel A. Light and dark gray surfaces indicate individual P domain molecules of the dimer. Amino acid exchanges in the P domain between neuraminidase-sensitive strains and CR3 are labeled and shown as ball-and-stick representations. Amino acids of the putative glycan-binding site are highlighted in green. (C) Conservation of amino acids within the putative glycan-binding site. Amino acid alignments of two capsid regions (aa 331 to 360 and aa 421 to 450) flanking the putative glycan-binding site are shown. MNV-1 and CR3 are aligned to a consensus sequence derived from 45 MNV capsids. An asterisk below the alignment marks conserved amino acids. Amino acids of the putative glycan-binding site are emphasized in boldface. Frequencies of the CR3-specifc mutations are indicated above the alignment and marked in green. (D) Growth curve analysis of wild-type and recombinant MNV strains on cultivated murine macrophages. RAW 264.7 cells were infected with CR3, MNV-1.CW1 (MNV1), or MNV-1.RVSS (RVSS) at an MOI of 2.0 for 1 h on ice. Unbound virus was washed off, and infection was allowed to proceed at 37°C for 8 or 24 h. Viral titers were determined by plaque assay and are presented as means ± SEM from three independent experiments.
Functional characterization of a glycan-binding site in MNV-1.
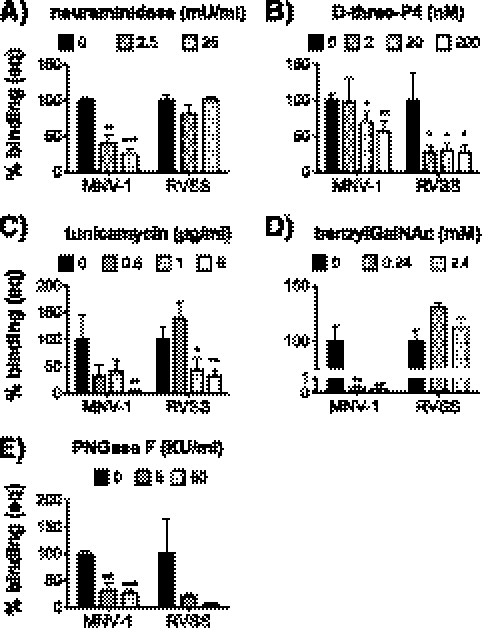
To investigate whether amino acids K345, I439, T441, and A442 constituted a functional host attachment site, glycan-binding profiles between the recombinant wild-type MNV-1.CW1 and mutant MNV-1.RVSS were determined. Depletion of terminal SA by pretreating RAW cells with neuraminidase resulted in a significant reduction (by 74% ± 7% at 25 mU/ml) of MNV-1.CW1 attachment, while no significant changes were observed in MNV-1.RVSS attachment (Fig. 4A). Recombinant viruses with partial mutations of the glycan-binding site, MNV-1.K345R and MNV-1.VSS, did not show significantly altered neuraminidase sensitivity compared to MNV-1.CW1 (data not shown). This suggested that all four amino acids contribute to SA binding and demonstrated that MNV-1.CW1, but not MNV-1.RVSS, requires terminal SA for host cell binding. A different pattern was observed following glycosphingolipid depletion (Fig. 4B). Macrophage binding of both viruses MNV-1.CW1 and MNV-1.RVSS was sensitive to d-threo-P4 treatment and was significantly reduced by 44% ± 15% and 72% ± 10%, respectively, at the highest concentration (Fig. 4B). This suggested that while MNV-1.RVSS binding is independent from terminal SA, it still binds glycosphingolipids, by binding either to internal SA or to the glycan backbone.
Fig 4.
