Abstract
HIV gene therapy has the potential to offer an alternative to the use of current small-molecule antiretroviral drugs as a treatment strategy for HIV-infected individuals. Therapies designed to administer HIV-resistant stem cells to an infected patient may also provide a functional cure, as observed in a bone marrow transplant performed with hematopoietic stem cells (HSCs) homozygous for the CCR5-Δ32-bp allele. In our current studies, preclinical evaluation of a combination anti-HIV lentiviral vector was performed, in vivo, in humanized NOD-RAG1−/− IL2rγ−/− knockout mice. This combination vector, which displays strong preintegration inhibition of HIV-1 infection in vitro, contains a human/rhesus macaque TRIM5α isoform, a CCR5 short hairpin RNA (shRNA), and a TAR decoy. Multilineage hematopoiesis from anti-HIV lentiviral vector-transduced human CD34+ HSCs was observed in the peripheral blood and in various lymphoid organs, including the thymus, spleen, and bone marrow, of engrafted mice. Anti-HIV vector-transduced CD34+ cells displayed normal development of immune cells, including T cells, B cells, and macrophages. The anti-HIV vector-transduced cells also displayed knockdown of cell surface CCR5 due to the expression of the CCR5 shRNA. After in vivo challenge with either an R5-tropic BaL-1 or X4-tropic NL4-3 strain of HIV-1, maintenance of human CD4+ cell levels and a selective survival advantage of anti-HIV gene-modified cells were observed in engrafted mice. The data provided from our study confirm the safety and efficacy of this combination anti-HIV lentiviral vector in a hematopoietic stem cell gene therapy setting for HIV and validates its potential application in future clinical trials.
INTRODUCTION
HIV gene therapy offers a potential alternative treatment strategy for HIV-infected individuals compared to the use of current antiretroviral drugs, which, after prolonged use, can become toxic and allow for the generation of escape mutants (7, 11, 20, 24, 26, 29). A recent hematopoietic stem cell (HSC) transplant for acute myeloid leukemia in an HIV-infected patient was performed utilizing allogeneic cells from an individual homozygous for the Δ32 CCR5 deletion (12, 14, 19). HIV-1 suppression has been observed in the recipient to date, even after halting antiretroviral drug therapy (16). The report of the success of this stem cell transplant is the first to describe a functional cure of an HIV-infected individual and brings about a realization that stem cell therapies for HIV-infected patients can have a dramatic impact on the outcome of their disease (15). Therefore, HIV stem cell gene therapy offers the possibility to mimic the results of this transplant by engineering a patient's autologous HSCs to express anti-HIV genes, thus conferring resistance to infection (27). Advantages in utilizing HSCs for HIV gene therapy include the reconstitution of an HIV-resistant immune system, the potential for lifelong protection from further HIV replication, and the possibility of a one-time treatment upon transplantation of anti-HIV gene-modified HSCs (27).
Numerous anti-HIV genes have been designed to inhibit HIV replication; however, the use of a single anti-HIV gene may not be sufficient to protect cells long-term from infection due to the high mutation rate of HIV (1, 3, 4, 13, 18, 21–23). This has been proven through the use of monotherapy with small-molecule antiretroviral drugs, which eventually select for viral escape mutants (7, 20, 26). Therefore, similar to combination approaches with small-molecule drugs, multiple anti-HIV genes inserted into a single gene therapy vector could potentially confer stronger protection from HIV infection in the long term while also preventing the generation of viral resistance (2, 5, 9, 10).
Anti-HIV genes targeted to block the early stages of HIV infection, including attachment and entry, reverse transcription, and integration, offer a number of advantages over molecules which act at later stages of infection, including preventing the generation of provirus and the continued replenishment of viral reservoirs, which are major reasons for the failure to cure HIV-infected individuals (1, 3, 4, 25, 28). In this regard, by combining multiple preintegration anti-HIV genes into a single vector, potent preintegration protection from HIV infection could be conferred (5, 17). In a previous report by our group, strong preintegration protection from HIV-1 infection, in vitro, was established by a triple-combination anti-HIV lentiviral vector containing a human/rhesus macaque TRIM5α isoform, a CCR5 short hairpin RNA (shRNA), and a TAR decoy (5). This vector not only prevented HIV integration in challenged cells but also blocked the generation of escape mutants.
For the preclinical analysis of anti-HIV genes and vectors, it is necessary to utilize an appropriate in vivo model capable of demonstrating safety and efficacy of the novel therapy (6, 8). The NOD-RAG1−/− IL2rγ−/− double mutant (NRG) mouse model offers the potential to evaluate multilineage human hematopoiesis from intrahepatic injection of human CD34+ HSCs into newborn mice. Three months after transplantation, functional human T cells, B cells, and macrophages can be detected in lymphoid organs, including the spleen, thymus, and bone marrow (6). Mice successfully engrafted with a human immune system can be infected with HIV and display normal HIV disease characteristics, including CD4+ cell depletion and an increase in plasma viremia (6). This mouse model offers a unique preclinical in vivo system to evaluate anti-HIV gene therapy molecules in human cells at a level acceptable to regulatory agencies.
In our current studies, the preclinical safety and efficacy of a combination anti-HIV lentiviral vector was evaluated, in vivo, in a humanized NRG mouse model. Here we demonstrate multilineage human hematopoiesis from anti-HIV lentiviral vector-transduced CD34+ HSCs in the peripheral blood and in various lymphoid organs, including the thymus, spleen, and bone marrow. After in vivo challenge with either an R5-tropic BaL-1 or an X4-tropic NL4-3 strain of HIV-1, maintenance of human CD4+ cells and a selective survival advantage were observed in mice containing the anti-HIV vector-transduced cells. The data provided here confirm the utility of this combination anti-HIV lentiviral vector in inhibiting HIV infection in a stem cell gene therapy setting and validate its potential for application in a future human clinical trial.
MATERIALS AND METHODS
Lentiviral vector design and production.
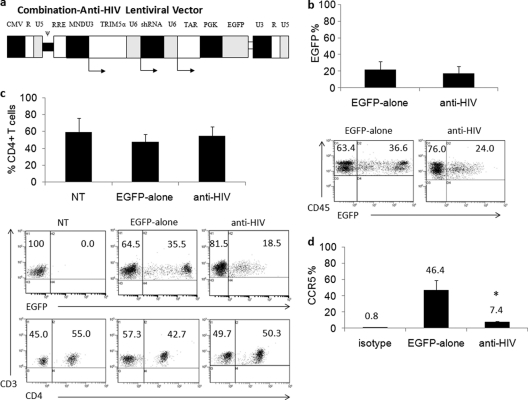
The construction of the combination anti-HIV lentiviral vector has been described previously (5). Briefly, a third-generation self-inactivating lentiviral vector, CCLc-MNDU3-x-PGK-EGFP (control vector), which contains an enhanced green fluorescent protein (EGFP) reporter gene and was used as the EGFP-alone control vector, was utilized to construct the combination anti-HIV vector (5). The chimeric human/rhesus macaque TRIM5α gene under the control of the MNDU3 promoter, a CCR5 shRNA under the control of a human polymerase III U6 promoter, and a TAR decoy under the control of a human polymerase III U6 promoter were inserted upstream of the phosphoglycerate kinase (PGK)-driven EGFP reporter gene to derive CCLc-combination-anti-HIV (Fig. 1a) (3–5, 22, 25).
Fig 1.
