Abstract
Viral drug toxicity, resistance, and an increasing immunosuppressed population warrant continued research into new avenues for limiting diseases associated with human cytomegalovirus (HCMV). In this study, a small interfering RNA (siRNA), siX3, was designed to target coding sequences within shared exon 3 of UL123 and UL122 transcripts encoding IE1 and IE2 immediate-early proteins of HCMV. Pretreatment of cells with siX3 reduced the levels of viral protein expression, DNA replication, and progeny virus production compared to control siRNA. Two siRNAs against UL54 and overlapping transcripts (UL55-57) were compared to siX3 in HCMV infection and were also found to be effective at inhibiting HCMV replication. Further investigation into the effects of the siRNAs on viral replication showed that pretreatment with each of the siRNAs resulted in an inhibition in the formation of mature replication compartments. The ability of these siRNAs to prevent or reduce certain cytopathic effects associated with HCMV infection was also examined. Infected cells pretreated with siX3, but not siUL54, retained promyelocytic leukemia (PML) protein in cellular PML bodies, an essential component of this host intrinsic antiviral defense. DNA damage response proteins, which are localized in nuclear viral replication compartments, were reduced in the siX3- and siUL54-treated cells. siX3, but not siUL54, prevented DNA damage response signaling early after infection. Therapeutic efficacy was demonstrated by treating cells with siRNAs after HCMV replication had commenced. Together, these findings suggest that siRNAs targeting exon 3 of the major IE genes or the UL54-57 transcripts be further studied for their potential development into anti-HCMV therapeutics.
INTRODUCTION
Human cytomegalovirus (HCMV) is a ubiquitous herpesvirus that can cause life-threatening diseases in immunocompromised individuals, such as AIDS patients and organ transplant recipients (56). HCMV-associated pneumonitis and retinitis are the most prevalent problems detected following reactivation of latent virus (57). HCMV is also the leading cause of infection-associated birth defects, which can culminate in hearing and vision loss, along with various mental disabilities (6, 65).
Although there have been numerous attempts to develop an effective HCMV vaccine, a successful formulation has not yet been clinically approved (40). A limited number of drugs are available for the treatment of HCMV infection, including ganciclovir (GCV); its orally available derivative, valganciclovir; cidofovir (CDV); foscarnet (FOS); and fomivirsen (35, 59). Among these drugs, GCV is the most widely used to treat most HCMV infections. Long-term treatment with any of these drugs, however, is frequently followed by toxic side effects and the emergence of drug-resistant mutants. Furthermore, GCV, valganciclovir, CDV, and FOS have similar mechanisms of action by targeting the viral DNA polymerase, an early (E) gene product (5, 23, 27–29, 41, 66). Another target drug, fomivirsen, is an antisense oligonucleotide that inhibits IE2 expression (1), but it has had limited clinical use (50). Clearly, additional safe therapeutic agents that limit HCMV replication are desirable.
RNA interference (RNAi) is an evolutionarily conserved mechanism of sequence-specific gene silencing that reduces the levels of protein products translated from a targeted mRNA (34). Using RNAi to reduce the levels of specific proteins not only aids the elucidation of their function but also provides the opportunity to consider potential therapeutic targets that could be used to treat various diseases (21). Multiple biotechnology companies are involved in developing RNAi agents as potent therapeutics for various human diseases, such as cardiovascular disease, neurological diseases, viral infections, cancer, etc. (21). With the ongoing efforts in RNAi-based therapeutic development, small interfering RNAs (siRNAs) and their derivatives have been designed to inhibit the expression of several genes related to virus infections in humans, including those with human immunodeficiency virus type 1, hepatitis C virus, hepatitis B virus, poliovirus, human papillomavirus, and influenza virus (9, 31, 32, 39, 52, 81), which provides evidence that RNAi has the potential to be an effective strategy to control viral diseases. There are also reports on RNAi-based targeting of HCMV genes (4, 22, 24, 68, 74, 79).
Cytomegaloviruses have species-specific tropisms. HCMV infects only humans and replicates in a limited number of human cell types in vitro. HCMV infection progresses through a well-characterized sequential process of immediate-early (IE), early (E), and late (L) viral gene expression. IE gene products transactivate the expression of E genes, which are required for viral DNA replication, and replicating viral DNA is associated with L gene expression. IE and E gene products also regulate late gene expression (14, 48, 82). The 72-kDa IE1 protein and 86-kDa IE2 protein are the first and most abundantly expressed proteins during HCMV infection. They are produced from differentially spliced transcripts under the control of the strong IE1/IE2 promoter-enhancer element known as the major immediate-early promoter (MIEP). The IE1 gene (UL123) produces a 1.9-kb mRNA consisting of exons 1, 2, 3, and 4, whereas the IE2 gene (UL122) produces multiple transcripts, including the predominant 2.25- to 1.4-kb mRNAs consisting of exons 1, 2, 3, and 5. The 1.4-kb transcript carries a spliced exon 5 (72). The first translated exon, exon 3, encodes 85 amino acids that are shared at their N termini of IE1 and IE2. This domain is responsible for controlling viral growth and altering cell cycle patterns through its ability to transactivate gene expression and control apoptosis (78).
HCMV IE proteins have multiple activities, including vital roles in viral and host cell activities and endogenous tumor suppressor inactivation (11–13, 26, 33, 37, 38, 49, 51, 70, 76, 84, 85). IE1 can specifically alter the host environment by inactivating tumor suppressors, transcriptionally activating host gene expression, and altering the nuclear architecture of promyelocytic leukemia (PML) bodies of infected cells within a few hours of infection (80). Similarly, IE2 also manipulates tumor suppressor functions and alters host and viral gene expression. The regulation of IE1 and IE2 gene expression is thought to be a critical component of latency and active replication due to transactivation and transcriptional repressive functions of these proteins. In addition, there is growing evidence that pathogenic features of HCMV are due, in part, to the abilities of IE1 and IE2 to alter the expression of cellular genes and host cell functions (17). Reducing or inhibiting the expression of IE1 and IE2 greatly represses the replication of HCMV (75, 86), and studies of mutant viruses have demonstrated that IE2 expression is essential for virus replication (77).
HCMV infection activates multiple markers of the cellular DNA damage response (DDR) pathway, including H2AX and p53 (11, 44, 67). Phosphorylated histone H2AX (γH2AX) and the p53 transcription factor play important roles in HCMV replication, and both proteins localize to viral replication compartments (RCs) (26, 44). Viral RCs are distinct architectural regions within the nucleus where many events associated with viral replication take place, including viral gene expression, DNA replication, and capsid packaging and maturation. Viral RCs mature into discrete structures at E times postinfection (p.i.) that can be detected by immunofluorescent staining of the DNA polymerase accessory factor pUL44. It has been proposed that the activation of p53 helps to elicit the cell cycle arrest in HCMV-infected fibroblasts by modulating p21 levels (12) and by facilitating viral gene expression (10).
Because IE1 and IE2 share the same first three exons, our strategy was to inhibit these most immediate-early gene expression products to block the initial step of virus replication using a novel siRNA (siX3) targeted to a shared sequence of UL122/UL123. We hypothesized that interfering with the production of the major IE proteins will likely have severe consequences for viral gene expression and replication while minimizing the detrimental effects of infection on the host cell. In the present study, we have investigated the inhibitory potential of siX3 at the level of HCMV viral gene expression, infectious virus production, and markers of host cell changes. We then compared these results to the inhibitory effects of siRNAs against UL54 and overlapping transcripts with UL55-UL57 (69), with the long-term goal of developing an informed RNAi-based therapeutic for HCMV diseases.
MATERIALS AND METHODS
Cells and viruses.
Human embryonic lung (HEL) fibroblasts were obtained from the Coriell Institute for Medical Research (Camden, NJ). The cells were cultured in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin. The human astrocytoma cell line (U373MG) was a generous gift from Eng-Shang Huang (University of North Carolina, Chapel Hill, NC) and cultured in DMEM supplemented with 5% FBS and 1% penicillin-streptomycin. All media, FBS, and antibiotics were from Gibco.
HCMV strain AD169 was obtained from the American Type Culture Collection (ATCC; Manassas, VA). HEL fibroblasts or U373MG cells were infected with HCMV AD169 at various multiplicities of infection (MOIs). Viral infections were performed in growth medium with 2% FBS for 2 h. The viral inoculum was removed and replaced with normal growth medium.
siRNA and transfections.
All of the siRNAs were synthesized by Qiagen (Foster, CA). For U373MG cells, siRNAs were transfected at 50 to 100 nM using Oligofectamine (Invitrogen Corporation, Carlsbad, CA). For HEL fibroblasts, transfections were performed by electroporation in siPort transfection buffer (Ambion, Austin, TX). The nonspecific siRNA (siCON) is a nonsense sequence and had no effect on parameters tested relative to mock transfection. Transfection conditions for individual siRNAs were optimized, and infections were usually performed 24 h posttransfection. For treatment experiments, transfections were performed at 24 hours postinfection (hpi). The sequences of the siRNAs used in this study are as follows: siCON, GATGCTGCATATAAGCAGC; siX3, CTATGTTGAGGAAGGAGGT; siUL54A, CTGCTCAACAAGTGGGTTT; siUL54B, CTTTTCAGAGCCGTGTTTT.
RNA isolation and Northern blot analysis.