Glycan-binding profiles of recombinant MNV-1 and RVSS on murine macrophages. Surface glycans on RAW 264.7 cells were depleted with neuraminidase (A), d-threo-P4 (B), tunicamycin (C), benzylGalNAc (D), or PNGase F (E) prior to incubation with MNV. Cell viability was monitored using WST-1 reagent and remained above 85% during all treatments (data not shown). Cells were infected with 200 genome equivalents of MNV-1.CW1 (MNV-1) and MNV-1.RVSS (RVSS) for 1 h on ice to prevent internalization. The number of bound genomes was determined by qRT-PCR. Results are shown as percent binding normalized to the mock treatment. Results of all assays are presented as means ± SEM from three independent experiments. Statistical analysis was performed using the two-tailed unpaired t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001. KU, kilo units.
In addition, the binding patterns of both viruses upon depletion of N- or O-linked glycoproteins were analyzed (Fig. 4C, D, and E). Tunicamycin treatment reduced binding of MNV-1.CW1 by 95% ± 1% at 5 μg/ml (Fig. 4C), while benzylGalNAc treatment almost completely abolished binding, even at a lower concentration (by 99% ± 0.1% at 0.24 mM) (Fig. 4D). In addition, PNGase F treatment reduced MNV-1.CW1 binding by 74% ± 8% and MNV-1.RVSS binding by 95% ± 4% (Fig. 4E). This indicated that recombinant MNV-1.CW1 also required glycoproteins for binding to macrophages.
Of note is that MNV-1.CW1 binding compared to MNV-1.CW3 binding was significantly more reduced following benzylGalNAc treatment at all concentrations (P < 0.001) (compare Fig. 1D and 4D) and after a 5-μg/ml tunicamycin treatment (P = 0.0124) (compare Fig. 1C and 4C). The capsid proteins of MNV-1.CW3 and MNV-1.CW1 differ in only one amino acid position (K296E) (61). The change of K (positive) to E (negative) altered the charge of this single residue, while the surrounding amino acids Phe297, Glu298, and Val304 were uncharged and not altered. This indicated that the increased glycoprotein dependency of MNV-1.CW1 is linked to amino acid 296 and suggested that this residue contributes to the overall glycan-binding epitope. Interestingly, binding to cultured macrophages by MNV-1.RVSS, which harbors the glutamic acid residue at position 296 like MNV-1.CW1, was only reduced by N-linked glycan depletion (by 69% ± 12% at 5 μg/ml tunicamycin and by 74% ± 8% with 50 kilo units [KU]/ml PNGAse F) but not benzylGalNAc treatment (Fig. 4C, D, and E). This suggested that the RVSS mutations may contribute more strongly to overall N-glycan binding and may override the contribution of E296 to O-linked glycoprotein binding.
Overall, these data supported our hypothesis that amino acids K345, I439, T441, and A442 in the MNV-1 capsid constitute a functional host attachment site linked to terminal SA binding to which amino acid 296 contributes.
Glycan binding contributes to MNV's tissue tropism.
To investigate, whether the interaction of MNV with surface glycans contributes to the pathogenesis of the virus, the tissue tropism of recombinant MNV-1.CW1 and MNV-1.RVSS was analyzed (Fig. 5). Seven-week-old C57BL/6 mice were infected with MNV-1.CW1 or MNV-1.RVSS as described above (Fig. 2A). At 2 dpi, significant changes in viral titers between MNV-1.CW1 and MNV-1-RVSS were observed in the MLN (6-fold) and the DI (9-fold). Similar virus loads were observed in the PI, while all other sites had no or only sporadically detectable titers. At 35 dpi, the MNV-1.CW1 and MNV-1-RVSS genomes were only sporadically detected (data not shown), suggesting that persistence is not linked to glycan binding. Furthermore, the glycan-binding site comprising amino acids 345, 439, 441, and 442 is not linked to observed differences in the large intestinal tropism of MNV-1 and CR3. Taken together, these data indicated for the first time that glycan binding may contribute to the tissue tropism of MNV.
Fig 5.
Comparison of the tissue tropisms of recombinant neuraminidase-sensitive MNV-1 and neuraminidase-resistant RVSS in vivo. Infection of 7-week-old C57BL/6 mice with recombinant MNV-1.CW1 (MNV-1) or MNV-1.RVSS (RVSS) was performed as described in the legend to Fig. 2. The MNV-1 data shown in Fig. 2 were regraphed for clarity. Titers from each mouse are represented by a circle, and medium titers are indicated by a horizontal line. Significant differences in titers between MNV-1 and RVSS were determined using the nonparametric Mann-Whitney test. *, P < 0.05; **, P < 0.01.