Combination anti-HIV lentiviral vector, peripheral blood engraftment of transduced cells, and CCR5 downregulation. A third-generation lentiviral vector, CCLc-x-PGK-EGFP, was utilized to generate the combination anti-HIV construct. (a) A human/rhesus macaque TRIM5α isoform was driven under the control of the MNDU3 promoter, and a CCR5 shRNA and a TAR decoy were driven under separate human polymerase III U6 small RNA promoters. These three anti-HIV genes were inserted upstream from the EGFP reporter gene. (b) CD34+ HSCs were transduced with the control EGFP-alone vector or the anti-HIV vector and transplanted into NRG pups. Transplanted mice were screened for human CD45 and EGFP expression in the peripheral blood for engraftment of transduced cells, either EGFP-alone (n = 12) or anti-HIV (n = 14). (c) The peripheral blood of nontransduced (NT) (n = 14), EGFP-alone (n = 12), and anti-HIV (n = 14) cell-engrafted mice was analyzed for human T cells with antibodies specific for CD3 and CD4 and also for expression of EGFP. (d) CD4+ human splenocytes were analyzed for the expression of CCR5 (n = 5 for EGFP-alone and n = 11 for anti-HIV). Bar graphs display averages and standard deviations for each cohort. Statistical significance (P < 0.05) is represented by an asterisk.
Lentiviral vectors were generated in HEK-293T packaging cells. Twenty-five micrograms of the packaging construct, pΔ8.9 (containing gag and pol genes), 25 μg of CCLc-MNDU3-x-PGK-EGFP (EGFP-alone control empty vector [EGFP-alone]) or CCLc-combination-anti-HIV (transfer vector), and 5 μg of vesicular stomatitis virus G (VSVG) (envelope) were transfected into HEK-293T packaging cells in T225 flasks by lipofection. Vector supernatants were collected at 48 h posttransfection and concentrated 100-fold by ultrafiltration. Titers of vectors were subsequently determined on HEK-293T cells, and the titers obtained ranged from 2 × 109 to 6 × 109 transducing units/ml.
Transduction of primary human CD34+ HSCs.
CD34+ hematopoietic stem cells (HSCs) were isolated from umbilical cord blood (NDRI, Philadelphia, PA) with Ficoll-Paque (GE Healthcare, Piscataway, NJ) and purified by magnetic bead column separation (Miltenyi Biotec, Auburn, CA). CD34+ cell isolation purity of >93% was routinely obtained. Total CD34+ cells were cultured in complete Iscove modified Dulbecco medium (IMDM) containing 10% fetal bovine serum (FBS) and supplemented with 50 ng/ml stem cell factor (SCF), Flt-3 ligand, and thrombopoietin (TPO). Cells were transduced with the EGFP-alone lentiviral vector or the anti-HIV combination vector (multiplicity of infection [MOI] of 10) for 3 h at 37°C with 8 μg/ml protamine sulfate.
Transplantation and screening of NRG mice.
NOD-RAG1−/− IL2rγ−/− double mutant (NRG) mice (stock number 007799) were obtained from The Jackson Laboratory (Sacramento, CA) and were used in compliance with institutional and IACUC guidelines and regulations. Two- to 5-day-old newborn NRG pups were sublethally irradiated with 200 cGy of gamma irradiation. Nontransduced, EGFP-alone-transduced (control), or anti-HIV vector-transduced HSCs (3 × 105 total cells/mouse) were injected intrahepatically into irradiated pups. At 3 months posttransplantation, mice were bled retro-orbitally and the peripheral blood was analyzed by fluorescence-activated cell sorting (FACS) for EGFP and human leukocytes with a phycoerythrin (PE)-Cy7-conjugated anti-human CD45 antibody (clone HI30), an allophycocyanin (APC)-conjugated CD3 antibody (clone HIT3A), and a PE-conjugated CD4 antibody (clone RPA-T4) (BD Biosciences, San Jose, CA).
FACS analysis of engrafted human immune cells.
To evaluate multilineage hematopoiesis in transplanted NRG mice, cells from the peripheral blood and various lymphoid organs, including the thymus, spleen, and bone marrow, were stained with anti-human antibodies and analyzed by FACS. T cells were stained with an APC-conjugated CD3 antibody (clone HIT3A), a PE-conjugated CD4 antibody (clone RPA-T4), or an APC-conjugated CD8 antibody (clone RPA-T8) (BD Biosciences, San Jose, CA). B cells were stained with a PE-conjugated CD19 antibody (clone HIB19) (BD Biosciences, San Jose, CA). Macrophages were stained with a PE-conjugated CD14 antibody (clone M5E2) (BD Biosciences, San Jose, CA). To detect cell surface expression of CCR5, cells were stained with a PE-conjugated anti-human antibody (clone 2D7) (BD Biosciences, San Jose, CA). Cells were also evaluated for EGFP expression to determine the levels of engraftment of vector-transduced cells. Isotype controls used were a PE-conjugated mouse IgG1 (clone MOPC-21) (BD Biosciences, San Jose, CA), a PE-conjugated mouse IgG2a (clone IC003P) (RND Systems, Minneapolis, MN), and an APC-conjugated mouse IgG1 (clone MOPC-21) (BD Biosciences, San Jose, CA). FACS analysis was performed on a Beckman Coulter FC-500 instrument.
In vivo HIV-1 challenge of engrafted NRG mice.
To determine whether the anti-HIV gene-modified cells were resistant to HIV-1 infection, engrafted mice were challenged in vivo with either an R5-tropic BaL-1 or an X4-tropic NL4-3 strain of HIV-1. BaL-1 virus was obtained from the NIH AIDS Research Reference and Reagent Program and grown in CD34+ cell-derived macrophages. To grow the stock virus, macrophages were infected with an MOI of 1.0 and cell culture supernatants were collected on various days postinfection. Viral titers were obtained using the Ghost cell assay. Briefly, Ghost-R5-X4-R3 cells were infected with serial dilutions of stock virus. At 48 hours postinfection, the Ghost cells were analyzed by flow cytometry for EGFP expression to obtain an infectious viral titer. For NL43, an infectious clone was obtained from the NIH AIDS Research Reference and Reagent Program and transfected into 293T cells. At 72 hours posttransfection, cell culture supernatants were collected, and titers were determined on Ghost-R5-X4-R3 cells by the Ghost cell assay.
Mice were infected intravenously (i.v.) with 200,000 total infectious units. On various weeks postinfection, peripheral blood draws were taken and analyzed for total human CD4+ cell percentage by FACS and for HIV plasma viremia by quantitative PCR (Q-PCR). For FACS analysis, the cells were stained with a PE-Cy7-conjugated CD45 antibody (clone HI30), an APC-conjugated CD3 antibody (clone HIT3A), and a PE-conjugated CD4 antibody (clone RPA-T4) (BD Biosciences, San Jose, CA). The EGFP percentage was also analyzed to determine the levels of vector-transduced cells. FACS analysis was performed on a Beckman Coulter FC-500 instrument.
To determine the levels of plasma viremia, viral RNA was extracted from plasma specimens from infected mice using a Qiagen viral RNA extraction kit (Qiagen, Valencia, CA). Reverse transcription with oligo(dT) primers was then performed using TaqMan RT reagents (Applied Biosystems, Carlsbad, CA). Quantitative PCR was then performed using SYBR green (Applied Biosystems, Carlsbad, CA) and a primer/probe set specific for the HIV pol gene: primers 5′-CTGGCTACTATTTCTTTTGCTA-3′ and 5′-TGGCATGGGTACCAGCACA-3′ and probe 5′-TTTATCTACTTGTTCATTTCCTCCAATTCCTT-3′ (IDT DNA Technologies, Coralville, IA). Q-PCR was performed on an Applied Biosystems 7200 analyzer.