Total RNA was isolated from mock- or HCMV-infected HEL fibroblasts using TRIzol reagent (Invitrogen Corporation, Carlsbad, CA) according to the manufacturer's instructions. Total RNA was electrophoresed through 1% formaldehyde-agarose gels and blotted onto Zeta-probe GT Genomic Tested blotting membranes (Bio-Rad, Hercules, CA). Biotin-labeled glyceraldehyde-3-phosphate dehydrogenase (GAPDH), IE1 (exon 4), and IE2 (exon 5) probes were generated by PCR using the following primers: GAPDH forward primer (5′-CAAGGTCATCCATGACAAC-3′), GAPDH reverse primer (5′-TGGTCGTTGAGGGCAATG-3′), IE1 forward primer (5′-ATAAGCGGGAGATGTGGATGGCTT-3′), IE1 reverse primer (5′-TCAATGCGGCGCTTCATTACACTG-3′), IE2 forward primer (5′-AGTCCGAGGAGATGAAATGCAGCA-3′), and IE2 reverse primer (5′-GTGGGTGTGCAATTCTTTGAGGCT-3′).
Blots were hybridized with the individual probes. Hybridization signals were visualized using a chemiluminescence kit as described by the manufacturer (KPL, Gaithersburg, MD). Blots were reprobed for GAPDH following stripping of previously bound probe.
DNA slot blot analysis.
Whole-cell DNA was prepared from HEL fibroblasts at the times indicated using a DNA isolation kit (Promega Corporation, Madison, WI). DNAs were quantified using a UV spectrophotometer and equally slotted onto a blotting membrane (Bio-Rad, Hercules, CA) using the Slot Blotter system (Bio-Rad, Hercules, CA). Biotin-labeled UL70 probes were generated by PCR using the following primers: UL70 forward primer (5′-CTCTTTCCCGTCTTCGTCTTCC-3′) and UL70 reverse primer (5′-GCATCAACACGCTTTCCGAGTC-3′).
The membranes were hybridized with UL70 probe, and hybridization signals were visualized with a chemiluminescence blotting kit as described by the manufacturer (KPL, Gaithersburg, MD).
Immunoblot analysis.
Infected cells were harvested at the indicated time points, and cell pellets were stored at −80°C. Thawed cell pellets were resuspended in radioimmunoprecipitation assay buffer (RIPA), which has been described previously (26), and incubated on ice for 1 h. Samples were sonicated for 15 s, and soluble proteins were collected by centrifugation for 10 min at 13,000 rpm in a microcentrifuge. Proteins were resolved by SDS-PAGE, and the proteins were transferred to a polyvinylidene difluoride (PVDF) membrane (Perkin-Elmer, Waltham, MA) by electroblotting. Detection of proteins was performed with antibodies specific for IE1-72 and IE2-86 (MAB810; Chemicon, Temecula, CA), pp65 (CA003-100; Virusys Corporation, Sykesville, MD), gB55 (Shan Lu, University of Massachusetts Medical School, Worcester, MA), and actin (A5316; Sigma, Saint Louis, MO) and horseradish peroxidase (HRP)-conjugated secondary antibodies. Protein bands were visualized by chemiluminescence with ECL reagent (Amersham, Piscataway, NJ).
Plaque reduction assay.
HEL fibroblasts were transfected with a serial dilution of siRNA and seeded as confluent monolayers in 24-well plates. Twenty-four hours later, the cells were infected with HCMV with a dilution sufficient to result in the formation of approximately 40 plaques per well in untreated cells. After a 2-h adsorption, virus was removed, and cells were washed twice with phosphate-buffered saline (PBS) and overlaid with DMEM containing 0.3% SeaPlaque GTG agarose (Lonza, Rockland, ME) and 0.2% FBS mixture. After 7 days of incubation at 37°C, the agarose overlays were removed and cell monolayers were fixed by the addition of 100% methyl alcohol and stained with Giemsa stain (Sigma-Aldrich, Saint Louis, MO). Viral titers were determined by counting plaques. The average titers were derived from triplicate samples for each concentration of siRNA. Error bars represent 1 standard deviation (SD).
Viral growth curves.
Cells were seeded and infected at the listed MOI for each experiment. At the indicated times postinfection, a small aliquot (200 μl) of supernatant was harvested from each dish and stored at −80°C. Viral titers were then determined on HEL fibroblasts by standard plaque assay techniques. Plaques were counted at 7 days p.i. using Giemsa stain (Sigma-Aldrich, Saint Louis, MO) to enhance the visualization of plaques. For U373MG cells, both intracellular and extracellular virus were harvested and pooled, and titers were determined by plaque assay on monolayers of HEL fibroblasts (53). A typical viral growth curve was shown in each experiment.
Immunofluorescence analysis.
Cells were plated on glass coverslips that were pretreated with 40% HCl for 2 min followed by a 5-min wash in 70% ethanol. Cells were infected with HCMV at the indicated MOIs. Cells were washed three times with PBS and fixed with 2% paraformaldehyde. Fixed cells were blocked in 10% FBS for 1 h at room temperature and incubated with antibodies against IE (MAB810; Chemicon, Temecula, CA), pUL44 (Virusys Corporation, Sykesville, MD), PML (Chemicon, Temecula, CA), p53 (Ab-6; Thermo, Fremont, CA), and γH2AX (Upstate Biotechnology, Waltham, MA). Fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA), Texas Red-conjugated goat anti-mouse IgG1 or IgG2a secondary antibodies, and FITC-conjugated goat anti-mouse IgG1 secondary antibodies (Southern Biotechnology Associates, Inc., Birmingham, AL) were used to detect bound primary antibody by immunofluorescence. Images were captured on a Nikon microscope and analyzed using Improvision software. Over 200 cells were counted per sample when quantifying cell staining. Triplicate samples were analyzed in each experiment.
Quantitative reverse transcription-PCR (qRT-PCR).
Quantitative analyses of UL54 transcripts were performed using the SYBR GreenER qPCR kit (Qiagen) on a DNA Engine Opticon 3 continuous fluorescence detection system (MJ Research, Incorporated, Waltham, MA). Total RNA was extracted from the infected cells using TRIzol (Invitrogen), and 100 ng of purified total RNA was reverse transcribed to cDNA using a cDNA synthesis kit (Invitrogen). The PCR was set according to the manufacturer's recommendations. Briefly, after an initial 10 min of denaturation at 95°C, 30 cycles of amplification were performed at 95°C for 30 s and 60°C for 1 min followed by a melting curve analysis. The amount of template RNA was normalized with the quantified GAPDH in each sample. The primer sets for amplification of UL54 were as follows: forward, 5′-CGTGCCGCGAGGTGTCATGT-3′; reverse, 5′-CACCAGGGTCTCGCGCCAAG-3′. The primer sets for GAPDH were as follows: forward, 5′-GAAGGTGAAGGTCGGAGTC-3′; reverse, 5′-GAAGATGGTGATGGGATTTC-3′. Quantitative experiments were performed at least three times, including a no-template control reaction. Relative expression was calculated using a modified comparative threshold cycle method (2−ΔΔCT) (42), in which CT was defined as the cycle number at which fluorescence reached a set threshold value. The differences between the CT values of the target genes and those of the corresponding internal control GAPDH gene, ΔCT (CTUL54 − CTGAPDH), were calculated. The changes between ΔCT of the treated group and that of the control group, ΔΔCT (ΔCTsiRNA − ΔCTinfection only), were computed. The expression level of the treated group relative to that of the control group was described by using the equation 2−ΔΔCT.
Statistical analysis.
Statistical analyses were performed using unpaired t tests. Values are expressed as the means ± standard deviations (SDs) of three independent experiments. A P value of ≤0.05 was considered statistically significant.
RESULTS
siRNA design.
The major IE (MIE) genes (UL123/122) were chosen for inhibition by RNAi to limit both the replication of HCMV and cytopathic effects associated with infection. The MIE gene products are required for or influence the expression of most HCMV genes (15, 36, 45, 58). To maximize the impact of siRNA transduction on viral gene expression, we designed siRNAs to target exon 3 of the MIE locus. Exon 3 contains the first protein-coding sequences of the MIE region and is shared by IE1 and several IE2 transcripts (Fig. 1A). A shared coding region was chosen with the notion that this would limit the opportunity for escape mutants to emerge. Initially, two siRNAs were compared for their abilities to reduce viral gene expression alone and in combination. Because introduction of one siRNA, siX3 (or X3), into cells prior to infection was much more effective at reducing viral gene expression than was introduction of the other siRNA or combinations of the two (data not shown), it was chosen for subsequent studies.
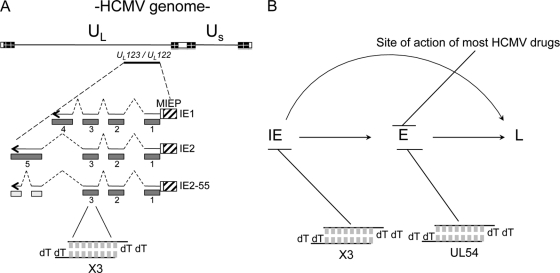
Fig 1.
Strategy for inhibiting human cytomegalovirus replication with siRNAs. (A) Schematic diagram representing the organization of the HCMV genome and the relative position of ORFs encoding the UL123/UL122 gene products, IE1 and IE2. Gray bars, exonic structure (exons 1 through 5) of the three major transcripts generated at IE times of infection. The X3 siRNA was designed to target exon 3, which is shared by these transcripts. UL, unique long, and US, unique short (regions of the HCMV genome); MIEP, major IE promoter. (B) Temporal targeting of siRNAs characterized in this study along with most anti-HCMV drugs. UL54, siRNA against UL54-57 transcripts.