DISCUSSION
Virus-receptor interactions are important for a successful infection and a major determinant for cell, tissue, and species tropism (48). Understanding these interactions at a molecular level is critical in elucidating how a virus infects its host and may provide insights into new antiviral targets. Thus, in this study, we present a comparative analysis of glycan binding profiles of two MNV strains in vitro and infection in vivo and report the first functional glycan-binding epitope for a GV norovirus. These experiments came to several conclusions.
First, MNV binding to permissive murine macrophages was strain dependent (summarized in Table 2). Our previous observations demonstrate that MNV strains MNV-1, S99, and WU11 use terminal SA on the ganglioside GD1a as an attachment receptor on murine macrophages (56). However, since neither terminal SA nor glycosphingolipid depletion completely abolished macrophage binding, we investigated whether MNV binds to additional host cell components. Here, we showed that MNV-1 also adhered to glycoproteins on the surface of macrophages. Furthermore, binding of the fecally isolated MNV strain (CR3) to macrophages was resistant to terminal SA and ganglioside depletion or benzylGalNAc treatment but depended on N-linked glycoproteins. Our data indicated that MNV, similarly to human norovirus (13, 52), may have evolved multiple strategies to bind distinct glycan receptors on the cell surface. Interestingly, a strain-dependent SA-binding phenotype is also observed in rotaviruses, where animal rotaviruses bind terminal SA in a neuraminidase-sensitive manner, while the neuraminidase-insensitive human rotaviruses attach to internal SA, which are inaccessible to neuraminidase cleavage (12, 17, 54). Additional studies are required to determine whether the neuraminidase-insensitive CR3 strain can utilize internal SA moieties during attachment.
Second, MNV strains showed different peak titers in the intestine and exhibited different tissue tropisms, particularly in the large intestine. Recent reports demonstrate that several MNV strains, including CR3 and MNV-3, exhibit differences in tissue tropism and persistence compared to MNV-1 (29, 58). Our data support these observations and demonstrated that a tropism for the large intestine is not unique to MNV-3, but also is observed with CR3. In addition, similar to MNV-3 (29), CR3 also reached significantly higher peak titers within the intestine compared to MNV-1. Previous work concluded that MLN are a site for persistent replication because persistent strains such as CR3, but not acute strains (i.e., MNV-1), were present in the MLN at 35 dpi (58). Interestingly, we were unable to detect CR3 genomes in the MLN despite high genome titers throughout the intestine (Fig. 2B). This may be due to a different passage virus stock or inoculation dose but suggested that while replication in the MLN may contribute to persistence, it is not a prerequisite.
Third, we identified a novel functional glycan-binding epitope in the MNV-1 capsid comprising amino acids 345, 439, 441, and 442 using mutagenesis. A comparison of the capsid protein amino acid sequences of the neuraminidase-sensitive strains MNV-1.CW3, S99, and WU11 with the neuraminidase-resistant CR3 strain identified unique amino acid residues within the CR3 capsid protein. Using reverse genetics, we generated recombinant MNV-1 viruses with amino acid residues from CR3, specifically MNV-1.VSS (I439V, T441S, and A442S), MNV-1.K345R, or the double mutant MNV-1.RVSS (K345R or I439V, T441S, or A442S), to test whether the CR3 residues could confer neuraminidase resistance to MNV-1-macrophage binding. This was indeed the case, and the combined mutations of all four amino acids in MNV-1.RVSS changed the terminal SA-binding phenotype of the neuraminidase-sensitive MNV-1.CW1 to the neuraminidase-resistant CR3 phenotype (Fig. 4). Therefore, the amino acids K345, I439, T441, and A442 form a conformational epitope that contains a functional glycan-binding site with a role in terminal SA-dependent host cell attachment. Interestingly, the same pattern did not repeat after d-threo-P4 depletion, and MNV-1.RVSS binding was reduced, while CR3 binding was increased. However, since these two viruses differ in additional amino acids in the capsid (although not surface exposed), we cannot rule out subtle changes in the overall protein structure. In the future, structural analysis of the protein-ligand interaction is needed to determine whether this epitope directly interacts with terminal SA moieties or whether structural differences exist between the MNV-1 and CR3 capsids.