In vitro challenge of anti-HIV gene-modified cells.
To determine the level of HIV-1 resistance in a purified population of anti-HIV gene-modified cells, splenocytes were collected from engrafted mice and sorted for EGFP expression. The sorted cells were further purified for human CD3+ T cells by magnetic bead separation (Miltenyi Biotec, Auburn, CA). CD3+ T cells, either control nontransduced or EGFP-positive (EGFP+) anti-HIV gene-modified cells (5 × 105 cells/well) were stimulated with 1 μg/ml interleukin-2 (IL-2) and 1 μg/ml phytohemagglutinin (PHA). On day 3 poststimulation, the cells were challenged at an MOI of 0.05 with either an R5-tropic BaL-1 or an X4-tropic NL43 strain of HIV-1. On various days postinfection, culture supernatants were collected and analyzed by p24 antigen enzyme-linked immunosorbent assay (ELISA).
Cytokine secretion from in vivo-derived T cells.
Splenocytes from engrafted mice were isolated and sorted based on EGFP expression. These sorted cells were further purified for human CD3+ T cells by magnetic bead separation (Miltenyi Biotec, Auburn, CA). CD3+ T cells, either control nontransduced or EGFP+ anti-HIV gene-modified cells (1 × 106 cells/well) were stimulated with 1 μg/ml IL-2 and 1 μg/ml phytohemagglutinin. On day 3 poststimulation, culture supernatants were collected and analyzed by FACS for expression of IL-4, IL-6, IL-10, tumor necrosis factor alpha (TNF-α), and gamma interferon (IFN-γ) using a BD cytokine bead array kit (BD Biosciences, San Jose, CA).
Karyotyping.
To determine if the anti-HIV vector-transduced mobilized peripheral blood CD34+ HSCs maintained their chromosomal and genetic stability, karyotyping was performed. Upon transduction, EGFP-positive cells were sorted based on EGFP expression and further cultured for 5 days in Methocult semisolid methylcellulose medium enriched with cytokines (Stem Cell Technologies, Vancouver, Canada) to promote robust proliferation. The cells were washed twice with complete Dulbecco modified Eagle medium (DMEM) with 10% FBS to dissolve the methylcellulose and then treated with colcemid, a mitotic inhibitor, for 30 min at 37°C to arrest the cells in metaphase. Cells were subsequently treated with cell stripper for 10 min at 37°C, followed by treatment with a KCl hypotonic solution and 3:1 methanol-acetic acid fixative solutions. Karyotyping slides were made and Giemsa banded. Karyotyping was performed on an Olympus Bx41 microscope with a DP20 camera. Analysis was performed with an Applied imaging system.
Statistical analysis.
t tests were used to compare the differences between different treatments of the study interest. The statistical analyses were conducted in R (version 2.10.1) for Windows. A significance level of 0.05 was used in hypothesis testing.
RESULTS
Successful engraftment of anti-HIV vector-transduced CD34+ HSCs.
A third-generation self-inactivating combination anti-HIV lentiviral vector expressing a human/rhesus macaque chimeric TRIM5α, a CCR5 shRNA, a TAR decoy, and an EGFP reporter gene was utilized in our experiments (Fig. 1a). Expression of all three anti-HIV genes has been previously demonstrated in vector-transduced cells (5). The control empty vector, EGFP-alone, does not contain any anti-HIV genes but does contain the PGK-EGFP reporter gene (5). To evaluate the potential of combination anti-HIV vector-transduced CD34+ HSCs to engraft NRG mice, FACS analysis was performed on transplanted mice. As displayed in Fig. 1b, successful engraftment of transduced cells was observed in the peripheral blood. The average engraftment of EGFP-alone vector-transduced cells was 21.9%, with a standard deviation of 9.4%, and the average engraftment of anti-HIV vector-transduced cells was 17.5%, with a standard deviation of 8.0%. The percentages obtained were from total human leukocytes stained with an anti-human CD45 antibody. No significant difference (P = 0.213) between engraftment of EGFP-alone and anti-HIV vector-transduced cells was observed.
To determine the levels of CD4+ T cell development in anti-HIV vector-transduced cell-engrafted mice, peripheral blood was stained with anti-human CD3 and CD4 antibodies (Fig. 1c). No significant difference (P = 0.063 and P = 0.420, respectively) in CD4+/CD3+ T cell development in the peripheral blood was observed in anti-HIV vector-transduced engrafted cells compared to control EGFP-alone vector-transduced and nontransduced engrafted cells. The average level of anti-HIV vector-transduced CD4+ T cells was 55.0% of CD3+ T cells (standard deviation of 10.3%) compared to nontransduced cells (average of 59.2% CD4+ T cells with a standard deviation of 16.4%) and EGFP-alone vector-transduced cells (average of 47.6% CD4+ T cells with a standard deviation of 9.0%). Representative flow cytometry plots are displayed below the bar graphs in Fig. 1. Table 1 displays peripheral blood engraftment results from nontransduced and EGFP-alone- and anti-HIV vector-transduced cell-transplanted mice. These results demonstrate that no cytotoxic effects were observed with the anti-HIV vector-transduced cells in the peripheral blood of engrafted mice.
Table 1.
Engraftment of the peripheral blood in transplanted NRG mice
| Transduction and mouse | Peripheral blood engraftment (%) |
|
|---|---|---|
| EGFP | CD4+/CD3+ | |
| Nontransduced | ||
| 1 | NAa | 59.9 |
| 2 | NA | 51.9 |
| 3 | NA | 56.7 |
| 4 | NA | 46.9 |
| 5 | NA | 55.2 |
| 6 | NA | 31.6 |
| 7 | NA | 62.0 |
| 8 | NA | 55.0 |
| EGFP-alone | ||
| 1 | 14.8 | 58.9 |
| 2 | 29.1 | 44.9 |
| 3 | 36.6 | 42.7 |
| 4 | 30.7 | 63.2 |
| 5 | 16.1 | 41.2 |
| 6 | 13.2 | 47.7 |
| 7 | 16.1 | 44.7 |
| 8 | 12.5 | 45.0 |
| Anti-HIV | ||
| 1 | 13.9 | 52.0 |
| 2 | 22.0 | 50.3 |
| 3 | 15.9 | 69.8 |
| 4 | 12.6 | 70.5 |
| 5 | 32.0 | 50.3 |
| 6 | 34.3 | 49.0 |
| 7 | 16.8 | 34.5 |
| 8 | 20.7 | 49.2 |
NA, not applicable.
To determine the levels of CCR5 downregulation of anti-HIV vector-transduced cells (cells which express the CCR5 shRNA), flow cytometry was performed on total human (CD4+/EGFP+) splenocytes from mice engrafted with EGFP-alone or anti-HIV vector-transduced cells. As displayed in Fig. 1d, anti-HIV vector-transduced cells expressed significantly (P = 0.002) decreased levels of cell surface CCR5 (7.4% positive) compared to control EGFP-alone vector-transduced cells (46.4% positive).
Multilineage human hematopoiesis from anti-HIV vector-transduced cells in lymphoid organs.