UL54 encodes the HCMV DNA polymerase, an enzyme essential for replication of the viral genome and the target of most anti-HCMV drugs (Fig. 1B). It is well established that inhibition of DNA polymerase activity, and predictably expression, will prevent HCMV replication. As with the strategy used to inhibit HCMV IE gene expression, an in silico and visual search of the UL54 sequence was used to identify potential target sequences for siRNA development. From this analysis, two candidate siRNAs, named siUL54A and siUL54B, were identified and tested for their antiviral activity compared to that of siX3. Given the overlap of transcripts among UL54, UL55, UL56, and UL57 (69), both siUL54 siRNAs should also target transcripts from UL55, which encodes gB55; UL56, which encodes pUL56; and UL57, which encodes pUL57.
siX3 alters the levels of IE transcripts containing exon 3 sequence.
To determine whether siX3 specifically targets sequences shared by mRNAs encoding IE1 and IE2, HEL fibroblasts were transfected with siX3 and infected with HCMV 24 h later. Northern blots of RNA from infected HEL fibroblasts were probed for sequences specific to exon 4 of UL123 (IE1) or exon 5 of UL122 (IE2) to measure IE1 and IE2 mRNA levels, respectively. Transfection of siX3 prior to HCMV infection reduced IE1 mRNA levels throughout the infection time course relative to cells that were either transfected with a control siRNA (CON) or untransfected (Fig. 2; data quantified in Fig. S1 in the supplemental material). These trends were observed at both low and high MOIs, respectively (Fig. 2A and B, respectively). The levels of the 2.25-kb IE2 transcript were also repressed throughout most of the infection time course. At E times (24 to 48 hpi), the levels of the 1.7-kb IE2 mRNA were reduced to ones similar to those of the 2.25-kb IE2 transcript. However, at L times p.i., there was an accumulation of a 1.7-kb transcript that hybridized with the IE2 exon 5 probe (Fig. 2; data quantified in Fig. S1). Given that accumulation of the 2.25-kb IE2 transcript was still reduced by siX3, this increase in the 1.7-kb RNA at late times was most likely due to a separate late transcript composed of exon 5 (60). This late transcript lacks exon 3 and would not be targeted by siX3.
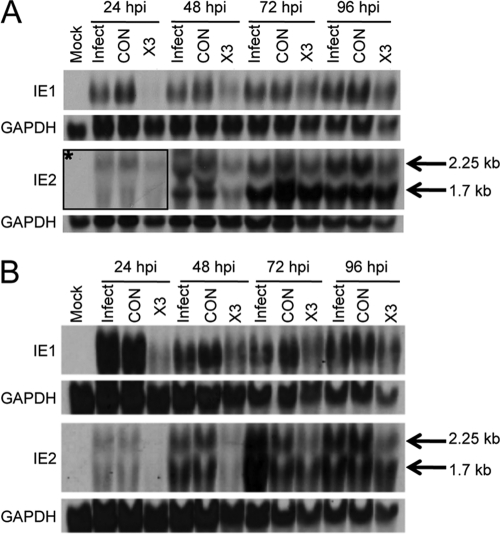
Fig 2.
Inhibition of IE gene expression by siX3. HEL fibroblasts were untransfected or transfected with X3 or CON siRNAs. Twenty-four hours after transfection, the cells were infected with HCMV AD169 at an MOI of 0.1 (A) or an MOI of 1.0 (B). At 24, 48, 72, and 96 hpi, total RNAs were extracted and analyzed for IE1- and IE2-specific transcripts by Northern blot hybridization with biotin-labeled probes to IE1 (exon 4) or IE2 (exon 5). Arrows mark the 1.7- and 2.25-kb IE2 transcripts. GAPDH is shown as a loading control. The boxed region labeled with an asterisk shows a longer exposure of that region of the membrane.
Sustained inhibition of cell-to-cell spread of HCMV by siX3.
A plaque reduction assay was used to determine if treatment with siX3 could sustain the inhibition of HCMV replication under conditions where superinfection of virus produced by untransfected cells is minimized. In a plaque reduction assay, only a small percentage of cells are infected and the samples are overlaid with agar. This semisolid overlay allows virus to spread only through cell-to-cell contact. HEL fibroblasts were transfected with various doses of siRNAs, infected with HCMV, and then overlaid with agar. The conditions used would generate ∼40 plaques/well in untreated infections by the 168-h (7-day) endpoint (Fig. 3). The transfection process appears to have improved the replication of HCMV because siCON-treated samples consistently produced more plaques than did infection alone, although this difference was generally within the error range. Wells containing cells transfected with siX3 resulted in 5- to 10-fold-lower plaque numbers at all siX3 doses examined.
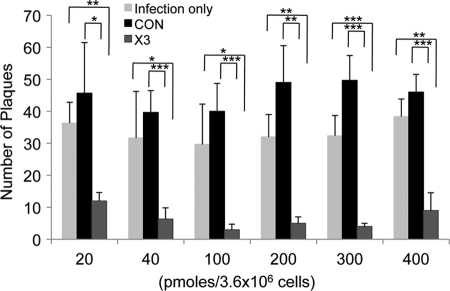
Fig 3.
Inhibition of cell-to-cell spread of HCMV by siX3 as measured by plaque reduction assay. HEL fibroblasts were transfected with X3 or CON siRNA at a dose of 20, 40, 100, 200, 300, or 400 pmol per 3.6 × 106 cells. The cells were seeded at 100% confluence and infected with ∼40 PFU of virus. After adsorption, cells were overlaid with agarose. After 7 days of incubation, viral titers were determined by staining and counting plaques. Histograms show the averages of three independent experiments, and the error bars denote the standard deviations. *, P < 0.022; **, P < 0.002; ***, P < 0.001.
Decreased viral protein expression and inhibition of infectious virus production in siX3-treated cells.
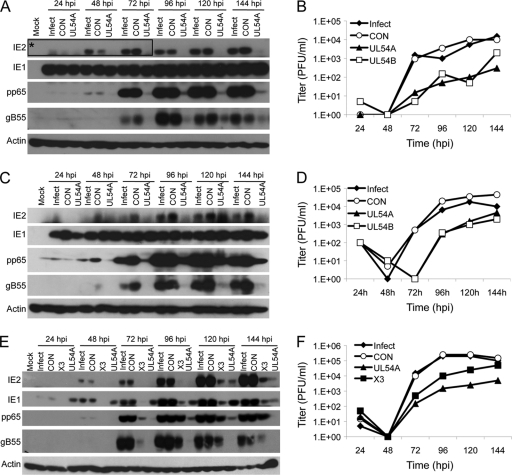
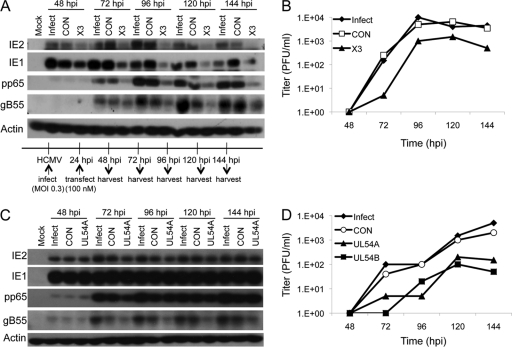
Immunoblots of infected HEL fibroblast lysates were sequentially probed with antibodies against representative viral IE, E, and E/L proteins to determine the consequences of siX3 transfection for the different stages of viral gene expression. As anticipated from the results obtained by Northern blotting, siX3 transfection reduced the levels of IE1 and IE2 proteins through 96 hpi at both MOIs tested (Fig. 4A and C). Early and late protein accumulation was also affected by siX3 at these time points as evidenced by the reduced levels of the E protein, pp65, and the E/L protein, gB55. At very late times p.i. (120 to 144 hpi), the levels of IE1 and pp65 appeared to recover in the siX3-transfected samples, whereas IE2 and gB55 protein levels remained reduced or undetectable depending on the amount of input virus (Fig. 4A and C). Thus, treatment of cells with siX3 results in a profound and sustained effect on the expression of all three classes of HCMV proteins.
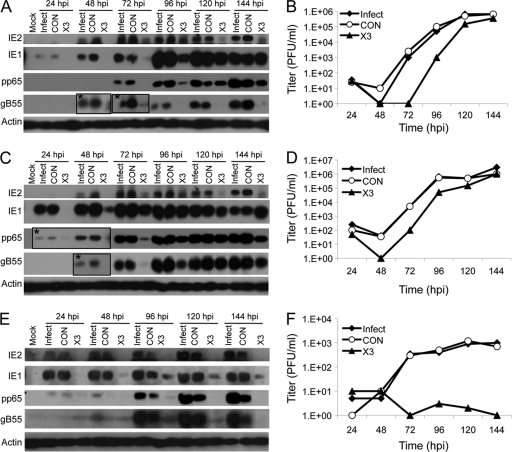
Fig 4.
siX3 pretreatment compromises HCMV replication. HEL fibroblasts were untransfected or transfected with X3 or CON siRNAs 24 h prior to infection with HCMV at an MOI of 0.1 (A and B) or 1.0 (C and D). (A and C) The levels of viral IE, E, and E/L protein expression were assessed by immunoblot analysis for IE1/IE2, pp65, and gB55, respectively. (B and D) Culture supernatants were assayed for infectious virus production by plaque assay. (E and F) U373MG cells were untransfected or transfected with X3 or CON siRNAs 24 h prior to infection with HCMV at an MOI of 0.3. (E) The levels of viral IE, E, and E/L protein expression were assessed by immunoblot analysis for IE1/IE2, pp65, and gB55, respectively. (F) Culture supernatants and clarified cell lysates were combined and assayed for infectious virus by plaque assay. Boxed areas in panels A and C that are labeled with asterisks show longer exposures of those regions of the immunoblotted membranes.