Fourth, the conformational glycan-binding epitope comprising amino acids 345, 439, 441, and 442 contributed to MNV tissue tropism in vivo. The generation of two closely related viruses with differences in the in vitro glycan-binding profile enabled studies investigating a link between glycan binding and MNV tropism in vivo. The neuraminidase-sensitive wild-type MNV-1 and the neuraminidase-resistant mutant MNV-1.RVSS differed in peak titers in the DI and MLN during acute infection (Fig. 5). While this correlation suggested a potential link between the in vitro phenotype and the in vivo tropism, further studies are needed to determine whether changes in tropism are a direct result of a difference in glycan binding between MNV strains or whether this is an indirect effect, e.g., replication efficiency of these viruses in tissues. Recent work suggests that the capsid is required for viral replication but did not identify specific amino acid residues (27). Furthermore, future studies are needed to investigate whether other parameters of pathogenesis, such as histopathology, are also changed. However, the glycan-binding epitope does not account for the large intestinal tropism, since MNV-1.RVSS did not mirror the CR3 tissue tropism and was absent in the large intestine. This indicated that determinants outside the four residues of the glycan-binding site account for the ability to infect the large intestine. Identification of such determinants will need to await the creation of a CR3 infectious clone.
Fifth, binding of MNV-1 to terminal sialic acids was not solely determined by amino acids K345, I439, T441, and A442 because CR3 and MNV-1.RVSS, which share the same amino acids in this epitope, differed in their terminal sialic acid dependency (summarized in Table 2). Our results also indicated a role for amino acid 296 in glycan binding. The capsid proteins of MNV-1.CW3 and MNV-1.CW1 differ in only one amino acid, K296E (61). MNV-1.CW3 binding to macrophages was significantly less sensitive to benzylGalNAc treatment than MNV-1.CW1, and a similar trend was observed for N-glycan depletions. This indicated that residue K296 directly or indirectly affects cell surface glycan binding via the viral glycan binding epitope (K345, I439, T441, and A442). While identification of specific glycans followed by detailed structural analysis is needed in the future to determine whether residue 296 directly interacts with a glycan, recent structural data demonstrate the flexibility of surface-exposed loops in MNV-1 (57) and provide a potential explanation for an indirect effect. Residue 296 is part of the surface exposed A′-B′ loop, which stabilizes the larger E′-F′ loop in an open conformation (57). In a closed formation, the E′-F′ loop relocates toward the canyon of the VP1 dimer and comes within 10 Å of amino acid A442 in the glycan binding epitope. This relocation is close enough to potentially mask the glycan-binding epitope or even take part in binding activity. In addition, the change in charge at position 296 (positive Lys versus negative Glu) may also affect binding since a single charged mutation at specific positions can impair O-glycosylation of a single acceptor site (42).
Interestingly, MNV-1.CW3 and MNV-1.CW1 differ significantly in their virulence in STAT1−/− mice (4, 61). Thus, the observed increased dependence of MNV-1.CW1 on binding to glycoproteins on the macrophage cell surface may contribute to attenuation in STAT1−/− mice, although further experiments are required to directly test this hypothesis.