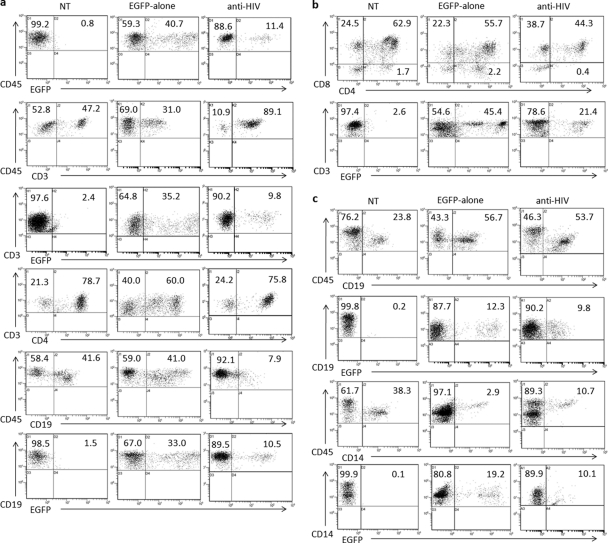
To determine if normal engraftment and multilineage hematopoiesis of anti-HIV vector-transduced cells had occurred in the lymphoid organs of transplanted NRG mice, flow cytometry was performed (Fig. 2). Single cells were isolated from the thymus, spleen, and bone marrow of engrafted mice and stained with respective antibodies as described in Materials and Methods. Table 2 displays lymphoid organ engraftment results from nontransduced and EGFP-alone- and anti-HIV vector-transduced cell-transplanted mice. No significant difference (P > 0.05) was observed with engraftment or development of anti-HIV gene-modified T cells (CD3+) in the peripheral blood, thymus, or spleen compared to that in nontransduced or EGFP-alone cell-engrafted mice. No observed differences in the size of the thymus grafts were noted between the engrafted mice. No significant difference (P > 0.05) was observed with engraftment or development of anti-HIV gene-modified B cells in the spleen or bone marrow compared to that in nontransduced or EGFP-alone cell-engrafted mice. The only significant difference observed in lymphoid organ engraftment of anti-HIV gene-modified cells (average, 11.88%; standard deviation, 1.78%) compared to nontransduced cells (average, 29.17%; standard deviation, 3.95%) was with CD14+/CD45+ cells in the bone marrow (P = 0.005). However, no significant difference (P = 0.112) in engraftment of bone marrow CD14+/CD45+ cells was observed when comparing the levels of EGFP-alone (average, 6.3%; standard deviation, 2.43%) and anti-HIV vector-transduced cells. These results demonstrated that no cytotoxic effects were observed with the anti-HIV vector-transduced HSCs, as they were capable of engrafting and undergoing multilineage hematopoiesis in various lymphoid organs of transplanted NRG mice at levels equivalent to those for control HSCs. Representative flow cytometry plots are displayed in Fig. 2.
Fig 2.
Engraftment of lymphoid organs in transplanted NRG mice. NRG mice were transplanted with CD34+ HSCs that were either nontransduced (NT) or transduced with a control EGFP-alone vector or the anti-HIV lentiviral vector. Upon engraftment, various lymphoid organs, including the spleen (a), thymus (b), and bone marrow (c), were analyzed for human cell engraftment. Flow cytometry was performed to detect EGFP expression along with total human leukocytes (CD45), T cells (CD3, CD4, and CD8), B cells (CD19), and macrophages (CD14). Data are representative of mice for each cohort. Complete data sets for each cohort of mice are included in Table 2.
Table 2.
Engraftment of lymphoid organs in transplanted NRG mice
| Transduction and mouse | Engraftment (%) |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Blood |
Thymus |
Spleen |
Bone marrow |
|||||||||||
| CD4/CD3 | EGFP/CD45 | CD4/CD8 | EGFP/CD3 | EGFP/CD45 | CD3/CD45 | EGFP/CD3 | CD4/CD3 | CD19/CD45 | EGFP/CD19 | CD19/CD45 | EGFP/CD19 | CD14/CD45a | EGFP/CD14 | |
| Nontransduced | ||||||||||||||
| 9 | 76.9 | NAb | 89.5 | NA | NA | 89.9 | NA | 94.7 | 6.7 | NA | 28.8 | NA | 34.3 | NA |
| 10 | 79.0 | NA | 95.4 | NA | NA | 38.3 | NA | 85.0 | 58.8 | NA | 56.9 | NA | 16.3 | NA |
| 11 | 60.0 | NA | 86.8 | NA | NA | 58.0 | NA | 83.8 | 37.2 | NA | 29.9 | NA | 33.5 | NA |
| 12 | 31.8 | NA | 89.2 | NA | NA | 28.7 | NA | 72.3 | 57.6 | NA | 37.3 | NA | 35.2 | NA |
| 13 | 86.7 | NA | 65.5 | NA | NA | 27.0 | NA | 45.6 | 56.2 | NA | 67.0 | NA | 17.4 | NA |
| 14 | 75.5 | NA | 72.5 | NA | NA | 47.2 | NA | 78.7 | 41.6 | NA | 23.8 | NA | 38.3 | NA |
| EGFP-alone | ||||||||||||||
| 9 | 63.2 | 39.1 | 72.2 | 45.4 | 40.7 | 31.0 | 35.2 | 60.0 | 41.0 | 33.0 | 81.3 | 36.3 | 13.5 | 32.5 |
| 10 | 40.7 | 17.2 | 87.0 | 26.3 | 16.3 | 21.8 | 24.5 | 46.2 | 74.0 | 15.5 | 56.7 | 12.3 | 2.9 | 19.2 |
| 11 | 37.3 | 21.5 | 79.6 | 12.0 | 17.0 | 35.4 | 17.3 | 52.9 | 59.3 | 20.2 | 24.2 | 24.2 | 4.3 | 25.2 |
| 12 | 41.2 | 16.1 | 85.6 | 23.4 | 19.9 | 30.6 | 18.8 | 54.1 | 66.2 | 23.3 | 47.0 | 22.6 | 4.5 | 21.7 |
| Anti-HIV | ||||||||||||||
| 9 | 66.1 | 10.9 | 61.7 | 8.9 | 16.6 | 14.4 | 7.6 | 64.5 | 84.1 | 8.9 | 83.6 | 11.3 | 8.5 | 10.5 |
| 10 | 52.0 | 13.9 | 83.2 | 17.4 | 8.8 | 19.9 | 9.0 | 54.2 | 62.9 | 10.3 | 53.7 | 9.8 | 10.7 | 10.1 |
| 11 | 50.3 | 7.7 | 85.7 | 9.0 | 5.6 | 59.7 | 3.6 | 64.8 | 25.2 | 7.9 | 54.6 | 7.7 | 16.4 | 5.9 |
| 12 | 60.8 | 12.3 | 65.1 | 5.8 | 6.6 | 68.1 | 6.0 | 33.1 | 29.6 | 8.3 | 9.6 | 7.8 | 9.7 | 8.5 |
| 13 | 47.9 | 9.5 | 81.5 | 5.7 | 5.7 | 20.6 | 4.6 | 57.7 | 66.0 | 8.0 | 14.6 | 10.2 | 7.8 | 7.8 |
| 14 | 66.8 | 22.2 | 53.6 | 21.4 | 11.4 | 89.1 | 9.8 | 75.8 | 7.9 | 10.5 | 77.9 | 11.4 | 18.2 | 14.0 |
There was a statistically significant difference (P < 0.05) between the average levels of CD14/CD45-positive cells in the bone marrow of anti-HIV vector-transduced cell-engrafted mice and those in nontransduced cell-engrafted mice.