The production of infectious virus following siRNA transfection was measured by plaque assay. The generation of progeny virus was not affected by prior transfection of control siRNA but was blocked for 2 to 3 days in HEL fibroblasts treated with siX3 (Fig. 4B and D). Interference with virus production by siX3 lasted through 96 h, during which there was a 1- to 3-log reduction in virus yield. This ability of siX3 to antagonize virus maturation was alleviated from 120 hpi onward at both MOIs. The gradual attenuation of siX3 to inhibit HCMV replication and maturation could have been due to turnover of siX3, given that only a single treatment of the siRNA was administered. Another possibility for this decay in activity is that infected but untransfected cells generated enough progeny viruses to overwhelm the effects of the siRNA by superinfection.
Human fibroblasts, including HEL fibroblasts, are notorious for their poor-to-modest transfection efficiency, and so it remains possible that a significant percentage of cells do not functionally take up siX3 under any condition. A cell line that can be transfected more efficiently and is also receptive to productive HCMV infection might serve as an alternate model for determining whether HCMV replication can be sustainably inhibited by siX3 transfection. U373MG cells are receptive to efficient lipid-based transfections and are permissive to HCMV replication. One drawback to using these cells is that HCMV replication results in lower titers. Thus, a direct comparison to results obtained in the different cell types is not feasible. Lipid-mediated transfection of siRNAs into U373MG cells prior to HCMV infection resulted in little or no accumulation of the two IE gene products, IE1 and IE2, as well as E (pp65) and E/L (gB55) proteins over the 144-h time course (Fig. 4E). This apparent block in viral gene expression is consistent with the absence of detectable progeny virus being generated in siX3-treated cells throughout the infection time course (Fig. 4F). Together, the results suggest that HCMV gene expression and replication can be inhibited with an siRNA specific to IE1/IE2 and that effective siRNA delivery is essential to prevent the drug from being overwhelmed by superinfection of treated cells.
The observation that siX3 treatment blocked virus production for at least 7 days in the plaque reduction assay raises the possibility that improvements in transfection efficiency might enhance the effectiveness of siX3 to inhibit HCMV replication. Multiple dosing of the siRNA could, in principle, increase the percentage of cells transfected with siRNAs. HEL fibroblasts were sequentially transfected (by electroporation or lipid-based transfection) two times before infection. However, this treatment did not improve the results obtained by a single transfection (data not shown). Why multiple dosing of siRNAs was only as effective as a single transfection is unclear. One possibility is that there was a subset of cells in the culture that were resistant to transfection or functionally incorporating siRNAs.
Comparison of siRNAs targeting the UL54 E gene with siX3 targeting the UL123/122 IE genes.
Two siRNAs that target the UL54 E gene were designed for comparison with siX3. The effectiveness of siUL54A and siUL54B in reducing UL54 expression is shown in Fig. S2 in the supplemental material. Transfection of either siRNA directed against UL54 expression into HEL fibroblasts prior to infection reduced the levels of E (pp65) and E/L (gB55) proteins throughout the experimental time course regardless of the MOI used (immunoblotting data for siUL54A are shown in Fig. 5A; additional immunoblotting data are included in Fig. S3A, C, and D in the supplemental material). The inhibitory effect was perhaps most pronounced for gB55, where little protein was detected for samples treated with either siUL54. This observation is likely due to dual effects of these siRNAs on gB55 expression. First, gB55 expression should be downstream of true E genes and be affected by E gene expression and viral DNA replication. Second, some of the UL54 and UL55 transcripts overlap, and so the siRNAs should also affect gB55 accumulation (69). In contrast, IE1 expression was not affected by siUL54 RNAs. This was expected given that IE1 expression begins prior to and is independent of expression of the viral DNA polymerase. The fact that IE1 expression was unaffected also suggests that the effect of the siUL54 RNAs on viral gene expression was specific and not a result of global changes in gene expression due to an intrinsic host response to the transfected siRNAs (e.g., interferon signaling). The effect that these siRNAs have on IE2 expression reflects a pattern that is characteristic of viral E and L genes. This is likely due to the fact that while IE2 is initially expressed at IE times, its rise in expression is coincident with viral DNA replication (30, 71), thus resulting in an accumulation pattern more akin to E and L gene products. Likewise, viral growth curves showed that both siUL54 RNAs inhibited the generation of infectious virus in HEL fibroblasts at both MOIs tested (Fig. 5B; see also Fig. S3B).
Fig 5.
Comparison of siRNAs targeting the UL54-57 locus with siX3 targeting the UL123/122 IE genes. (A and B) HEL fibroblasts were untransfected or transfected with UL54A, UL54B, or CON siRNAs 24 h prior to infection with HCMV at an MOI of 0.1. Results from a similar experiment done at an MOI of 1.0 are shown in Fig. S3B to D in the supplemental material. (A) The levels of viral IE, E, and E/L protein expression were assessed by immunoblot analysis for IE1/IE2, pp65, and gB55, respectively. Results for the UL54B siRNA are shown in Fig. S3A in the supplemental material. (B) Culture supernatants were assayed for infectious virus production by plaque assay. (C and D) U373MG cells were untransfected or transfected with UL54A, UL54B, or CON siRNAs 24 h prior to infection with HCMV at an MOI of 0.3. (C) The levels of viral IE, E, and E/L protein expression were assessed by immunoblot analysis for IE1/IE2, pp65, and gB55, respectively. Results for the UL54B siRNA are shown in Fig. S4 in the supplemental material. (D) Culture supernatants and clarified cell lysates were combined and assayed for infectious virus by plaque assay. (E and F) Comparison of siUL54A with siX3 pretreatment in compromising HCMV replication. HEL fibroblast cells were untransfected or transfected with UL54A, X3, or CON siRNAs 24 h prior to infection with HCMV at an MOI of 0.1. (E) The levels of viral IE, E, and E/L protein expression were assessed by immunoblot analysis for IE1/IE2, pp65, and gB55, respectively. (F) Culture supernatants were assayed for infectious virus production by plaque assay. The boxed region in panel A labeled with an asterisk shows a longer exposure of that region of the membrane.
To determine whether more efficient siRNA transfection would improve the inhibitory effects of siUL54A and siUL54B on viral replication, the siRNAs were tested in U373MG cells. As observed in HEL fibroblasts, IE1 expression was only moderately affected by either UL54 siRNA (Fig. 5C; see also Fig. S4 in the supplemental material). E (pp65) and E/L (gB55) gene expression was similarly reduced over the infection time course, and IE2 levels remained low on immunoblots derived from the siUL54-treated samples. The reduction in viral gene expression correlated with reduced yields of virus during this infection time course as well (Fig. 5D). No production of infectious virus was detected until 96 hpi in the siUL54-treated cells (the scored virus at the 24-hpi time point is unabsorbed input virus from the initial infection). From 96 hpi onward, the UL54 siRNAs inhibited virus yield by approximately 1 to 1.5 logs. Thus, decreasing the expression of the viral DNA polymerase and UL55-57 delayed virus production and reduced virus yields. However, the presumed increase in transfection efficiency of U373MG did not improve the effectiveness of these siRNAs.
Since the results that targeting IE gene expression or the E/L transcripts from UL54-57 with siRNAs inhibited HCMV replication were observed in separate experiments, siX3 and siUL54s were directly compared for efficacy. Because the two siUL54 RNAs produced similar results, only one, siUL54A, was included in this experiment. Immunoblots of HEL fibroblasts pretreated with siX3 or siUL54A showed reduced levels of E and E/L gene expression (Fig. 5E), though siUL54A-treated cells had lower levels of pp65 and gB55 throughout the time course. Both siRNAs also reduced IE2, with siUL54A having a more dramatic impact on IE2 levels. This was in contrast to the analysis of IE1 expression, where siX3 treatment resulted in lower IE1 levels at all times except 144 hpi. For the high-MOI infection (data not shown), a similar pattern of viral protein accumulation emerges, with siUL54A reducing viral protein accumulation to a greater extent than siX3. Direct comparison of virus yields in infected HEL fibroblasts showed that both siX3 and siUL54A reduced virus production. However, siUL54A had a greater impact on virus yield (Fig. 5F). Taken together, these results suggest that an siRNA targeting UL54-57 is effective at inhibiting HCMV replication in HEL fibroblasts.
To determine whether a combination of siX3 and siUL54 can control viral replication better than either siRNA alone, individual or both siRNAs were transfected into HEL fibroblasts at different siRNA doses (see Fig. S5A and B in the supplemental material for the low siRNA dose; see Fig. S5C and D for the high siRNA dose). Immunoblots show similar patterns of E and E/L gene expression for siUL54 alone and when combined with siX3 to transfect cells (see Fig. S5A and C), although siX3-plus-siUL54-transfected cells had lower levels of IE1 expression than did cells transfected with just siUL54. The combined siRNA transfections had effects on viral yields similar to that of the siUL54 transfection. Transfections with siUL54 or siUL54 plus siX3 performed better than did that with siX3 alone in reducing the production of infectious virus (see Fig. S5B and D). Taken together, these results suggest that the combination of siX3 and siUL54 is no better at inhibiting viral replication than is siUL54, although IE1 expression is reduced when the two siRNAs are used together.
Inhibition of viral DNA replication by siX3 and siUL54 pretreatment.