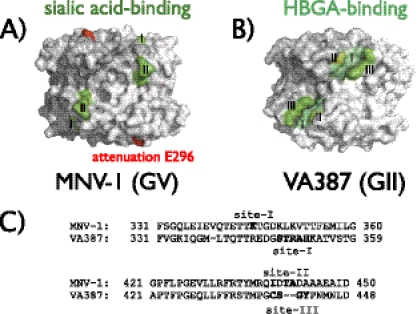
Sixth, the relative location of the glycan-binding epitope is conserved between human and murine noroviruses (Fig. 6). Crystal structure analysis of different GII strains, including VA387 and TCH05 (GII.4), VA207 (GII.9), Vietnam026 (GII.10), and Hiro (GII.12) demonstrate that the HBGA glycan-binding epitope is located in a conserved elevated region in the P domain dimer (8, 22) (Fig. 6B). The same was true for MNV-1 (Fig. 6A). Furthermore, a striking resemblance was observed when comparing the locations of the VA387 (GII) and MNV-1 (GV) glycan-binding sites, which are positioned in similar locations on opposite sides of the P dimer interface and at similar points within the amino acid sequence (Fig. 6). This suggested that while human and murine noroviruses bind different glycan receptors, the necessity for glycan binding in a specific surface-exposed region of the capsid protein is conserved beyond species barriers. Furthermore, it also raised the intriguing possibility that the choice of specific glycans contributes to species specificity. In support of this, no binding of MNV-1 to available synthetic HBGA has so far been observed (13) despite the ability of MNV RNA to replicate in human cells (60), and the human ABO HBGA system, which is pivotal for human norovirus infection, does not exist in mice or any other small animal model (16).
Fig 6.
Comparison of glycan-binding sites between GII and GV noroviruses. P domain dimers of MNV-1.CW1 (GV) and VA387 (GII) (PDB no. 3LQ6 and 2OBR, respectively) are shown as 3D-reconstructed surface models. Light and dark gray surfaces distinguish the individual P domain monomers. (A) Locations of the glycan-binding sites of MNV identified herein. The SA-binding epitope in MNV-1.CW1 (site I represents K345, and site II represents I439, T441, or A442) is highlighted in green, and the attenuation mutation K296E is highlighted in red. (B) The locations of three HBGA-binding sites in the human norovirus GII strain VA387 are highlighted in shades of green: site I (aa 343 to 347), site II (aa 374), and site III (aa 441 to 444) (23, 53). (C) Amino acid residues encompassing the glycan-binding sites in VA387 and MNV-1. Shown are amino acid alignments of the two glycan-binding regions in MNV-1 compared to those in VA387.
In summary, the results presented in this study provide data on the types of receptor molecules involved in MNV binding of macrophages and indicate that the preference of different strains for specific glycans contributes to the tissue tropism of the virus.
ACKNOWLEDGMENTS
This work was funded by NIH R01 AI080611 to C.E.W. J.W.P. was funded by a University of Michigan Human Genetics training grant (NIH T32 GM 07544), a Molecular Mechanisms of Microbial Pathogenesis training grant (NIH T32 AI 007528), and an American Heart Association predoctoral fellowship (grant 10PRE3650036).
We thank J. Shayman (University of Michigan) for the generous gift of d-threo-P4 and M. Jiang (University of Michigan) for critical reading of the manuscript.