NA, not applicable.
Maintenance of CD4+ T cell levels in anti-HIV cell-engrafted mice upon in vivo HIV-1 infection.
To determine whether resistance to HIV-1 infection was conferred in anti-HIV vector-transduced cell-engrafted mice, the levels of total human CD4+ cells in the peripheral blood were analyzed by flow cytometry at various weeks postinfection. Maintenance of normal human CD4+ levels was observed in anti-HIV cell-engrafted mice (solid lines) upon challenge with either an R5-tropic BaL-1 (Fig. 3a) or an X4-tropic NL43 (Fig. 3b) strain of HIV-1. In the anti-HIV gene-modified cell-engrafted mice (solid lines), total human CD4+ T cell percentages gated on human CD45+ leukocytes ranged between 46.2 and 65.4% and between 37.5 and 79.2% at the end of the challenge experiment with BaL-1 or NL43 HIV-1, respectively. This was in comparison to control EGFP-alone vector-transduced cell-engrafted mice (dashed lines), where total human CD4+ T cell percentages gated on human CD45+ cells declined over the course of infection and ranged from 19.1 to 29.1% and from 5.9 to 15.1% at the end of the challenge experiment with BaL-1 or NL43 HIV-1, respectively. The average levels of CD4+ T cells preinfection and at the end of the challenge experiments are displayed in Fig. 3c and d for BaL-1 and NL4-3 infections, respectively. In EGFP-alone cell-engrafted mice infected with BaL-1, CD4+ T cell levels decreased to an average of 25.8%, which was significantly different (P = 0.002) from those in anti-HIV cell-engrafted mice, where average CD4+ T cell levels were 55.1% (Fig. 3c). A significant difference (P = 0.001) was also observed in the average level of CD4+ T cells in EGFP-alone cell-engrafted mice (9.1%) infected with NL4-3 compared to the average level of anti-HIV CD4+ T cells (58.4%) (Fig. 3d). Representative flow cytometry plots demonstrating CD4+ cell levels in the infected mice are displayed below the bar graphs in Fig. 3. These results demonstrated that the combination anti-HIV vector significantly enhanced survival of human T cells in the face of an HIV infection.
Fig 3.
Detection of human CD4+ T cells in the peripheral blood of HIV-1-infected NRG humanized mice. (a and b) NRG mice successfully engrafted with either control EGFP-alone or combination anti-HIV vector-transduced cells were infected i.v. with either an R5-tropic BaL-1 (a) or an X4-tropic NL4-3 (b) strain of HIV-1. At various weeks postinfection, mice were bled and analyzed by FACS for total human CD4+ cell percentage. Solid lines represent anti-HIV cell-engrafted mice. Dashed lines represent control EGFP-alone cell-engrafted mice. (c and d) Comparisons between CD4+ T cell level averages preinfection and postinfection were performed for both the BaL-1 (c)- and NL4-3 (d)-infected mice engrafted with either EGFP-alone or anti-HIV vector-transduced cells. Bar graphs display averages and standard deviations from four mice for each cohort for each set of infections. Statistical significance (P < 0.05) is represented by an asterisk. Representative flow cytometry plots are displayed.
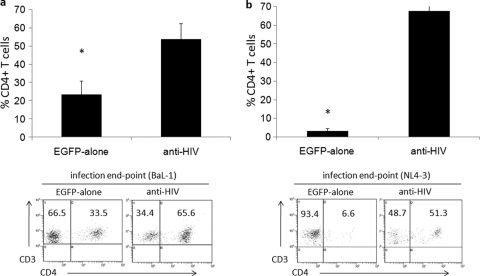
To evaluate the levels of human CD4+ cells in the lymphoid organs of infected mice, mice were sacrificed at the end of the challenge experiments and human CD4+ T cells from the spleen were analyzed. As displayed in Fig. 4a, upon infection with BaL-1, human CD4+ T cell levels were maintained in the spleen (average of 53.8% of human CD3+ T cells, with a standard deviation of 8.6%) in anti-HIV vector-transduced cell-engrafted mice. This was in comparison to mice engrafted with control EGFP-alone vector-transduced cells, which displayed a significantly (P = 0.002) decreased level of CD4+ human T cells in the spleen (average of 23.3%, with a standard deviation of 7.5%). As displayed in Fig. 4b, upon infection with NL4-3, human CD4+ T cell levels were maintained in the spleen (average of 67.4% of human CD3+ T cells, with a standard deviation of 9.3%) in anti-HIV vector-transduced cell-engrafted mice. This was in comparison to mice engrafted with control EGFP-alone vector-transduced cells, which displayed a significantly (P = 0.001) decreased level of CD4+ human CD3+ T cells in the spleen (average of 3.2%, with a standard deviation of 1.4%). Representative flow cytometry plots demonstrating CD4+ cell levels in the infected mice are displayed below the bar graphs in Fig. 4. These results demonstrate that normal CD4+ cell levels could be maintained during the course of an HIV infection in a cell population which contained CD4+ cells derived from anti-HIV gene-modified HSCs.
Fig 4.
Detection of human CD4+ T cells in the spleens of HIV-1-infected NRG humanized mice. NRG mice successfully engrafted with either control EGFP-alone or combination anti-HIV vector-transduced cells were infected i.v. with either an R5-tropic BaL-1 (a) or an X4-tropic NL4-3 (b) strain of HIV-1. After completion of the in vivo challenge experiments, infected mice were sacrificed and the spleens were analyzed by flow cytometry for CD4+ T cell (CD3+) levels. Bar graphs display averages and standard deviations from four mice for each cohort for each infection. Statistical significance (P < 0.05) is represented by an asterisk. Representative flow cytometry plots are displayed.
Selective survival advantage of anti-HIV vector-transduced cells upon in vivo HIV-1 infection.
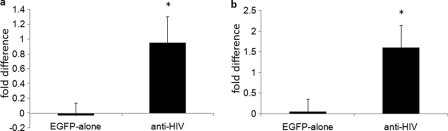
During HIV-1 infection of a mixed population of protected anti-HIV gene-expressing cells and unprotected nontransduced cells, a selective survival advantage should be observed with the anti-HIV gene-modified cells. This selective advantage would be demonstrated with an increase in the number of anti-HIV vector-transduced cells during the course of infection due to killing of the infected nontransduced cells. To evaluate whether this had occurred in vivo in HIV-1-infected mice, the total percentages of EGFP+ human CD4+ cells were analyzed by flow cytometry at various weeks postinfection in the peripheral blood. As demonstrated in Fig. 5, anti-HIV vector-transduced cell-engrafted mice displayed an increase in total EGFP+/CD4+ cells when challenged in vivo with either a BaL-1 (Fig. 5a) or an NL43 (Fig. 5b) strain of HIV-1 from preinfection to the time of euthanizing the mice. During infection with BaL-1 (Fig. 5a), the levels of anti-HIV gene-modified CD4+ cells increased, on average, 0.95-fold, with a standard deviation of 0.35-fold. This was a significant increase (P = 0.006) compared to those in control EGFP-alone vector-transduced cells, which decreased 0.03-fold, with a standard deviation of 0.17-fold. In mice infected with NL4-3 (Fig. 5b), the levels of anti-HIV gene-modified CD4+ cells increased 1.60-fold, with a standard deviation of 0.54-fold. This was a significant increase (P = 0.005) compared to those in control EGFP-alone vector-transduced cells, which increased 0.05-fold, with a standard deviation of 0.30-fold. These results demonstrate that in the presence of an HIV-1 infection, in vivo, cells expressing anti-HIV genes have a selective survival advantage and an increase in their percentage of total cells due to their ability to resist infection and the killing of unprotected cells.