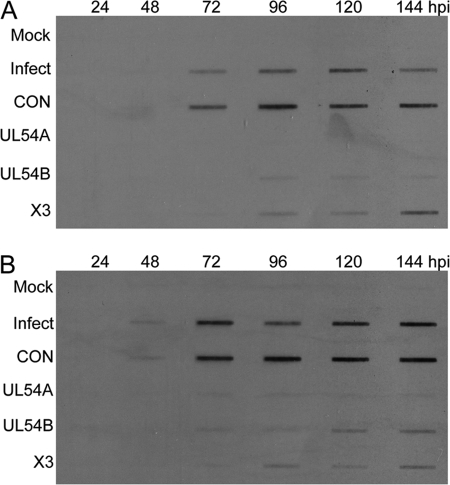
Total DNA was extracted from infected cells, and the accumulation of viral DNA was monitored by slot blot hybridization as a measure of viral DNA replication. Detectable levels of viral DNA became apparent at 48 to 72 hpi in the untransfected and siCON-treated samples but not in the siX3- and siUL54-transfected samples (Fig. 6). This pattern was observed at the two MOIs tested and suggests that treatment with the siRNAs resulted in the inhibition of viral DNA replication. At 96 to 120 hpi, viral DNA could be detected in the siX3-treated cells, with lower levels appearing in siUL54-treated cells. However, transfection with either siX3 or the siUL54 siRNAs greatly reduced levels of viral DNA relative to those for the control treatments. By 144 hpi, the levels of viral DNA in siX3-transfected cells were approaching those observed in the controls at both MOIs. Surprisingly, there is little to no detectable viral DNA in UL54A siRNA-treated cells at any time point. One possible explanation for the different effects of siX3 and siUL54s on viral DNA levels is that the siUL54 molecules prevented the accumulation of the UL54-encoded DNA polymerase below levels necessary for logarithmic amplification of the viral genome. The result of interfering with viral DNA polymerase accumulation would then be the observed, more-linear amplification of viral DNA.
Fig 6.
Inhibition of viral DNA replication by siX3 and siUL54 pretreatment. HEL fibroblast cells were untransfected or transfected with UL54A, UL54B, X3, or CON siRNAs 24 h prior to infection with HCMV at an MOI of 0.1 (A) or 1.0 (B). The cell pellets were harvested at the indicated times p.i. Total DNA was extracted from infected cells, and equivalent concentrations of DNA were slotted in each well of the slot blot as a measure of viral DNA replication by hybridization with a biotin-labeled probe to UL70.
siX3 and siUL54 alter the formation of viral RCs.
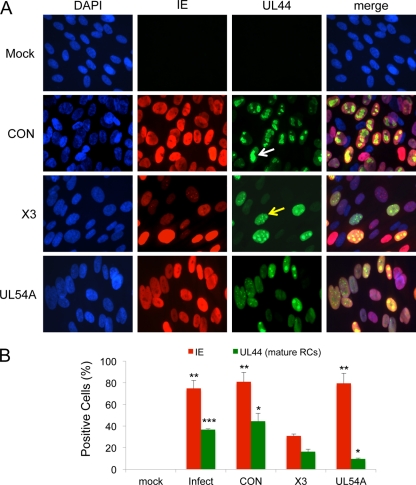
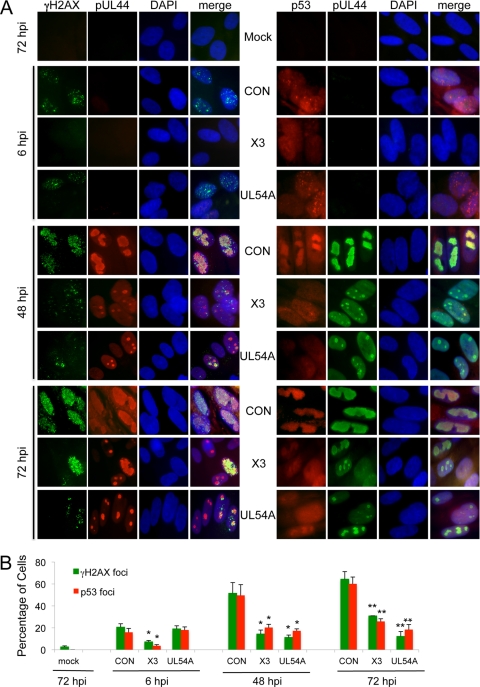
Monitoring viral replication compartments (RCs) offers a qualitative and temporal measure of HCMV replication. Viral RCs begin as multiple, discrete structures early in infection, which, based on studies in herpes simplex virus (HSV) (16), fuse into a single, large structure that can be referred to as a “mature” RC. Both immature and mature RCs can be detected by immunofluorescent staining for pUL44, an accessory factor involved in the replication of the HCMV genome (Fig. 7). Samples treated with a control siRNA or not treateddisplayed enlarged nuclei and IE staining typical of HCMV infections (Fig. 7A). The formation of mature RCs in cells as detected by pUL44 immunostaining was apparent in most of the infected cells (Fig. 7B). Cells treated with siX3 showed reduced IE staining in most cells, whereas siUL54A had no effect on IE staining. Cells treated with either siX3 or siUL54A rarely displayed mature RCs. Rather, some infected cells showed high levels of diffuse nuclear pUL44 staining, while others contained immature, “pre-RC” structures. Quantitation of the percentage of cells staining positive for merged, mature RCs was reduced by treatment with siX3 or siUL54A relative to that for control siRNA-treated cells (Fig. 7B). These results suggest that IE expression and UL54 expression are important for the formation of viral RCs.
Fig 7.
Pretreatment with X3 and UL54 siRNAs alters the formation of viral replication compartments (RCs). HEL fibroblast cells were untransfected or transfected with X3, UL54A, or CON siRNAs 24 h prior to infection with HCMV at an MOI of 1.0. Cells were fixed at 48 hpi, and pUL44 and IE proteins were detected by immunostaining. (A) Localization of IE and pUL44 proteins. Cells with “immature” RCs were defined as those with multiple, small pUL44 compartments (for example, see the yellow arrow), and cells with “mature” RCs were identified as those composed of single, larger pUL44 compartments typically representing more than two-thirds of the nucleus (for example, see the white arrow). 4′,6-Diamidino-2-phenylindole (DAPI) staining is used to define nuclei. (B) The percentage of fibroblasts with mature RCs was plotted relative to the percentage of those lacking RCs or having immature RCs. The percentage of fibroblasts with IE staining was also determined. Over 200 cells were scored per sample. Histograms show the averages of three independent experiments, and the error bars denote the standard deviations.*, P < 0.005 relative to X3; **, P < 0.0005 relative to X3; ***, P < 0.0001 relative to X3.
Only siX3-pretreated cells retain PML in cellular PML bodies following infection.
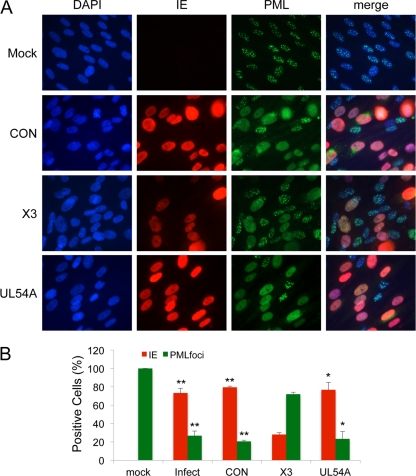
We next determined the consequences of siRNA transfection for host cell function starting with an analysis of nuclear, promyelocytic leukemia protein (PML) bodies (also known as promyelocytic oncogenic domains [PODs] or nuclear domains 10 [ND-10]). Nuclear PML bodies are important sites of host gene expression and splicing, and alteration of PML bodies impacts the cellular gene expression program. PML body integrity is also a component of the host intrinsic antiviral defense. Displacement of PML protein from PML bodies occurs during infection of nuclear replicating DNA viruses, including HCMV (62). Uninfected cells retain PML within PML bodies, which were apparent as discrete foci by immunostaining for the PML protein (Fig. 8A). PML protein was observed as diffuse nuclear staining in HCMV-infected cells treated with siCON, indicating dispersal of the protein from PML bodies (Fig. 8A). When quantified, nearly all cells staining positive for IE protein in the infection controls lacked clearly defined PML foci (Fig. 8B). By reducing the levels of cells that were positive for IE protein, which is responsible for PML dispersal, most of the siX3-treated cells retained the normal punctate PML staining pattern. This effect is different from that on cells treated with siUL54A, where the IE and PML focal patterns are similar to those of infected cells that were treated with the control siRNA. Because PML is involved in cellular growth regulation, transcription, DNA replication and repair, and posttranscriptional regulation of gene expression (7), the different effects of siX3 and siUL54A on PML status should have a significant impact on the expression of host genes and cell function.
Fig 8.
Cells pretreated with X3 siRNA retain PML in cellular PML bodies following infection. HEL fibroblast cells were untransfected or transfected with X3, UL54A, or CON siRNAs 24 h prior to infection with HCMV at an MOI of 1.0. Cells were fixed at 48 hpi, and PML was detected by immunostaining. (A) Images show the localization of IE and PML proteins. Cells with typical PML bodies are seen in normal cells (Mock). PML protein was observed as diffuse nuclear staining in HCMV-infected cells pretreated with siCON. DAPI staining is used to define nuclei. (B) The percentages of fibroblasts with IE-positive staining or PML bodies were plotted. Over 200 cells were scored per sample. Histograms show the averages of three independent experiments, and the error bars denote the standard deviations. *, P < 0.0004 relative to X3; **, P < 0.0001 relative to X3.
DDR proteins fail to localize to viral RCs in siX3- and siUL54-treated cells.
It has been shown elsewhere that certain DDR proteins accumulate and localize to RCs in infected cells (26, 44). Given that siX3 and siUL54 alter the formation of viral RCs, we speculated that they would also affect the localization of DDR proteins. As shown in Fig. 9A, γH2AX and p53 coalesced into RCs as detected by pUL44 coimmunostaining by 48 hpi in control siRNA-treated cells. However, only small percentages of cells showed RC localization for γH2AX and p53 in siX3- and siUL54-treated cells at 48 and 72 hpi (Fig. 9A; data quantified in Fig. 9B). Because host DNA damage responses can be induced by HCMV at IE times (26), we also determined whether the siRNAs can also affect this event very early in infection. As expected, siX3, but not siUL54, reduced γH2AX and p53 focus formation at 6 hpi.