Footnotes
Published ahead of print 21 March 2012
REFERENCES
- 1. Apweiler R, Hermjakob H, Sharon N. 1999. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim. Biophys. Acta 1473:4–8 [DOI] [PubMed] [Google Scholar]
- 2. Armstrong GD. 2000. Carbohydrate receptor mimicry as a basis for antibacterial therapy. Curr. Opin. Drug Discov. Dev. 3:191–202 [PubMed] [Google Scholar]
- 3. Atmar RL. 2010. Noroviruses—state of the art. Food Environ. Virol. 2:117–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bailey D, Thackray LB, Goodfellow IG. 2008. A single amino acid substitution in the murine norovirus capsid protein is sufficient for attenuation in vivo. J. Virol. 82:7725–7728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bomsel M, Alfsen A. 2003. Entry of viruses through the epithelial barrier: pathogenic trickery. Nat. Rev. Mol. Cell Biol. 4:57–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brockhausen I, et al. 1992. Control of O-glycan synthesis: specificity and inhibition of O-glycan core 1 UDP-galactose:N-acetylgalactosamine-alpha-R beta 3-galactosyltransferase from rat liver. Biochem. Cell Biol. 70:99–108 [DOI] [PubMed] [Google Scholar]
- 7. Bull RA, White PA. 2011. Mechanisms of GII.4 Norovirus evolution. Trends Microbiol. 19:233–240 [DOI] [PubMed] [Google Scholar]
- 8. Cao S, et al. 2007. Structural basis for the recognition of blood group trisaccharides by norovirus. J. Virol. 81:5949–5957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chachu KA, et al. 2008. Antibody is critical for the clearance of murine norovirus infection. J. Virol. 82:6610–6617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chaudhry Y, Skinner MA, Goodfellow IG. 2007. Recovery of genetically defined murine norovirus in tissue culture by using a fowlpox virus expressing T7 RNA polymerase. J. Gen. Virol. 88:2091–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen Y, et al. 2011. Crystallography of a Lewis-binding norovirus, elucidation of strain-specificity to the polymorphic human histo-blood group antigens. PLoS Pathog. 7:e1002152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ciarlet M, Estes MK. 1999. Human and most animal rotavirus strains do not require the presence of sialic acid on the cell surface for efficient infectivity. J. Gen. Virol. 80:943–948 [DOI] [PubMed] [Google Scholar]
- 13. Donaldson EF, Lindesmith LC, Lobue AD, Baric RS. 2010. Viral shape-shifting: norovirus evasion of the human immune system. Nat. Rev. 8:231–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Elbein AD. 1991. Glycosidase inhibitors: inhibitors of N-linked oligosaccharide processing. FASEB J. 5:3055–3063 [DOI] [PubMed] [Google Scholar]
- 15. Estes MK, Prasad BV, Atmar RL. 2006. Noroviruses everywhere: has something changed? Curr. Opin. Infect. Dis. 19:467–474 [DOI] [PubMed] [Google Scholar]
- 16. Fan X, et al. 2010. Induction of human blood group A antigen expression on mouse cells, using lentiviral gene transduction. Hum. Gene Ther. 21:877–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fukudome K, Yoshie O, Konno T. 1989. Comparison of human, simian, and bovine rotaviruses for requirement of sialic acid in hemagglutination and cell adsorption. Virology 172:196–205 [DOI] [PubMed] [Google Scholar]
- 18. Gahmberg CG, Tolvanen M. 1996. Why mammalian cell surface proteins are glycoproteins. Trends Biochem. Sci. 21:308–311 [PubMed] [Google Scholar]
- 19. Glass RI, Parashar UD, Estes MK. 2009. Norovirus gastroenteritis. N. Engl. J. Med. 361:1776–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Green KY. 2007. Caliciviridae, p 949–980 In Knipe DM, et al. (ed), Fields virology, 5th ed, vol 1 Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 21. Hansen S, et al. 2007. Closure of medical departments during nosocomial outbreaks: data from a systematic analysis of the literature. J. Hosp. Infect. 65:348–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hansman GS, et al. 2011. Crystal structures of GII.10 and GII.12 norovirus protruding domains in complex with histo-blood group antigens reveal details for a potential site of vulnerability. J. Virol. 85:6687–6701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hansman GS, Jiang XJ, Green KY. (ed). 2010. Caliciviruses molecular and cellular virology, vol 1 Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