Fig 5.
Selective survival advantage of anti-HIV gene-modified cells in HIV-1-infected NRG mice. Mice successfully engrafted with either control EGFP-alone or combination anti-HIV vector-transduced cells were infected i.v. with either an R5-tropic BaL-1 (a) or an X4-tropic NL4-3 (b) strain of HIV-1. At various weeks postinfection, mice were bled and analyzed by FACS for EGFP+/CD4+ human cell percentage. Fold difference in EGFP+/CD4+ T cell level averages preinfection and postinfection are displayed for both the EGFP-alone and anti-HIV vector-transduced cells. Bar graphs display averages and standard deviations from four mice for each cohort for each set of infections. Statistical significance (P < 0.05) is represented by an asterisk.
Detection of plasma viremia and resistance of anti-HIV gene-modified cells upon ex vivo HIV-1 challenge.
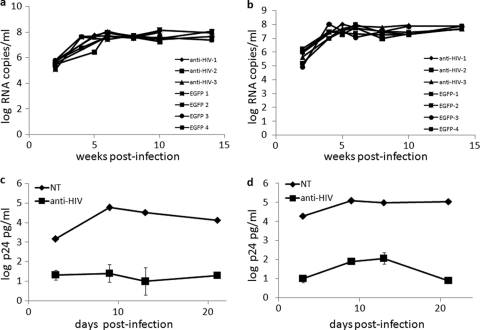
A hallmark of successfully engrafted NRG mice is their ability to display plasma viremia upon infection with HIV-1 (6). Therefore, to determine if the levels of plasma viremia had decreased in HIV-1-infected NRG mice engrafted with anti-HIV vector-transduced HSCs, Q-PCR was performed on plasma specimens from mice at various weeks postinfection. As demonstrated in Fig. 6, the mice engrafted with anti-HIV vector-transduced cells did not display a decrease in HIV-1 plasma viremia over the course of infection. Levels of HIV-1 RNA copies per milliliter remained between 7 and 8 log units in the anti-HIV cell-engrafted mice, which were similar to those in control cell-engrafted mice (Fig. 6). These results were observed for both the R5-tropic BaL-1 (Fig. 6a) and X4-tropic NL43 (Fig. 6b) challenge experiments.
Fig 6.
Detection of in vivo plasma viremia and in vitro HIV-1 challenge of sorted spleen T cells. (a and b) NRG mice successfully engrafted with either control EGFP-alone or combination anti-HIV vector-transduced cells were infected i.v. with either an R5-tropic BaL-1 (a) or an X4-tropic NL4-3 (b) strain of HIV-1. At various weeks postinfection, mice were bled and the plasma was analyzed by Q-PCR using a primer/probe pair specific for the HIV pol gene. (c and d) In vitro HIV-1 challenge experiments were performed on human CD3+ T cells, both nontransduced (EGFP−) and anti-HIV vector transduced (EGFP+), with an R5-tropic BaL-1 (c) or an X4-tropic NL4-3 (d) strain of HIV-1. At various days postinfection, culture supernatants were collected and analyzed for p24 by antigen ELISA. p24 ELISAs were performed in triplicate.
To evaluate whether HIV-1 resistance could be conferred when a pure population of anti-HIV vector-transduced cells was challenged with HIV-1, EGFP+ and EGFP− human CD3+ T cells were sorted from the spleens of successfully engrafted mice. Upon stimulation with IL-2 and PHA for 3 days postsorting, the cells were challenged with either R5-tropic BaL-1 (Fig. 6c) or X4-tropic NL43 (Fig. 6d) at an MOI of 0.05. At various days postinfection, cell culture supernatants were collected and analyzed for HIV-1 p24 by antigen ELISA. As displayed in Fig. 6, a strong reduction in HIV output (>4 log units) was observed in the sorted anti-HIV vector-transduced cell culture compared to the nontransduced cell culture. These results demonstrate that pure populations of anti-HIV gene-transduced cells isolated from engrafted animals, when faced with an HIV-1 viral load, display potent resistance to infection, coupled with strongly diminished HIV-1 production.
Anti-HIV gene-modified cells are functional and retain a normal karyotype.
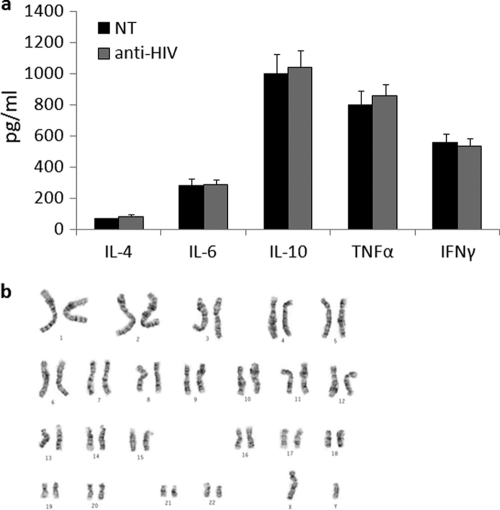
To determine whether the anti-HIV gene-modified cells were functionally normal, the levels of secretion of specific cytokines were measured. T cells from the spleens of engrafted mice were purified and sorted based on EGFP and human CD3 expression. The T cells were stimulated with IL-2 and PHA and cultured for 3 days. At day 3 poststimulation, culture supernatants were collected and analyzed by FACS for expression of IL-4, IL-6, IL-10, TNF-α, and IFN-γ using a BD cytokine bead array. As displayed in Fig. 7a, no significant difference (P > 0.05) in the secretion of any of the cytokines measured was detected in the anti-HIV gene-modified cell cultures compared to control nontransduced cells. These data show that transduction had not affected normal cytokine secretion from the T cells.
Fig 7.
Cytokine expression and karyotypic analysis of anti-HIV vector-transduced cells. (a) Anti-HIV gene-modified T cells from spleen were sorted based on EGFP/CD3 expression and stimulated with IL-2 and PHA. At day 3 poststimulation, culture supernatants were analyzed by FACS for expression of IL-4, IL-6, IL-10, TNF-α, and IFN-γ. Cytokine expression experiments were performed in triplicate. (b) A representative karyotyping analysis of anti-HIV vector-transduced human CD34+ HSCs. Karyotypic analyses were performed in duplicate.
With the transduction of CD34+ HSCs with a combination anti-HIV lentiviral vector and their subsequent differentiation toward hematopoietic lineages, it is possible that chromosomal and genetic abnormalities could arise. Therefore, karyotyping analyses were performed on anti-HIV vector-transduced CD34+ HSCs. As displayed in Fig. 7b, the anti-HIV cells retained a normal chromosome profile. Normal banding was observed, and no translocations or other detectable chromosomal abnormalities were detected.
The results obtained from these experiments demonstrated that the anti-HIV T cells appear to be functionally normal and that the transduction and expression of the anti-HIV genes did not cause any gene rearrangements or abnormalities in chromosome organization.