Fig 9.
γH2AX and p53 fail to accumulate in viral RCs in cells pretreated with X3 or UL54 siRNAs. HEL fibroblast cells were transfected with X3, UL54A, or CON siRNAs 24 h prior to infection with HCMV at an MOI of 0.1. Cells were fixed at indicated times p.i.; γH2AX and pUL44, or p53 and pUL44, were detected by coimmunostaining. (A) Localization of γH2AX, p53, and pUL44 proteins. γH2AX and p53 coalesce into structures reminiscent of RCs as marked by pUL44 immunostaining in control siRNA-treated cells by 48 hpi. DAPI staining is used to define nuclei. (B) Plot of the percentage of HEL fibroblasts with γH2AX or p53 foci. Over 200 cells were scored per sample. Histograms show the averages of three independent experiments, and the error bars denote the standard deviations. *, P < 0.005 relative to CON, 6 hpi or 48 hpi; **, P < 0.0005 relative to CON, 72 hpi.
Inhibition of HCMV replication by therapeutic siRNA treatment.
The use of siRNAs in clinical practice would likely be therapeutic, where the treatment is given after infection has been established. The ability to efficiently deliver siRNAs to U373MG cells using lipid-based transfection makes it possible to determine whether siRNA treatment of infected cells will affect virus replication. U373MG cells were transfected with siRNAs at 24 hpi, and viral protein synthesis and progeny virus production were assessed (Fig. 10). As seen with pretreatment, transfection of infected U373MG cells with siX3 postinfection resulted in sustained reductions of IE, E, and E/L gene expression as measured by immunoblotting for IE1, IE2, pp65, and gB55 (Fig. 10A). Therapeutic transfection of infected U373MG cells with siX3 allowed for infectious progeny production, which differs from results obtained when cells were transfected prior to infection (compare Fig. 4F and Fig. 10B). Here, treating infected cells with siX3 reduced viral titers by ∼1 log (Fig. 10B). For comparison, siRNAs targeting UL54 expression were individually transfected into cells 24 hpi to determine their effect on viral gene expression and replication (Fig. 10C and D; see also Fig. S6 in the supplemental material). By 24 hpi, E gene expression, including UL54, should have already commenced, and so some DNA polymerase may have been available to initiate viral DNA replication by the time that the siRNAs were functional in cells. Similar to the results obtained following therapeutic treatment with siX3, viral yields were decreased by 1 to 2 logs when the UL54 siRNAs were introduced into infected cells (Fig. 10D), although there was only a detectable change in E/L gene expression as measured by immunoblotting for gB55 (Fig. 10C; see also Fig. S6). Together with the siX3 data, these results provide strong evidence that siRNAs that target essential viral genes are able to reduce virus production to 1 to 10% of normal replication levels even when cells are therapeutically treated (with a single dose) after the viral infection is established.
Fig 10.
Inhibition of HCMV replication by therapeutic treatment with X3 or UL54 siRNAs. U373MG cells were therapeutically transfected with X3 (A and B), UL54A (C and D) or UL54B (D; also see Fig. S6 in the supplemental material), or CON siRNA at 24 hpi with HCMV at an MOI of 0.3. (A and C) The levels of viral IE, E, and E/L protein expression were assessed by immunoblot analysis for IE1/IE2, pp65, and gB55, respectively. (B and D) Culture supernatants and clarified cell lysates were combined and assayed for infectious virus production by plaque assay.
DISCUSSION
Therapies to treat HCMV infections have been difficult to manage because of side effects associated with antiviral drugs. Effective vaccines to protect individuals from HCMV infection or protect them from infection-associated diseases do not exist. Silencing of gene expression by siRNAs is becoming a powerful tool for the development of new therapies. In this study, we demonstrated that an siRNA directed against a shared exon of UL122-UL123 reduces the expression of the targeted transcripts, which has the expected effects on IE, E, and L protein expression; DNA replication; viral propagation; and PML body integrity. Further, targeted depletion of IE1 and IE2 proteins or the viral DNA polymerase alters the formation of viral RCs and the recruitment of DDR proteins to the RCs at E and L time points. These events mark important hallmarks of productive replication, and their perturbation may explain the mechanism behind the efficacy of the siRNA treatments. Significantly, we also demonstrated therapeutic efficacy by treating cells with siRNAs after HCMV replication had commenced.
To our knowledge, no study has considered using one siRNA to simultaneously abolish the expression of multiple IE genes, even though reports have demonstrated that siRNA-mediated targeting of other open reading frames (ORFs) can affect HCMV replication (4, 24, 68, 74, 79). A result of this present study is the demonstration that targeting a conserved and shared region of IE genes can inhibit HCMV replication, thereby illustrating the potential of using this region as a new drug target for RNAi-based therapeutics. Consistent with this possibility, our observations are similar to those made for an exon 3-deleted virus, IE delta 30–77 (78). This mutant virus exhibits impaired growth at both high and low MOIs. Likewise, attempts to rescue virus from a ul122-deficient bacterial artificial chromosome (BAC) fail to induce cytopathic effects and the expression of several viral early genes is undetectable (46).
The similarity of the results obtained here with siX3 to those made with an exon 3-deleted virus (78) illustrates the potential of siRNA-based targeting of HCMV gene expression for performing quick and relatively inexpensive genetic studies of viral gene function. However, there are caveats to the siRNA approach, including the targeting of multiple viral genes due to overlapping transcription and incomplete suppression of gene expression. Complementary experiments such as the inclusion of multiple siRNAs that target the expression of the same gene and inclusion of appropriate controls can often mitigate these concerns.
The antiviral activity of siX3 and siUL54, which targets UL54-57, translates into a >1- to 3-log reduction of virus replication in HEL fibroblasts, with some conditions producing no detectable infectious virus. This level of antiviral activity compares favorably with that of siRNAs against herpes simplex virus, where a 1.5-log reduction in virus titer in cell culture experiments was shown to be sufficient to protect mice from lethal intravaginal challenge when animals were pretreated with an anti-HSV siRNA (55). Our results are tempered somewhat by the observations of HCMV replication recovering from siRNA treatment in HEL fibroblasts. Taken together with data obtained in U373MG cells, which are more efficiently transfected than HEL fibroblasts, it appears that efficient delivery of siRNAs is essential for efficacy. Still, these experiments demonstrate that a single prophylactic or therapeutic treatment of cells with anti-HCMV siRNA is sufficient to have a dramatic impact on viral replication and cytopathic effects.
The fact that siX3 effectively blocked both IE gene expression and nascent virus production in U373MG cells raises the potential for targeting exon 3 as an adjuvant therapy for malignant gliomas, which are often-lethal brain cancers. HCMV has been shown to be associated with malignant gliomas (18, 25, 47, 61, 63, 64; B. Bhattacharjee, N. Renzette, and T. F. Kowalik, submitted for publication), and the U373MG cell line is derived from an astrocytoma, which is a form of malignant glioma. While it is unclear whether viral replication occurs in these cancers, HCMV IE gene expression is a hallmark of this disease (18, 43, 47, 64, 73). IE1 expression has also been shown to stimulate both cell proliferation and signature signaling pathways in glioma cells (19). Thus, inhibiting IE gene expression may have therapeutic value as a component of an aggressive treatment regimen for this cancer.
One of the greatest challenges in the development of RNAi-based therapies has been delivery in vivo. Efficacy of siRNAs has been related to the targeted cell type and delivery strategy. Different modifications of siRNA/siRNA-like molecules have been used to stabilize the molecules, such as using sulfate in place of phosphate in vivo (54), or to improve uptake, such as tagging siRNAs with cholesterol (83). However, in certain circumstances, we have been able to deliver functional unmodified siRNAs in vivo (8). The effectiveness of siRNAs typically lasts for a few days to weeks. However, this effective window may be sufficient for antiviral applications with acute phenotypes. Others have recently developed a clever system that delivers an RNase P-based ribozyme to greatly reduce mouse CMV replication both in vitro and in vivo (3). An orally delivered carrier for delivery of siRNAs has also been shown to be effective in vitro and in vivo (2). It will be interesting to compare the effectiveness of delivery vehicles and RNA therapeutics (ribozyme versus siRNA) in treating CMV infections.
We attempted to improve the efficacy of the siRNAs through the use of multiple dosing of siRNAs, expecting that the siRNA effects would be more potent and sustained. However, the results of these experiments were no different than those of single-dose transfections. Why multiple dosing did not improve the effectiveness of the siRNAs is unclear. One possibility is that there is a subset of fibroblasts that are resistant to transfection. In contrast, we were successful in therapeutically reducing HCMV replication by introducing siRNAs postinfection. To our knowledge, this is the first study to demonstrate that cells previously infected with HCMV can be treated with siRNAs in this manner.
HCMV drug resistance, toxicity, and the limited repertoire of antiviral targets represent major challenges in managing HCMV infections in patients. The specificity and flexibility of siRNA approaches offer new ways for treating HCMV infection in place of conventional antiviral strategies, which are mainly based on chemical inhibitors of the HCMV polymerase (i.e., ganciclovir, foscarnet, or cidofovir). However, the potential for resistance to RNAi-based therapies exists, and given the genetic variability of viral genomes, it may be possible to select for point mutations that are resistant to infection, as has been observed in HIV-infected cells (20). In principle, selection for escape mutants can be minimized by directing siRNAs against highly conserved viral genes or by combining siRNAs that target conserved and transcribed regions of the viral genome. The X3 siRNA was designed with both approaches in mind. Still, it might be worthwhile to consider targeting the major IE locus together with other ORFs to further reduce the potential for resistance to develop. Such considerations await further study.