- 24. Higgins PD, et al. 2011. Transient or persistent norovirus infection does not alter the pathology of Salmonella typhimurium induced intestinal inflammation and fibrosis in mice. Comp. Immunol. Microbiol. Infect. Dis. 34:247–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hsu CC, Riley LK, Wills HM, Livingston RS. 2006. Persistent infection with and serologic cross-reactivity of three novel murine noroviruses. Comp. Med. 56:247–251 [PubMed] [Google Scholar]
- 26. Hsu CC, Wobus CE, Steffen EK, Riley LK, Livingston RS. 2005. Development of a microsphere-based serologic multiplexed fluorescent immunoassay and a reverse transcriptase PCR assay to detect murine norovirus 1 infection in mice. Clin. Diagn. Lab. Immunol. 12:1145–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hyde JL, Gillespie LK, Mackenzie JM. 1 February 2012. Mouse norovirus-1 utilizes the cytoskeleton network to establish localization of the replication complex proximal to the microtubule organizing center. J. Virol. doi:10.1128/JVI.05784-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Isa P, Arias CF, Lopez S. 2006. Role of sialic acids in rotavirus infection. Glycoconj. J. 23:27–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kahan SM, et al. 2011. Comparative murine norovirus studies reveal a lack of correlation between intestinal virus titers and enteric pathology. Virology 421:202–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Karst SM. 2010. Pathogenesis of noroviruses, emerging RNA viruses. Viruses 2:748–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Karst SM, Wobus CE, Lay M, Davidson J, Virgin HW., IV 2003. STAT1-dependent innate immunity to a Norwalk-like virus. Science 299:1575–1578 [DOI] [PubMed] [Google Scholar]
- 32. Kitajima M, et al. 2009. Development of a broadly reactive nested reverse transcription-PCR assay to detect murine noroviruses, and investigation of the prevalence of murine noroviruses in laboratory mice in Japan. Microbiol. Immunol. 53:531–534 [DOI] [PubMed] [Google Scholar]
- 33. Kuan SF, Byrd JC, Basbaum C, Kim YS. 1989. Inhibition of mucin glycosylation by aryl-N-acetyl-alpha-galactosaminides in human colon cancer cells. J. Biol. Chem. 264:19271–19277 [PubMed] [Google Scholar]
- 34. Kuhn P, Tarentino AL, Plummer TH, Jr, Van Roey P. 1994. Crystallization and preliminary crystallographic analysis of peptide-N4-(N-acetyl-beta-D-glucosaminyl)asparagine amidase PNGase F. J. Mol. Biol. 241:622–623 [DOI] [PubMed] [Google Scholar]
- 35. Lee L, Abe A, Shayman JA. 1999. Improved inhibitors of glucosylceramide synthase. J. Biol. Chem. 274:14662–14669 [DOI] [PubMed] [Google Scholar]
- 36. Le Pendu J. 2004. Histo-blood group antigen and human milk oligosaccharides: genetic polymorphism and risk of infectious diseases. Adv. Exp. Med. Biol. 554:135–143 [DOI] [PubMed] [Google Scholar]
- 37. Le Pendu J, Ruvoen-Clouet N, Kindberg E, Svensson L. 2006. Mendelian resistance to human norovirus infections. Semin. Immunol. 18:375–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lindesmith L, et al. 2003. Human susceptibility and resistance to Norwalk virus infection. Nat. Med. 9:548–553 [DOI] [PubMed] [Google Scholar]
- 39. Mahler M, Kohl W. 2009. A serological survey to evaluate contemporary prevalence of viral agents and Mycoplasma pulmonis in laboratory mice and rats in western Europe. Lab Anim. 38:161–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Maley F, Trimble RB, Tarentino AL, Plummer TH., Jr 1989. Characterization of glycoproteins and their associated oligosaccharides through the use of endoglycosidases. Anal. Biochem. 180:195–204 [DOI] [PubMed] [Google Scholar]
- 41. Mumphrey SM, et al. 2007. Murine norovirus 1 infection is associated with histopathological changes in immunocompetent hosts, but clinical disease is prevented by STAT1-dependent interferon responses. J. Virol. 81:3251–3263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nehrke K, Hagen FK, Tabak LA. 1996. Charge distribution of flanking amino acids influences O-glycan acquisition in vivo. J. Biol. Chem. 271:7061–7065 [DOI] [PubMed] [Google Scholar]