DISCUSSION
Stem cell gene therapy for HIV has the potential to offer an alternative therapeutic approach for HIV-infected individuals (27). With the possibility of a one-time treatment by transducing self-renewing and self-repopulating HSCs with potent anti-HIV genes, the removal of small-molecule antiretroviral drug therapy and the control of HIV replication could occur, with the development of a complete HIV-resistant immune system. This has been demonstrated with a bone marrow transplant in Berlin, Germany, where an HIV-infected individual received allogeneic HSCs from a donor who was homozygous for the CCR5-Δ32-bp allele (14). The recipient is currently free from HIV replication while also having discontinued antiretroviral drug therapy (16). The results from this study highlight the potential use of HIV-resistant stem cells to provide a functional cure for HIV-infected patients (15).
In our current study, we evaluated the in vivo safety and efficacy of triple-combination anti-HIV lentiviral vector-transduced CD34+ HSCs in a NOD-RAG1−/− IL2rγ−/− double mutant (NRG) mouse model. This anti-HIV vector contains three highly potent anti-HIV genes which individually confer strong resistance to HIV-1 infection. However, due to the high mutation rate of HIV, its ability to generate escape variants, and the various tropisms of the virus, it is critical to develop combination therapies which act at multiple stages of the HIV life cycle. This has been demonstrated with the use of monotherapy small-molecule antiretroviral drugs, which eventually give rise to viral escape mutants (7, 20, 26). Therefore, similar to combination approaches with antiretroviral drugs, multiple anti-HIV genes inserted into a single gene therapy vector may offer stronger protection from HIV infection and will have a greater chance of preventing viral resistance.
The use of multiple HIV resistance genes, however, is not the only aspect to consider when designing HIV gene therapies. The specific stage of the life cycle targeted and the mechanism of action of the anti-HIV gene also need to be taken into consideration. Anti-HIV genes which block early stages of HIV infection, including attachment and entry (CCR5 shRNA) and reverse transcription and integration (TRIM5α), will prevent the generation of provirus and viral reservoirs, which are main reasons for the failure to cure HIV-infected individuals (1, 3–5, 25, 28). In our previous study evaluating the in vitro efficacy of this combination vector (CCR5 shRNA, human/rhesus TRIM5α, and a TAR decoy), strong preintegration protection from HIV-1 infection was observed (5). This anti-HIV vector not only prevented viral integration but also blocked the generation of viral escape variants, which led to the next step in evaluating the in vivo safety and efficacy of vector-transduced CD34+ HSCs in the current studies.
After transduction of CD34+ HSCs with the anti-HIV vector and injection into newborn NRG pups, successful engraftment of combination anti-HIV vector-transduced cells was achieved (Fig. 1). In vivo development of transduced CD45+ human leukocytes (Fig. 1b) and human T cells (Fig. 1c) was observed in the peripheral blood, similar to the case for control vector-transduced cells. To further evaluate the safety of anti-HIV vector-transduced CD34+ HSCs, engraftment and multilineage hematopoiesis were analyzed in the spleen, thymus, and bone marrow. Successful development of mature immune cells, including T cells, B cells, and macrophages, was observed in the lymphoid organs of anti-HIV vector-transduced cell-engrafted mice (Fig. 2). Therefore, transduction with the anti-HIV vector did not have any detrimental effects on the engraftment or multilineage hematopoiesis of the gene-modified CD34+ HSCs.
The major goal of HIV stem cell gene therapy is to transplant anti-HIV gene-modified HSCs into infected patients, which would further develop into HIV-resistant immune cells capable of blocking HIV infection and thriving in the face of a viral load. Therefore, to evaluate the levels of protection from HIV-1 infection of the anti-HIV gene-modified immune cells which developed in the NRG mice, both total human CD4+ cell percentages and plasma viremia in HIV-1-infected mice were measured, as a decline in CD4+ cells and a rise in plasma viremia are hallmark characteristics of HIV disease progression. Our results demonstrated that anti-HIV gene-modified human CD4+ cells indeed were protected from HIV-1 infection in the face of a viral load. Maintenance of normal human CD4+ T cell levels was observed in mice engrafted with the anti-HIV vector-transduced CD34+ HSCs. Even though a portion of the nontransduced cell population was being killed during HIV-1 infection, the anti-HIV gene-modified cells were able to thrive and maintain normal levels of CD4+ cells. This was in comparison to the case for control EGFP-alone vector-transduced CD34+ HSC-engrafted mice, which upon infection demonstrated a steady decline of human CD4+ T cells due to their inability to resist infection (Fig. 3). The ability of the anti-HIV gene-modified cells to survive during HIV-1 infection was also observed in the spleens of infected mice. The maintenance of human CD4+ T cells in mice engrafted with the anti-HIV CD34+ HSCs was due to a selective survival advantage of the protected cells. Upon challenge with either an R5- or an X4-tropic strain of HIV-1, an increase in EGFP+ cell percentages was observed in mice engrafted with the anti-HIV CD34+ HSCs. This was due to expansion of the anti-HIV resistant cell population during infection and also to the killing of the nontransduced population in the same mice, which acted as an internal control. When plasma viremia was measured, however, the levels of HIV-1 RNA in the blood were found to be similar for all mice studied no matter whether they were engrafted with anti-HIV or control EGFP-alone vector-transduced CD34+ HSCs. This was due to unprotected nontransduced cells which were cotransplanted with the anti-HIV vector-transduced cells. The mice were transplanted with a mixed population of both vector-transduced and nontransduced cells, and hence, upon challenge, HIV-1 will be able to infect the unprotected cells and establish infection. The selective pressure of HIV-1 on the anti-HIV gene-modified cells enabled them to be selected for expansion and proliferation; however, there were still unprotected cells for the virus to continually infect and produce plasma viremia (Fig. 6). Therefore, higher transduction efficiencies and in vivo engraftment percentages may need to be achieved to reach an optimal level of HIV-resistant immune cells to suppress HIV-1 replication and decrease plasma viremia. This was observed when we challenged a sorted and pure population of anti-HIV gene-modified human T cells from the spleens of engrafted mice. Upon ex vivo challenge, a pure population of anti-HIV T cells was capable of resisting HIV-1 infection and displayed a significant decrease in HIV-1 p24 output compared to nontransduced human T cells (Fig. 6). Also, in addition to the higher transduction efficiencies needed, the human immune system of an infected individual may come to the aid to decrease the viral load. If HIV-specific immune cells survive and are not killed off by HIV, over the long term (a time period that cannot be easily tested in a short-term animal model) the viral load may be substantially reduced, since HIV-producing cells from the viral reservoirs can be safely eliminated by immune cells. Even though we did not observe a decrease in viral load, we were able to demonstrate that the cells expressing the anti-HIV genes were capable of resisting infection in vivo. These cells were able to maintain normal human CD4+ cell levels due to a selective survival advantage upon HIV challenge in vivo. Therefore, efficacy was observed with the population of anti-HIV gene-expressing cells. If a higher transduction efficiency and in vivo engraftment of HIV-resistant cells can be achieved, a marked reduction in plasma viremia could occur. However, if a pure population of anti-HIV gene expressing HSCs could be transplanted into patients, complete suppression of HIV replication could be accomplished, as demonstrated with the Berlin patient. One promising method to achieve this scenario is to use clonal induced pluripotent stem cells (iPSCs) which express anti-HIV genes. This was demonstrated in a recent study where complete protection from HIV-1 infection was observed due to the expression of a combination of anti-HIV genes in every macrophage derived from the iPSC line (17).
Here we have demonstrated the safety and efficacy of this combination anti-HIV lentiviral vector in a humanized mouse model which is capable of demonstrating multilineage hematopoiesis from engrafted human CD34+ HSCs. The subsequent protection and expansion of the HIV-resistant immune cells in the face of an HIV load establish the utility of this vector for use in future clinical trials.