In conclusion, our investigation shows that an siRNA designed against a shared exon of the major IE ORFs or siRNAs against a locus encompassing UL54-57, which encodes the viral DNA polymerase, among other proteins, are effective at inhibiting HCMV replication and reducing viral gene expression when used in prophylactic or therapeutic contexts. Moreover, both siRNAs prevent cellular cytotoxicity at the level of DNA damage signaling. Delivery of the siRNA against the major viral IE gene transcripts also results in retention of PML body integrity, which influences host gene expression and is an important component of the intrinsic immune defense against HCMV. The combined effects of reduced viral replication and cytotoxicity support a proposition for the continued development of siRNA therapies for the treatment of HCMV infections.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Kowalik laboratory for commenting on the manuscript.
This work was supported by the NIH (AI076189 to T.F.K.). T.F.K. is a member of the UMass DERC (DK32520).
The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Footnotes
Published ahead of print 21 March 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Anderson KP, Fox MC, Brown-Driver V, Martin MJ, Azad RF. 1996. Inhibition of human cytomegalovirus immediate-early gene expression by an antisense oligonucleotide complementary to immediate-early RNA. Antimicrob. Agents Chemother. 40:2004–2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aouadi M, et al. 2009. Orally delivered siRNA targeting macrophage Map4k4 suppresses systemic inflammation. Nature 458:1180–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bai Y, et al. 2011. Oral delivery of RNase P ribozymes by Salmonella inhibits viral infection in mice. Proc. Natl. Acad. Sci. U. S. A. 108:3222–3227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bai Z, et al. 2010. Inhibition of human cytomegalovirus infection by IE86-specific short hairpin RNA-mediated RNA interference. Biosci. Biotechnol. Biochem. 74:1368–1372 [DOI] [PubMed] [Google Scholar]
- 5. Baldanti F, et al. 2004. Human cytomegalovirus double resistance in a donor-positive/recipient-negative lung transplant patient with an impaired CD4-mediated specific immune response. J. Antimicrob. Chemother. 53:536–539 [DOI] [PubMed] [Google Scholar]
- 6. Blaho JA. 2010. Human cytomegalovirus infection in pregnant women and neonates: a new risk factor for cardiovascular disease? Arch. Clin. Microbiol. 1:1–5 [Google Scholar]
- 7. Borden KL. 2002. Pondering the promyelocytic leukemia protein (PML) puzzle: possible functions for PML nuclear bodies. Mol. Cell. Biol. 22:5259–5269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bradley SP, et al. 2005. Successful incorporation of short-interfering RNA into islet cells by in situ perfusion. Transplant. Proc. 37:233–236 [DOI] [PubMed] [Google Scholar]
- 9. Butz K, et al. 2003. siRNA targeting of the viral E6 oncogene efficiently kills human papillomavirus-positive cancer cells. Oncogene 22:5938–5945 [DOI] [PubMed] [Google Scholar]
- 10. Casavant NC, et al. 2006. Potential role for p53 in the permissive life cycle of human cytomegalovirus. J. Virol. 80:8390–8401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Castillo JP, et al. 2005. Human cytomegalovirus IE1-72 activates ataxia telangiectasia mutated kinase and a p53/p21-mediated growth arrest response. J. Virol. 79:11467–11475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Castillo JP, Kowalik TF. 2002. Human cytomegalovirus immediate early proteins and cell growth control. Gene 290:19–34 [DOI] [PubMed] [Google Scholar]
- 13. Castillo JP, Yurochko AD, Kowalik TF. 2000. Role of human cytomegalovirus immediate-early proteins in cell growth control. J. Virol. 74:8028–8037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chan G, Hemmings DG, Yurochko AD, Guilbert LJ. 2002. Human cytomegalovirus-caused damage to placental trophoblasts mediated by immediate-early gene-induced tumor necrosis factor-alpha. Am. J. Pathol. 161:1371–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chang CP, Malone CL, Stinski MF. 1989. A human cytomegalovirus early gene has three inducible promoters that are regulated differentially at various times after infection. J. Virol. 63:281–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chang L, et al. 2011. Herpesviral replication compartments move and coalesce at nuclear speckles to enhance export of viral late mRNA. Proc. Natl. Acad. Sci. U. S. A. 108:E136–E144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chiou SH, et al. 2006. The immediate early 2 protein of human cytomegalovirus (HCMV) mediates the apoptotic control in HCMV retinitis through up-regulation of the cellular FLICE-inhibitory protein expression. J. Immunol. 177:6199–6206 [DOI] [PubMed] [Google Scholar]
- 18. Cobbs CS, et al. 2002. Human cytomegalovirus infection and expression in human malignant glioma. Cancer Res. 62:3347–3350 [PubMed] [Google Scholar]
- 19. Cobbs CS, Soroceanu L, Denham S, Zhang W, Kraus MH. 2008. Modulation of oncogenic phenotype in human glioma cells by cytomegalovirus IE1-mediated mitogenicity. Cancer Res. 68:724–730 [DOI] [PubMed] [Google Scholar]
- 20. Das AT, et al. 2004. Human immunodeficiency virus type 1 escapes from RNA interference-mediated inhibition. J. Virol. 78:2601–2605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Devi GR. 2006. siRNA-based approaches in cancer therapy. Cancer Gene Ther. 13:819–829 [DOI] [PubMed] [Google Scholar]
- 22. Dittmer A, Bogner E. 2006. Specific short hairpin RNA-mediated inhibition of viral DNA packaging of human cytomegalovirus. FEBS Lett. 580:6132–6138 [DOI] [PubMed] [Google Scholar]
- 23. Drew WL, et al. 1991. Prevalence of resistance in patients receiving ganciclovir for serious cytomegalovirus infection. J. Infect. Dis. 163:716–719 [DOI] [PubMed] [Google Scholar]
- 24. Duan QJ, Tao R, Hu MF, Shang SQ. 2009. Efficient inhibition of human cytomegalovirus UL122 gene expression in cell by small interfering RNAs. J. Basic Microbiol. 49:531–537 [DOI] [PubMed] [Google Scholar]
- 25. Dziurzynski K, et al. 2011. Glioma-associated cytomegalovirus mediates subversion of the monocyte lineage to a tumor propagating phenotype. Clin. Cancer Res. 17:4642–4649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. E X, et al. 2011. An E2F1-mediated DNA damage response contributes to the replication of human cytomegalovirus. PLoS Pathog. 7:e1001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Eckle T, et al. 2002. Rapid development of ganciclovir-resistant cytomegalovirus infection in children after allogeneic stem cell transplantation in the early phase of immune cell recovery. Bone Marrow Transplant. 30:433–439 [DOI] [PubMed] [Google Scholar]
- 28. Erice A. 1999. Resistance of human cytomegalovirus to antiviral drugs. Clin. Microbiol. Rev. 12:286–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Erice A, et al. 1989. Progressive disease due to ganciclovir-resistant cytomegalovirus in immunocompromised patients. N. Engl. J. Med. 320:289–293 [DOI] [PubMed] [Google Scholar]
- 30. Fehr AR, Yu D. 2011. Human cytomegalovirus early protein pUL21a promotes efficient viral DNA synthesis and the late accumulation of immediate-early transcripts. J. Virol. 85:663–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ge Q, et al. 2003. RNA interference of influenza virus production by directly targeting mRNA for degradation and indirectly inhibiting all viral RNA transcription. Proc. Natl. Acad. Sci. U. S. A. 100:2718–2723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gitlin L, Karelsky S, Andino R. 2002. Short interfering RNA confers intracellular antiviral immunity in human cells. Nature 418:430–434 [DOI] [PubMed] [Google Scholar]
- 33. Hagemeier C, Walker SM, Sissons PJ, Sinclair JH. 1992. The 72K IE1 and 80K IE2 proteins of human cytomegalovirus independently trans-activate the c-fos, c-myc and hsp70 promoters via basal promoter elements. J. Gen. Virol. 73:2385–2393 [DOI] [PubMed] [Google Scholar]
- 34. Hannon GJ. 2002. RNA interference. Nature 418:244–251 [DOI] [PubMed] [Google Scholar]
- 35. Henderly DE, Freeman WR, Causey DM, Rao NA. 1987. Cytomegalovirus retinitis and response to therapy with ganciclovir. Ophthalmology 94:425–434 [DOI] [PubMed] [Google Scholar]
- 36. Hermiston TW, Malone CL, Witte PR, Stinski MF. 1987. Identification and characterization of the human cytomegalovirus immediate-early region 2 gene that stimulates gene expression from an inducible promoter. J. Virol. 61:3214–3221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hwang ES, et al. 2009. Human cytomegalovirus IE1-72 protein interacts with p53 and inhibits p53-dependent transactivation by a mechanism different from that of IE2-86 protein. J. Virol. 83:12388–12398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Johnson RA, Yurochko AD, Poma EE, Zhu L, Huang ES. 1999. Domain mapping of the human cytomegalovirus IE1-72 and cellular p107 protein-protein interaction and the possible functional consequences. J. Gen. Virol. 80:1293–1303 [DOI] [PubMed] [Google Scholar]
- 39. Kapadia SB, Brideau-Andersen A, Chisari FV. 2003. Interference of hepatitis C virus RNA replication by short interfering RNAs. Proc. Natl. Acad. Sci. U. S. A. 100:2014–2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Khanna R, Diamond DJ. 2006. Human cytomegalovirus vaccine: time to look for alternative options. Trends Mol. Med. 12:26–33 [DOI] [PubMed] [Google Scholar]