- 43. Olofsson S, Bergstrom T. 2005. Glycoconjugate glycans as viral receptors. Ann. Med. 37:154–172 [DOI] [PubMed] [Google Scholar]
- 44. Prasad BV, et al. 1999. X-ray crystallographic structure of the Norwalk virus capsid. Science 286:287–290 [DOI] [PubMed] [Google Scholar]
- 45. Rani CS, et al. 1995. Cell cycle arrest induced by an inhibitor of glucosylceramide synthase. Correlation with cyclin-dependent kinases. J. Biol. Chem. 270:2859–2867 [DOI] [PubMed] [Google Scholar]
- 46. Rockx B, Baric RS, de Grijs I, Duizer E, Koopmans MP. 2005. Characterization of the homo- and heterotypic immune responses after natural norovirus infection. J. Med. Virol. 77:439–446 [DOI] [PubMed] [Google Scholar]
- 47. Shanker S, et al. 2011. Structural analysis of histo-blood group antigen binding specificity in a norovirus GII.4 epidemic variant: implications for epochal evolution. J. Virol. 85:8635–8645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Smith AE, Helenius A. 2004. How viruses enter animal cells. Science 304:237–242 [DOI] [PubMed] [Google Scholar]
- 49. Smith DC, Lord JM, Roberts LM, Johannes L. 2004. Glycosphingolipids as toxin receptors. Semin. Cell Dev. Biol. 15:397–408 [DOI] [PubMed] [Google Scholar]
- 50. Tamura M, Natori K, Kobayashi M, Miyamura T, Takeda N. 2004. Genogroup II noroviruses efficiently bind to heparan sulfate proteoglycan associated with the cellular membrane. J. Virol. 78:3817–3826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tan M, et al. 2008. Noroviral P particle: structure, function and applications in virus-host interaction. Virology 382:115–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tan M, Jiang X. 2011. Norovirus-host interaction: multi-selections by human histo-blood group antigens. Trends Microbiol. 19:382–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tan M, Jiang X. 2010. Norovirus gastroenteritis, carbohydrate receptors, and animal models. PLoS Pathog. 6:e1000983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Taube S, Jiang M, Wobus CE. 2010. Glycosphingolipids as receptors for non-enveloped viruses. Viruses 2:1011–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Taube S, Kurth A, Schreier E. 2005. Generation of recombinant norovirus-like particles (VLP) in the human endothelial kidney cell line 293T. Arch. Virol. 150:1425–1431 [DOI] [PubMed] [Google Scholar]
- 56. Taube S, et al. 2009. Ganglioside-linked terminal sialic acid moieties on murine macrophages function as attachment receptors for murine noroviruses. J. Virol. 83:4092–4101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Taube S, et al. 2010. High-resolution X-ray structure and functional analysis of the murine norovirus 1 capsid protein protruding domain. J. Virol. 84:5695–5705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Thackray LB, et al. 2007. Murine noroviruses comprising a single genogroup exhibit biological diversity despite limited sequence divergence. J. Virol. 81:10460–10473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tkacz JS, Lampen O. 1975. Tunicamycin inhibition of polyisoprenyl N-acetylglucosaminyl pyrophosphate formation in calf-liver microsomes. Biochem. Biophys. Res. Commun. 65:248–257 [DOI] [PubMed] [Google Scholar]
- 60. Ward VK, et al. 2007. Recovery of infectious murine norovirus using pol II-driven expression of full-length cDNA. Proc. Natl. Acad. Sci. U. S. A. 104:11050–11055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wobus CE, et al. 2004. Replication of Norovirus in cell culture reveals a tropism for dendritic cells and macrophages. PLoS Biol. 2:e432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wobus CE, Thackray LB, Virgin HW., IV 2006. Murine norovirus: a model system to study norovirus biology and pathogenesis. J. Virol. 80:5104–5112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yang B, Chuang H, Yang KD. 2009. Sialylated glycans as receptor and inhibitor of enterovirus 71 infection to DLD-1 intestinal cells. Virol. J. 6:141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yunus MA, Chung LM, Chaudhry Y, Bailey D, Goodfellow I. 2010. Development of an optimized RNA-based murine norovirus reverse genetics system. J. Virol. Methods 169:112–118 [DOI] [PMC free article] [PubMed] [Google Scholar]