ACKNOWLEDGMENTS
This work was supported by University of California—Davis Health System start-up funds from the Dean's office for the Stem Cell Program and by the James B. Pendleton Charitable Trust. This work was also supported in part by the Gin and Imy Mar stem cell research fund. The NIH AIDS Research and Reference Reagent Program provided many reagents and cell lines used in this work. Biostatistics support for this publication was made possible by grant UL1 RR024146 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH), and the NIH Roadmap for Medical Research.
Information on NCRR is available at http://www.ncrr.nih.gov/. Information on Reengineering the Clinical Research Enterprise can be obtained from http://nihroadmap.nih.gov/clinicalresearch/overview-translational.asp.
The contents of this paper are solely the responsibility of the authors and do not necessarily represent the official view of NCRR or NIH.
Footnotes
Published ahead of print 7 March 2012
REFERENCES
- 1. An DS, et al. 2007. Stable reduction of CCR5 by RNAi through hematopoietic stem cell transplant in non-human primates. Proc. Natl. Acad. Sci. U. S. A. 104:13110–13115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anderson J, et al. 2007. Safety and efficacy of a lentiviral vector containing three anti-HIV genes—CCR5 ribozyme, tat-rev siRNA, and TAR decoy—in SCID-hu mouse-derived T cells. Mol. Ther. 15:1182–1188 [DOI] [PubMed] [Google Scholar]
- 3. Anderson J, Akkina R. 2007. Complete knockdown of CCR5 by lentiviral vector-expressed siRNAs and protection of transgenic macrophages against HIV-1 infection. Gene Ther. 14:1287–1297 [DOI] [PubMed] [Google Scholar]
- 4. Anderson J, Akkina R. 2008. Human immunodeficiency virus type 1 restriction by human-rhesus chimeric tripartite motif 5alpha (TRIM 5alpha) in CD34+(+) cell-derived macrophages in vitro and in T cells in vivo in severe combined immunodeficient (SCID-hu) mice transplanted with human fetal tissue. Hum. Gene Ther. 19:217–228 [DOI] [PubMed] [Google Scholar]
- 5. Anderson JS, Javien J, Nolta JA, Bauer G. 2009. Preintegration HIV-1 inhibition by a combination lentiviral vector containing a chimeric TRIM5alpha protein, a CCR5 shRNA, and a TAR decoy. Mol. Ther. 17:2103–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Berges BK, Rowan MR. 2011. The utility of the new generation of humanized mice to study HIV-1 infection: transmission, prevention, pathogenesis, and treatment. Retrovirology 8:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Blanco JL, Varghese V, Rhee SY, Gatell JM, Shafer RW. 2011. HIV-1 integrase inhibitor resistance and its clinical implications. J. Infect. Dis. 203:1204–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Denton PW, Garcia JV. 2009. Novel humanized murine models for HIV research. Curr. HIV/AIDS Rep. 6:13–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. DiGiusto DL, et al. 2010. RNA-based gene therapy for HIV with lentiviral vector-modified CD34(+) cells in patients undergoing transplantation for AIDS-related lymphoma. Sci. Transl. Med. 2:36ra43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ding SF, Lombardi R, Nazari R, Joshi S. 2002. A combination anti-HIV-1 gene therapy approach using a single transcription unit that expresses antisense, decoy, and sense RNAs, and transdominant negative mutant Gag and Env proteins. Front. Biosci. 7:15–28 [DOI] [PubMed] [Google Scholar]
- 11. Feeney ER, Mallon PW. 2010. Impact of mitochondrial toxicity of HIV-1 antiretroviral drugs on lipodystrophy and metabolic dysregulation. Curr. Pharm. Des. 16:3339–3351 [DOI] [PubMed] [Google Scholar]
- 12. Huang Y, et al. 1996. The role of a mutant CCR5 allele in HIV-1 transmission and disease progression. Nat. Med. 2:1240–1243 [DOI] [PubMed] [Google Scholar]
- 13. Humeau LM, et al. 2004. Efficient lentiviral vector-mediated control of HIV-1 replication in CD4 lymphocytes from diverse HIV+ infected patients grouped according to CD4 count and viral load. Mol. Ther. 9:902–913 [DOI] [PubMed] [Google Scholar]
- 14. Hutter G, et al. 2009. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N. Engl. J. Med. 360:692–698 [DOI] [PubMed] [Google Scholar]
- 15. Hutter G, Schneider T, Thiel E. 2009. Transplantation of selected or transgenic blood stem cells—a future treatment for HIV/AIDS? J. Int. AIDS Soc. 12:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hutter G, Thiel E. 2011. Allogeneic transplantation of CCR5-deficient progenitor cells in a patient with HIV infection: an update after 3 years and the search for patient no. 2. AIDS 25:273–274 [DOI] [PubMed] [Google Scholar]
- 17. Kambal A, et al. 2011. Generation of HIV-1 resistant and functional macrophages from hematopoietic stem cell-derived induced pluripotent stem cells. Mol. Ther. 19:584–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kohn DB, et al. 1999. A clinical trial of retroviral-mediated transfer of a Rev-responsive element decoy gene into CD34+ cells from the bone marrow of human immunodeficiency virus-1-infected children. Blood 94:368–371 [PubMed] [Google Scholar]
- 19. Liu R, et al. 1996. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply exposed individuals to HIV-1 infection. Cell 86:267–377 [DOI] [PubMed] [Google Scholar]
- 20. Martinez-Picado J, et al. 2000. Antiretroviral resistance during successful therapy of HIV type 1 infection. Proc. Natl. Acad. Sci. U. S. A. 97:10948–10953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Michienzi A, Li S, Zaia JA, Rossi JJ. 2002. A nucleolar TAR decoy inhibitor of HIV-1 replication. Proc. Natl. Acad. Sci. U. S. A. 22:14047–14052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mitsuyasu RT, et al. 2009. Phase 2 gene therapy trial of an anti-HIV ribozyme in autologous CD34++ cells. Nat. Med. 5:285–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Novina CD, et al. 2002. siRNA-directed inhibition of HIV-1 infection. Nat. Med. 8:681–686 [DOI] [PubMed] [Google Scholar]
- 24. Nunez M. 2010. Clinical syndromes and consequences of antiretroviral-related hepatotoxicity. Hepatology 52:1143–1155 [DOI] [PubMed] [Google Scholar]
- 25. Sawyer SL, Emerman M, Malik HS. 2005. Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proc. Natl. Acad. Sci. U. S. A. 102:2832–2837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Seclen E, et al. 2010. Primary resistance to maraviroc in a large set of R5-V3 viral sequences from HIV-1 infected patients. J. Antimicrob. Chemother. 65:2502–2504 [DOI] [PubMed] [Google Scholar]
- 27. Strayer DS, et al. 2005. Current status of gene therapy strategies to treat HIV/AIDS. Mol. Ther. 11:823–841 [DOI] [PubMed] [Google Scholar]
- 28. Stremlau M, et al. 2004. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature 427:848–853 [DOI] [PubMed] [Google Scholar]
- 29. Veloso S, et al. 2010. Pharmacogenetics of the metabolic disturbances and atherosclerosis associated with antiretroviral therapy in HIV-infected patients. Curr. Pharm. Des. 16:3379–3389 [DOI] [PubMed] [Google Scholar]