- 41. Knox KK, Drobyski WR, Carrigan DR. 1991. Cytomegalovirus isolate resistant to ganciclovir and foscarnet from a marrow transplant patient. Lancet 337:1292–1293 [DOI] [PubMed] [Google Scholar]
- 42. Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 43. Lucas KG, Bao L, Bruggeman R, Dunham K, Specht C. 2011. The detection of CMV pp65 and IE1 in glioblastoma multiforme. J. Neurooncol. 103:231–238 [DOI] [PubMed] [Google Scholar]
- 44. Luo MH, Rosenke K, Czornak K, Fortunato EA. 2007. Human cytomegalovirus disrupts both ataxia telangiectasia mutated protein (ATM)- and ATM-Rad3-related kinase-mediated DNA damage responses during lytic infection. J. Virol. 81:1934–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Malone CL, Vesole DH, Stinski MF. 1990. Transactivation of a human cytomegalovirus early promoter by gene products from the immediate-early gene IE2 and augmentation by IE1: mutational analysis of the viral proteins. J. Virol. 64:1498–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Marchini A, Liu H, Zhu H. 2001. Human cytomegalovirus with IE-2 (UL122) deleted fails to express early lytic genes. J. Virol. 75:1870–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mitchell DA, et al. 2008. Sensitive detection of human cytomegalovirus in tumors and peripheral blood of patients diagnosed with glioblastoma. Neuro Oncol. 10:10–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mocarski E. 1996. Cytomegaloviruses and their replication, p 2447–2492 In Fields BN, et al. (ed), Fields virology, 3rd ed Lippincott-Raven Publishers, Philadelphia, PA [Google Scholar]
- 49. Muganda P, Mendoza O, Hernandez J, Qian Q. 1994. Human cytomegalovirus elevates levels of the cellular protein p53 in infected fibroblasts. J. Virol. 68:8028–8034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mulamba GB, Hu A, Azad RF, Anderson KP, Coen DM. 1998. Human cytomegalovirus mutant with sequence-dependent resistance to the phosphorothioate oligonucleotide fomivirsen (ISIS 2922). Antimicrob. Agents Chemother. 42:971–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Murphy EA, Streblow DN, Nelson JA, Stinski MF. 2000. The human cytomegalovirus IE86 protein can block cell cycle progression after inducing transition into the S phase of permissive cells. J. Virol. 74:7108–7118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nekhai S, Jerebtsova M. 2006. Therapies for HIV with RNAi. Curr. Opin. Mol. Ther. 8:52–61 [PubMed] [Google Scholar]
- 53. Netterwald J, et al. 2005. Two gamma interferon-activated site-like elements in the human cytomegalovirus major immediate-early promoter/enhancer are important for viral replication. J. Virol. 79:5035–5046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Nykanen A, Haley B, Zamore PD. 2001. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell 107:309–321 [DOI] [PubMed] [Google Scholar]
- 55. Palliser D, et al. 2006. An siRNA-based microbicide protects mice from lethal herpes simplex virus 2 infection. Nature 439:89–94 [DOI] [PubMed] [Google Scholar]
- 56. Pass RF. 2001. Cytomegalovirus, p 2675–2706 In Knipe DM, et al. (ed), Fields virology, 4th ed Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 57. Peggs KS. 2004. Cytomegalovirus following stem cell transplantation: from pharmacologic to immunologic therapy. Expert Rev. Anti Infect. Ther. 2:559–573 [DOI] [PubMed] [Google Scholar]
- 58. Pizzorno MC, O'Hare P, Sha L, LaFemina RL, Hayward GS. 1988. Trans-activation and autoregulation of gene expression by the immediate-early region 2 gene products of human cytomegalovirus. J. Virol. 62:1167–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Polis MA. 1992. Foscarnet and ganciclovir in the treatment of cytomegalovirus retinitis. J. Acquir. Immune Defic. Syndr 5(Suppl. 1):S3–S10 [PubMed] [Google Scholar]
- 60. Puchtler E, Stamminger T. 1991. An inducible promoter mediates abundant expression from the immediate-early 2 gene region of human cytomegalovirus at late times after infection. J. Virol. 65:6301–6306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ranganathan P, Clark PA, Kuo JS, Salamat MS, Kalejta RF. 2012. Significant association of multiple human cytomegalovirus genomic loci with glioblastoma multiforme samples. J. Virol. 86:854–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Regad T, Chelbi-Alix MK. 2001. Role and fate of PML nuclear bodies in response to interferon and viral infections. Oncogene 20:7274–7286 [DOI] [PubMed] [Google Scholar]
- 63. Sampson JH, Mitchell DA. 2011. Is cytomegalovirus a therapeutic target in glioblastoma? Clin. Cancer Res. 17:4619–4621 [DOI] [PubMed] [Google Scholar]
- 64. Scheurer ME, Bondy ML, Aldape KD, Albrecht T, El-Zein R. 2008. Detection of human cytomegalovirus in different histological types of gliomas. Acta Neuropathol. 116:79–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Schleiss MR. 2008. Congenital cytomegalovirus infection: update on management strategies. Curr. Treat. Options Neurol. 10:186–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Scholz M, Doerr HW, Cinatl J. 2001. Inhibition of cytomegalovirus immediate early gene expression: a therapeutic option? Antiviral Res. 49:129–145 [DOI] [PubMed] [Google Scholar]
- 67. Shen YH, et al. 2004. Human cytomegalovirus causes endothelial injury through the ataxia telangiectasia mutant and p53 DNA damage signaling pathways. Circ. Res. 94:1310–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Shin MC, et al. 2006. Inhibition of UL54 and UL97 genes of human cytomegalovirus by RNA interference. Acta Virol. 50:263–268 [PubMed] [Google Scholar]
- 69. Smuda C, Bogner E, Radsak K. 1997. The human cytomegalovirus glycoprotein B gene (ORF UL55) is expressed early in the infectious cycle. J. Gen. Virol. 78:1981–1992 [DOI] [PubMed] [Google Scholar]
- 70. Speir E, et al. 1994. Potential role of human cytomegalovirus and p53 interaction in coronary restenosis. Science 265:391–394 [DOI] [PubMed] [Google Scholar]
- 71. Stenberg RM, Depto AS, Fortney J, Nelson JA. 1989. Regulated expression of early and late RNAs and proteins from the human cytomegalovirus immediate-early gene region. J. Virol. 63:2699–2708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Stenberg RM, Witte PR, Stinski MF. 1985. Multiple spliced and unspliced transcripts from human cytomegalovirus immediate-early region 2 and evidence for a common initiation site within immediate-early region 1. J. Virol. 56:665–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Straat K, et al. 2009. Activation of telomerase by human cytomegalovirus. J. Natl. Cancer Inst. 101:488–497 [DOI] [PubMed] [Google Scholar]
- 74. Tao R, Hu M, Duan Q, Shang S. 2008. Efficient inhibition of human cytomegalovirus DNA polymerase expression by small hairpin RNA in vitro. Curr. Microbiol. 57:395–400 [DOI] [PubMed] [Google Scholar]
- 75. Trang P, et al. 2000. Effective inhibition of human cytomegalovirus gene expression and replication by a ribozyme derived from the catalytic RNA subunit of RNase P from Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 97:5812–5817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Tsai HL, Kou GH, Chen SC, Wu CW, Lin YS. 1996. Human cytomegalovirus immediate-early protein IE2 tethers a transcriptional repression domain to p53. J. Biol. Chem. 271:3534–3540 [PubMed] [Google Scholar]
- 77. White EA, Clark CL, Sanchez V, Spector DH. 2004. Small internal deletions in the human cytomegalovirus IE2 gene result in nonviable recombinant viruses with differential defects in viral gene expression. J. Virol. 78:1817–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. White EA, Spector DH. 2005. Exon 3 of the human cytomegalovirus major immediate-early region is required for efficient viral gene expression and for cellular cyclin modulation. J. Virol. 79:7438–7452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wiebusch L, Truss M, Hagemeier C. 2004. Inhibition of human cytomegalovirus replication by small interfering RNAs. J. Gen. Virol. 85:179–184 [DOI] [PubMed] [Google Scholar]
- 80. Wilkinson GW, Kelly C, Sinclair JH, Rickards C. 1998. Disruption of PML-associated nuclear bodies mediated by the human cytomegalovirus major immediate early gene product. J. Gen. Virol. 79:1233–1245 [DOI] [PubMed] [Google Scholar]
- 81. Wu J, Nandamuri KM. 2004. Inhibition of hepatitis viral replication by siRNA. Expert Opin. Biol. Ther. 4:1649–1659 [DOI] [PubMed] [Google Scholar]
- 82. Yi L, Lin JY, Gao Y, Feng ZJ, Wang DX. 2008. Detection of human cytomegalovirus in the atherosclerotic cerebral arteries in Han population in China. Acta Virol. 52:99–106 [PubMed] [Google Scholar]
- 83. Yuan H, et al. 2008. Effects of cholesterol-tagged small interfering RNAs targeting 12/15-lipoxygenase on parameters of diabetic nephropathy in a mouse model of type 1 diabetes. Am. J. Physiol. Renal Physiol. 295:F605–F617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Yurochko AD, Kowalik TF, Huong SM, Huang ES. 1995. Human cytomegalovirus upregulates NF-kappa B activity by transactivating the NF-kappa B p105/p50 and p65 promoters. J. Virol. 69:5391–5400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Zhu H, Shen Y, Shenk T. 1995. Human cytomegalovirus IE1 and IE2 proteins block apoptosis. J. Virol. 69:7960–7970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Zou H, et al. 2003. Engineered RNase P ribozymes are efficient in cleaving a human cytomegalovirus mRNA in vitro and are effective in inhibiting viral gene expression and growth in human cells. J. Biol. Chem. 278:37265–37274 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.